Multiplexable, High-Throughput DNA-Based Technologies in Screening and Confirmatory Testing of Newborn Conditions: A Scoping Review
Abstract
1. Introduction
2. Methodology
2.1. Protocol
2.2. Literature Search, Abstract and Title Screening
2.3. Study Categorization
2.4. Eligibility Criteria
2.5. Data Extraction
3. Results
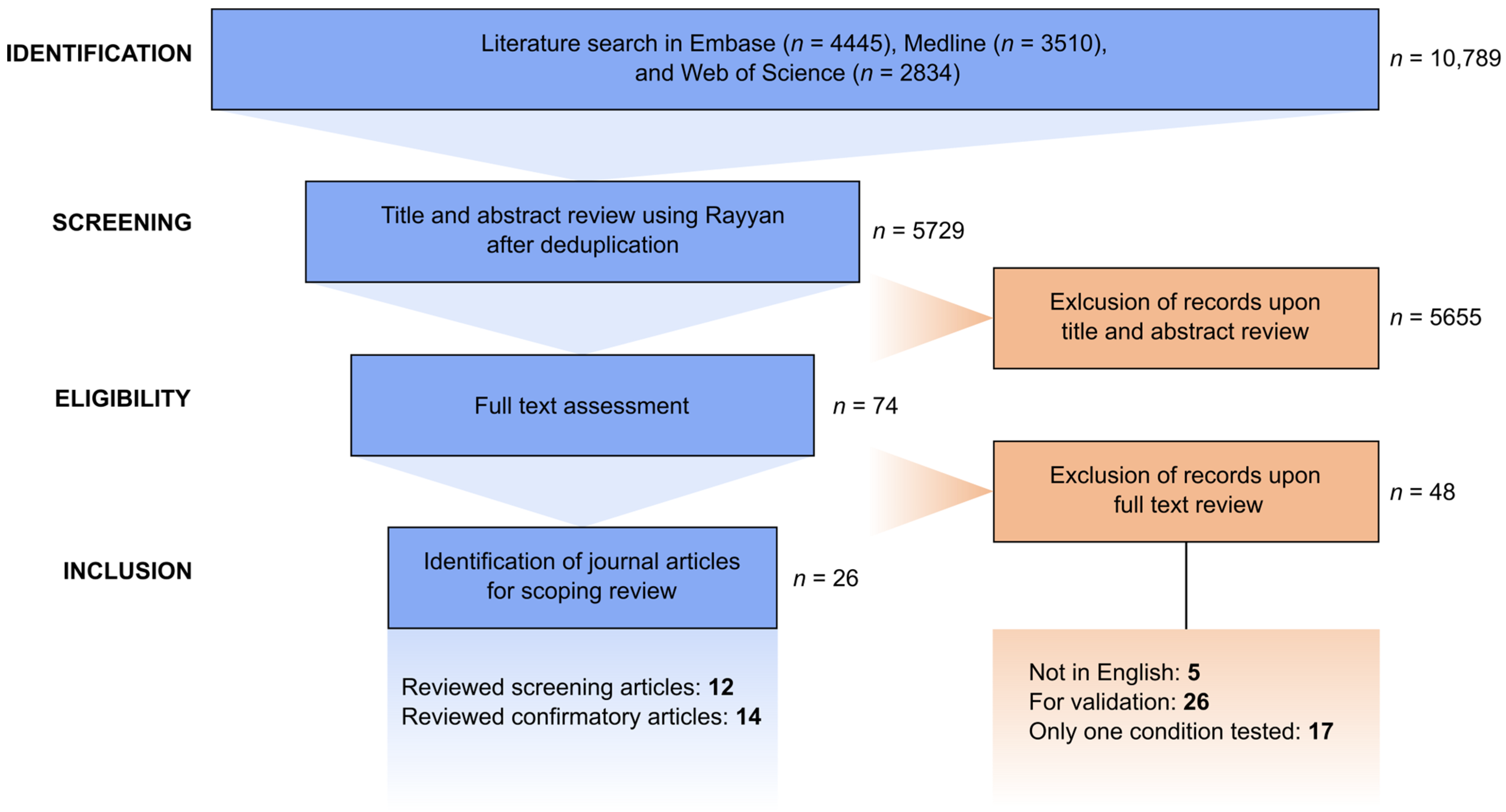
3.1. Literature Search and Selection Process
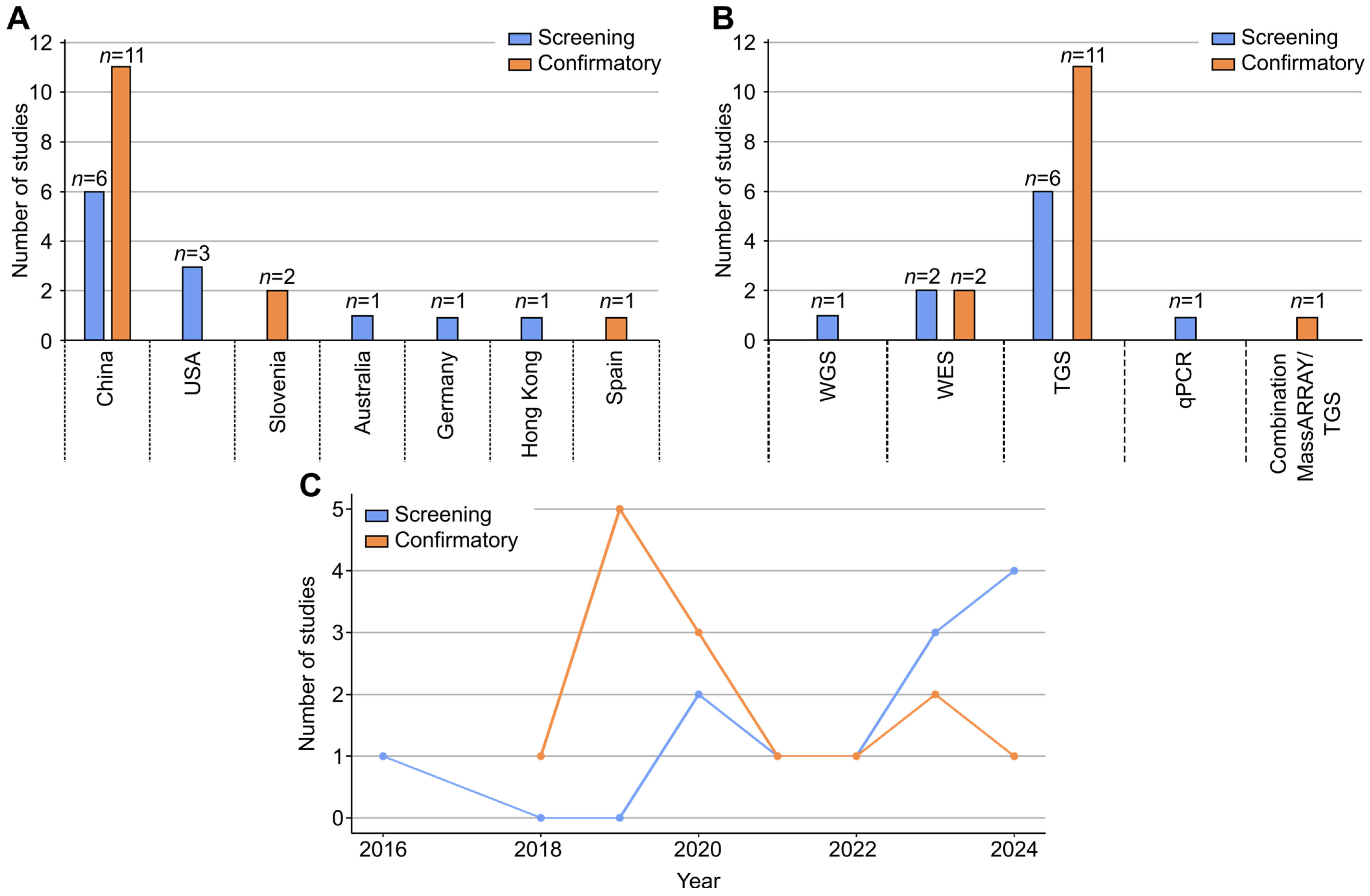
3.2. Study Characteristics
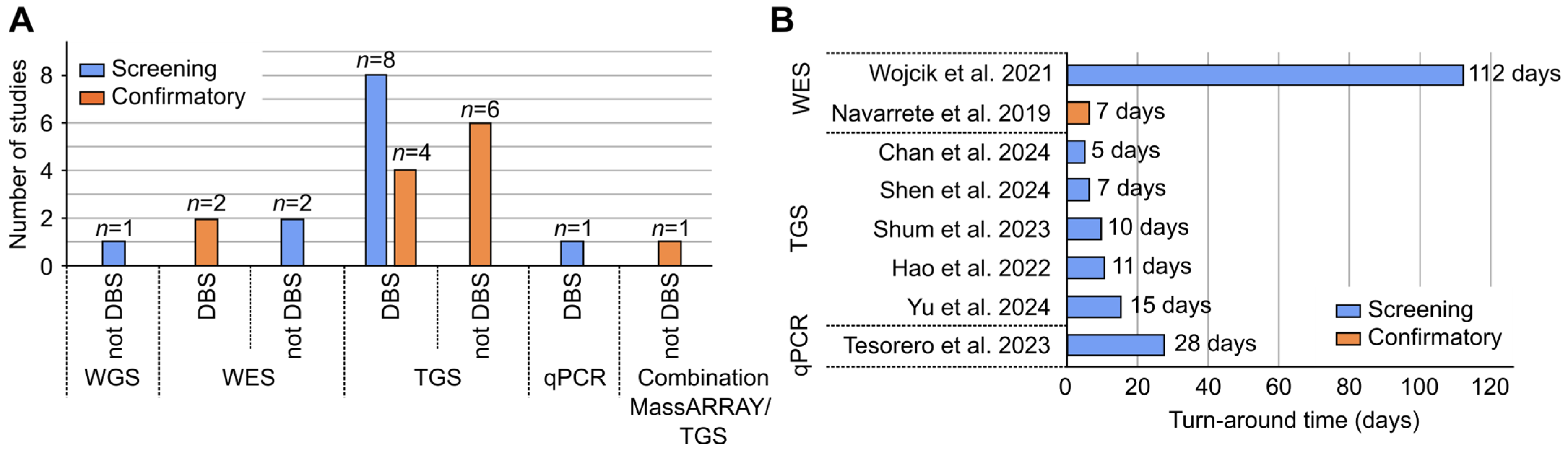
3.3. Dried Blood Spot (DBS) as a Source of gDNA
3.4. Turnaround Time (TAT)
3.5. Sensitivity, Specificity, Positive Predictive Value, and Negative Predictive Value
3.6. Technical Limitations of Identified Technologies in Terms of Variant Detection and Interpretation
3.7. Complementary Non-DNA-Based Tests Used in Screening and Confirmatory Articles
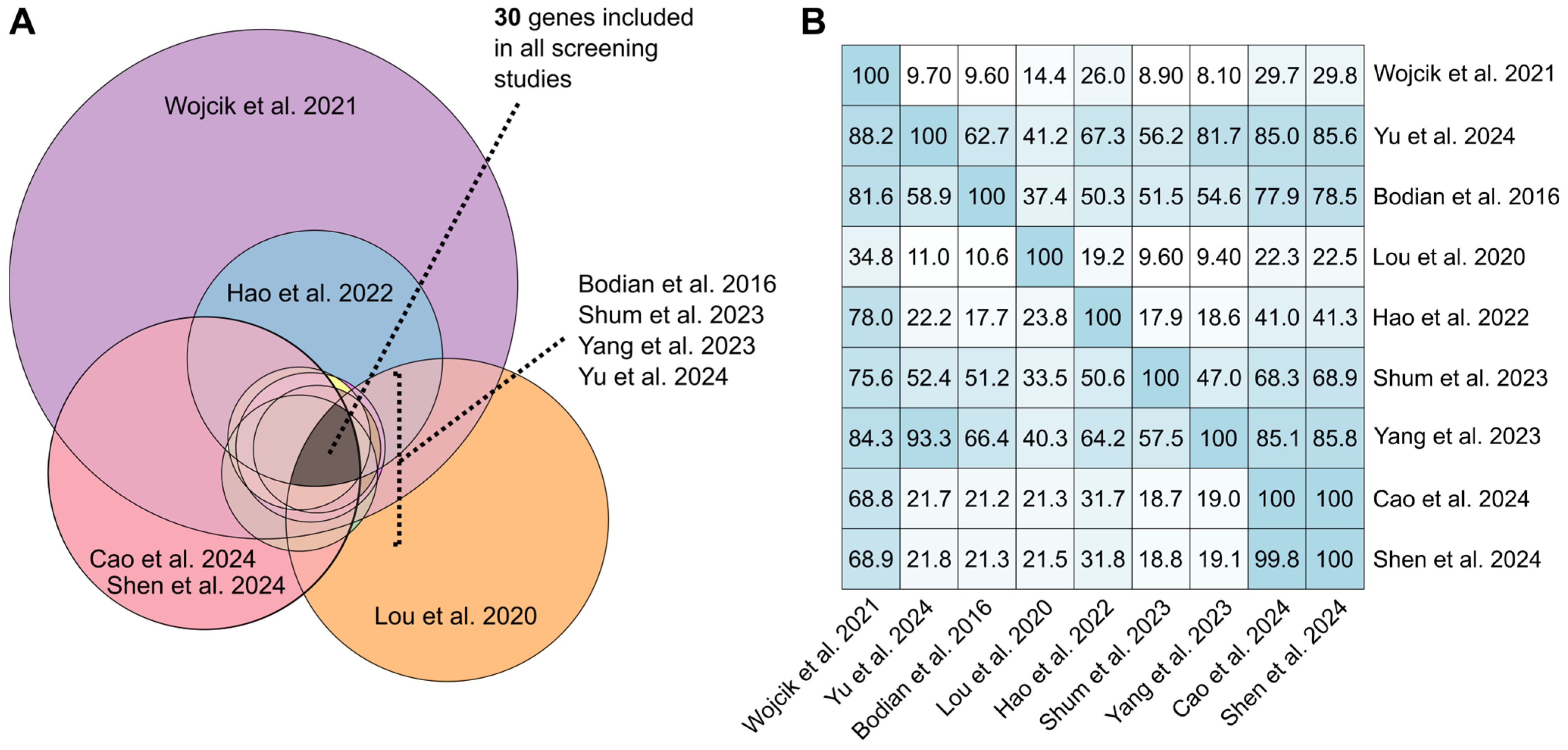
3.8. Genes and Conditions Included
3.9. Gene List Comparison Across Screening Articles
3.10. Detection of VUS, Low/Incomplete-Penetrance Variants, Carrier State of Technologies
3.11. Cost
4. Discussion
Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DBS | dried blood spot |
| NGS | next-generation sequencing |
| WES | whole-exome sequencing |
| TGS | targeted genome sequencing |
| qPCR | quantitative polymerase chain reaction |
| WGS | whole-genome sequencing |
| SNV | single-nucleotide variant |
| CNV | copy number variant |
| PCR | polymerase chain reaction |
| gDNA | genomic deoxyribonucleic acid |
| NBS | newborn screening |
| VUS | variant of uncertain significance |
| TAT | turnaround time |
References
- Remec, Z.I.; Podkrajsek, K.T.; Lampret, B.R.; Kovac, J.; Groselj, U.; Tesovnik, T.; Battelino, T.; Debeljak, M. Next-Generation Sequencing in Newborn Screening: A Review of Current State. Front. Genet. 2021, 12, 662254. [Google Scholar] [CrossRef]
- Ding, S.; Han, L.A.-O. Newborn screening for genetic disorders: Current status and prospects for the future. Pediatr. Investig. 2022, 6, 291–298. [Google Scholar] [CrossRef]
- Clark, R.H.; Kelleher, A.S.; Chace, D.H.; Spitzer, A.R. Gestational age and age at sampling influence metabolic profiles in premature infants. Pediatrics 2014, 134, e37–e46. [Google Scholar] [CrossRef]
- Cortés, E.; Roldán, A.M.; Palazón-Bru, A.; Rizo-Baeza, M.M.; Manero, H.; Gil-Guillén, V.F. Differences in immunoreactive trypsin values between type of feeding and ethnicity in neonatal cystic fibrosis screening: A cross-sectional study. Orphanet J. Rare Dis. 2014, 9, 166. [Google Scholar] [CrossRef]
- Kwan, A.; Abraham, R.S.; Currier, R.; Brower, A.; Andruszewski, K.; Abbott, J.K.; Baker, M.; Ballow, M.; Bartoshesky, L.E.; Bonilla, F.A.; et al. Newborn screening for severe combined immunodeficiency in 11 screening programs in the United States. J. Am. Med. Assoc. 2014, 312, 729–738. [Google Scholar] [CrossRef]
- Gutierrez-Mateo, C.; Timonen, A.; Vaahtera, K.; Jaakkola, M.; Hougaard, D.M.; Bybjerg-Grauholm, J.; Baekvad-Hansen, M.A.-O.; Adamsen, D.; Filippov, G.; Dallaire, S.; et al. Development of a Multiplex Real-Time PCR Assay for the Newborn Screening of SCID, SMA, and XLA. Int. J. Neonatal Screen. 2019, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.E.; Darras, B.T.; Swoboda, K.A.-O.; Estrella, E.; Chen, J.Y.H.; Abbott, M.A.; Hay, B.N.; Kumar, B.; Counihan, A.M.; Gerstel-Thompson, J.; et al. Massachusetts’ Findings from Statewide Newborn Screening for Spinal Muscular Atrophy. Int. J. Neonatal Screen. 2021, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Tesorero, R.A.-O.; Janda, J.A.-O.; Hörster, F.; Feyh, P.; Mütze, U.A.-O.; Hauke, J.; Schwarz, K.; Kunz, J.B.; Hoffmann, G.F.; Okun, J.G. A high-throughput newborn screening approach for SCID, SMA, and SCD combining multiplex qPCR and tandem mass spectrometry. PLoS ONE 2023, 18, e0283024. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Lin, C.H.; Yin, X.; Zhu, L.; Yang, J.; Shen, Y.; Yang, C.; Chen, X.; Hu, H.; Ma, Q.; et al. Newborn Screening for Spinal Muscular Atrophy in China Using DNA Mass Spectrometry. Front. Genet. 2019, 10, 1255. [Google Scholar] [CrossRef]
- Jurinke, C.; Oeth, P.; van den Boom, D. MALDI-TOF mass spectrometry: A versatile tool for high-performance DNA analysis. Mol. Biotechnol. 2004, 26, 147–164. [Google Scholar] [CrossRef]
- Pham, H.; Del Angel, S.; Xiong, W.; Astill, M.; Bethers, H.; Harris, T.; Case, K.; Sibio, S.; Mao, R. eP386: Cystic fibrosis 165 pathogenic variants genotyping by MassARRAY. Genet. Med. 2022, 24, S243–S244. [Google Scholar] [CrossRef]
- Svidnicki, M.C.; Silva-Costa, S.M.; Ramos, P.Z.; dos Santos, N.Z.; Martins, F.T.; Castilho, A.M.; Sartorato, E.L. Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iPLEX® technology. BMC Med. Genet. 2015, 16, 85. [Google Scholar] [CrossRef]
- Lu, Y.H.; Huang, P.H.; Wang, L.Y.; Hsu, T.R.; Li, H.Y.; Lee, P.C.; Hsieh, Y.P.; Hung, S.C.; Wang, Y.C.; Chang, S.K.; et al. Improvement in the sensitivity of newborn screening for Fabry disease among females through the use of a high-throughput and cost-effective method, DNA mass spectrometry. J. Hum. Genet. 2018, 63, 1–8. [Google Scholar] [CrossRef]
- Minten, T.; Bick, S.; Adelson, S.; Gehlenborg, N.; Amendola, L.M.; Boemer, F.; Coffey, A.J.; Encina, N.; Ferlini, A.; Kirschner, J.; et al. Data-driven consideration of genetic disorders for global genomic newborn screening programs. Genet. Med. 2025, 27, 101443. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chitnis, N.; Monos, D.; Dinh, A. Next-generation sequencing technologies: An overview. Hum. Immunol. 2021, 82, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Wetterstrand, K.A. DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available online: https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data (accessed on 17 August 2025).
- Friedman, J.M.; Cornel, M.C.; Goldenberg, A.J.; Lister, K.J.; Sénécal, K.; Vears, D.F. Genomic newborn screening: Public health policy considerations and recommendations. BMC Med. Genom. 2017, 10, 9. [Google Scholar] [CrossRef]
- Stark, Z.A.-O.; Scott, R.A.-O. Genomic newborn screening for rare diseases. Nat. Rev. Genet. 2023, 24, 755–766. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Institute, J.B. Scoping Reviews—Resources [Internet]. Available online: https://jbi.global/scoping-review-network/resources (accessed on 17 August 2025).
- Bodian, D.L.; Klein, E.; Iyer, R.K.; Wong, W.A.-O.; Kothiyal, P.; Stauffer, D.; Huddleston, K.C.; Gaither, A.D.; Remsburg, I.; Khromykh, A.; et al. Utility of whole-genome sequencing for detection of newborn screening disorders in a population cohort of 1,696 neonates. Genet. Med. 2016, 18, 221–230. [Google Scholar] [CrossRef]
- Roman, T.S.; Crowley, S.B.; Roche, M.I.; Foreman, A.K.M.; O’Daniel, J.M.; Seifert, B.A.; Lee, K.; Brandt, A.; Gustafson, C.; DeCristo, D.M.; et al. Genomic Sequencing for Newborn Screening: Results of the NC NEXUS Project. Am. J. Hum. Genet. 2020, 107, 596–611. [Google Scholar] [CrossRef]
- Wojcik, M.A.-O.; Zhang, T.; Ceyhan-Birsoy, O.; Genetti, C.A.; Lebo, M.S.; Yu, T.W.; Parad, R.B.; Holm, I.A.; Rehm, H.L.; Beggs, A.H.; et al. Discordant results between conventional newborn screening and genomic sequencing in the BabySeq Project. Genet. Med. 2021, 23, 1372–1375. [Google Scholar] [CrossRef]
- He, X.; Kuang, J.; Lai, J.; Huang, J.; Wang, Y.; Lan, G.; Xie, Y.; Shi, X. A retrospective analysis of MS/MS screening for IEM in high-risk areas. BMC Med. Genom. 2023, 16, 57. [Google Scholar] [CrossRef] [PubMed]
- Lampret, B.R.; Remec, Ž.I.; Torkar, A.D.; Tanšek, M.; Šmon, A.; Koračin, V.; Čuk, V.; Perko, D.; Ulaga, B.; Jelovšek, A.M.; et al. Expanded Newborn Screening Program in Slovenia using Tandem Mass Spectrometry and Confirmatory Next Generation Sequencing Genetic Testing. Zdr. Varsto 2020, 59, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Li, S.; Ma, Y.; Jing, M.; Wang, Y.; Wang, F.; Yang, M.; Miao, T.; Liu, J. Ethnic preference distribution of inborn errors of metabolism: A 4-year study in a multi-ethnic region of China. Clin. Chim. Acta 2020, 511, 160–166. [Google Scholar] [CrossRef]
- Smon, A.; Lampret, B.R.; Groselj, U.; Tansek, M.Z.; Kovac, J.; Perko, D.; Bertok, S.; Battelino, T.; Podkrajsek, K.T. Next generation sequencing as a follow-up test in an expanded newborn screening programme. Clin. Biochem. 2018, 52, 48–55. [Google Scholar] [CrossRef]
- Tan, J.; Chen, D.; Chang, R.; Pan, L.; Yang, J.; Yuan, D.; Huang, L.; Yan, T.; Ning, H.; Wei, J.; et al. Tandem Mass Spectrometry Screening for Inborn Errors of Metabolism in Newborns and High-Risk Infants in Southern China: Disease Spectrum and Genetic Characteristics in a Chinese Population. Front. Genet. 2021, 12, 631688. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Wang, B.; Liu, S.; Yu, B.; Wang, T. Application of Next-Generation Sequencing Following Tandem Mass Spectrometry to Expand Newborn Screening for Inborn Errors of Metabolism: A Multicenter Study. Front. Genet. 2019, 10, 86. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhou, C.; Xu, P.; Jin, X.; Liu, W.; Wang, W.; Huang, C.; Jiang, M.; Chen, X. Newborn screening and diagnosis of inborn errors of metabolism: A 5-year study in an eastern Chinese population. Clin. Chim. Acta 2020, 502, 133–138. [Google Scholar] [CrossRef]
- Navarrete, R.; Leal, F.; Vega, A.I.; Morais-López, A.; Garcia-Silva, M.T.; Martín-Hernández, E.; Quijada-Fraile, P.; Bergua, A.; Vives, I.; García-Jiménez, I.; et al. Value of genetic analysis for confirming inborn errors of metabolism detected through the Spanish neonatal screening program. Eur. J. Hum. Genet. 2019, 27, 556–562. [Google Scholar] [CrossRef]
- Chan, T.A.-O.; Mak, C.A.-O.X.; Yeung, M.C.W.; Law, E.C.; Cheung, J.; Wong, T.K.; Cheng, V.W.; Lee, J.A.-O.; Wong, J.C.L.; Fung, C.W.; et al. Harnessing Next-Generation Sequencing as a Timely and Accurate Second-Tier Screening Test for Newborn Screening of Inborn Errors of Metabolism. Int. J. Neonatal Screen. 2024, 10, 19. [Google Scholar] [CrossRef]
- Shen, G.; Li, W.; Zhang, Y.; Chen, L. Next-generation sequencing based newborn screening and comparative analysis with MS/MS. BMC Pediatr. 2024, 24, 230. [Google Scholar] [CrossRef]
- Shum, B.O.V.; Pretorius, C.J.; Sng, L.M.F.; Henner, I.; Barahona, P.; Basar, E.; McGill, J.; Wilgen, U.; Zournazi, A.; Downie, L.; et al. Feasibility of Targeted Next-Generation DNA Sequencing for Expanding Population Newborn Screening. Clin. Chem. 2023, 69, 890–900. [Google Scholar] [CrossRef]
- Hao, C.; Guo, R.; Hu, X.; Qi, Z.; Guo, Q.; Liu, X.; Liu, Y.; Sun, Y.; Zhang, X.; Jin, F.; et al. Newborn screening with targeted sequencing: A multicenter investigation and a pilot clinical study in China. J. Genet. Genom. 2022, 49, 13–19. [Google Scholar] [CrossRef]
- Yu, B.; Yang, Y.; Zhou, L.; Wang, Q. Evaluating a Novel Newborn Screening Methodology: Combined Genetic and Biochemical Screenings. Arch. Med. Res. 2024, 55, 102959. [Google Scholar] [CrossRef]
- Luo, H.; Wang, J.; Chen, J.; Yi, H.; Yang, X.; Peng, Y.; Ni, L.; Yang, Y.Q.; Zhang, X.M.; Huang, H. Prevalence of inherited metabolic disorders among newborns in Zhuzhou, a southern city in China. Front. Genet. 2024, 15, 1197151. [Google Scholar] [CrossRef]
- Cao, Z.; He, X.; Wang, D.; Gu, M.; Suo, F.; Qiang, R.; Zhang, R.; Song, C.; Wang, X.; Zhu, B.; et al. Targeted exome sequencing strategy (NeoEXOME) for Chinese newborns using a pilot study with 3423 neonates. Mol. Genet. Genom. Med. 2024, 12, e2357. [Google Scholar] [CrossRef]
- Luo, X.; Sun, Y.; Xu, F.; Guo, J.; Li, L.; Lin, Z.; Ye, J.; Gu, X.; Yu, Y. A pilot study of expanded newborn screening for 573 genes related to severe inherited disorders in China: Results from 1,127 newborns. Ann. Transl. Med. 2020, 8, 1058. [Google Scholar] [CrossRef]
- Yang, R.L.; Qian, G.L.; Wu, D.W.; Miao, J.K.; Yang, X.; Wu, B.Q.; Yan, Y.Q.; Li, H.B.; Mao, X.M.; He, J.; et al. A multicenter prospective study of next-generation sequencing-based newborn screening for monogenic genetic diseases in China. World J. Pediatr. 2023, 19, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zheng, Q.; Zheng, T.; Zheng, Z.; Lin, W.; Fu, Q. Expanded newborn screening for inherited metabolic disorders and genetic characteristics in a southern Chinese population. Clin. Chim. Acta 2019, 494, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Men, S.A.-O.; Liu, S.; Zheng, Q.; Yang, S.; Mao, H.; Wang, Z.; Gu, Y.; Tang, X.; Wang, L. Incidence and genetic variants of inborn errors of metabolism identified through newborn screening: A 7-year study in eastern coastal areas of China. Mol. Genet. Genom. Med. 2023, 11, e2152. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Ma, J.; Zhang, Q.; Gao, A.; Wang, Q.; Li, H.; Xiang, J.; Wang, B. Expanded Newborn Screening for Inborn Errors of Metabolism by Tandem Mass Spectrometry in Suzhou, China: Disease Spectrum, Prevalence, Genetic Characteristics in a Chinese Population. Front. Genet. 2019, 10, 1052. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Qiu, Y.; Zhang, C. Expanded newborn screening for inherited metabolic disorders by tandem mass spectrometry in a northern Chinese population. Front. Genet. 2022, 13, 801447. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Qiang, R.; Song, C.; Ma, X.; Zhang, Y.; Li, F.; Wang, R.; Yu, W.; Feng, M.; Yang, L.; et al. Spectrum analysis of inborn errors of metabolism for expanded newborn screening in a northwestern Chinese population. Sci. Rep. 2021, 11, 2699. [Google Scholar] [CrossRef]
- Tang, C.; Li, L.; Chen, T.; Li, Y.; Zhu, B.; Zhang, Y.; Yin, Y.; Liu, X.; Huang, C.; Miao, J.; et al. Newborn Screening for Inborn Errors of Metabolism by Next-Generation Sequencing Combined with Tandem Mass Spectrometry. Int. J. Neonatal Screen. 2024, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Yin, Y.; Chen, M.; Gong, N.; Peng, Y.; Liu, H.; Miao, J. Combined genetic screening and traditional newborn screening to improve the screening efficiency of congenital hypothyroidism. Front. Pediatr. 2023, 11, 1185802. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Fan, C.; Huang, Y.; Feng, J.; Zhang, Y.; Miao, J.; Wang, X.; Li, Y.; Huang, C.; Jin, W.; et al. Genomic Sequencing as a First-Tier Screening Test and Outcomes of Newborn Screening. JAMA Netw. Open 2023, 6, e2331162. [Google Scholar] [CrossRef]
- Rijksinstituut voor Volksgezondheid en Milieu (RIVM). Verwijstermijnen. Available online: https://draaiboekhielprikscreening.rivm.nl/proces/de-uitslag/verwijstermijnen (accessed on 1 July 2025).
- Health Resources and Services Administration (HRSA). Newborn Screening Timeliness Goals. Available online: https://www.hrsa.gov/advisory-committees/heritable-disorders/newborn-screening-timeliness (accessed on 3 September 2025).
- Vaillant, A.A.J.; Mohseni, M. Severe Combined Immunodeficiency. Available online: https://www.ncbi.nlm.nih.gov/books/NBK539762 (accessed on 17 August 2025).
- Yang, Y.; Muzny, D.M.; Reid, J.G.; Bainbridge, M.N.; Willis, A.; Ward, P.A.; Braxton, A.; Beuten, J.; Xia, F.; Niu, Z.; et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013, 369, 1502–1511. [Google Scholar] [CrossRef]
- Lumaka, A.A.-O.; Fasquelle, C.; Debray, F.G.; Alkan, S.; Jacquinet, A.; Harvengt, J.A.-O.; Boemer, F.A.-O.; Mulder, A.; Vaessen, S.; Viellevoye, R.; et al. Rapid Whole Genome Sequencing Diagnoses and Guides Treatment in Critically Ill Children in Belgium in Less than 40 Hours. Int. J. Mol. Sci. 2023, 24, 4003. [Google Scholar] [CrossRef]
- Reinstein, E. Challenges of using next generation sequencing in newborn screening. Genet. Res. 2015, 97, e21. [Google Scholar] [CrossRef]
- Bassaganyas, L.; Freedman, G.; Vaka, D.; Wan, E.; Lao, R.; Chen, F.; Kvale, M.; Currier, R.J.; Puck, J.M.; Kwok, P.A.-O. Whole exome and whole genome sequencing with dried blood spot DNA without whole genome amplification. Hum. Mutat. 2018, 39, 167–171. [Google Scholar] [CrossRef]
- Boemer, F.A.-O.; Fasquelle, C.; d’Otreppe, S.; Josse, C.A.-O.; Dideberg, V.; Segers, K.; Guissard, V.; Capraro, V.; Debray, F.G.; Bours, V. A next-generation newborn screening pilot study: NGS on dried blood spots detects causal mutations in patients with inherited metabolic diseases. Sci. Rep. 2017, 7, 17641. [Google Scholar] [CrossRef]
- Ding, Y.; Owen, M.; Le, J.; Batalov, S.A.-O.; Chau, K.; Kwon, Y.H.; Van Der Kraan, L.; Bezares-Orin, Z.; Zhu, Z.; Veeraraghavan, N.A.-O.; et al. Scalable, high quality, whole genome sequencing from archived, newborn, dried blood spots. NPJ Genom. Med. 2023, 8, 5. [Google Scholar] [CrossRef]
- McBride, D.A.-O.; Fielding, C.; Newington, T.; Vatsiou, A.; Fischl, H.; Bajracharya, M.; Thomson, V.S.; Fraser, L.J.; Fujita, P.A.; Becq, J.; et al. Whole-Genome Sequencing Can Identify Clinically Relevant Variants from a Single Sub-Punch of a Dried Blood Spot Specimen. Int. J. Neonatal Screen. 2023, 9, 52. [Google Scholar] [CrossRef]
- Agrawal, P.; Katragadda, S.; Hariharan, A.K.; Raghavendrachar, V.G.; Agarwal, A.; Dayalu, R.; Awasthy, D.; Sharma, S.C.; Sivasamy, Y.K.; Lakshmana, P.; et al. Validation of whole genome sequencing from dried blood spots. BMC Med. Genom. 2021, 14, 110. [Google Scholar] [CrossRef]
- Slatko, B.E.; Gardner, A.F.; Ausubel, F.M. Overview of Next-Generation Sequencing Technologies. Curr. Protoc. Mol. Biol. 2018, 122, e59. [Google Scholar] [CrossRef]
- Dietzen, D.J.; Rinaldo, P.; Whitley, R.J.; Rhead, W.J.; Hannon, W.H.; Garg, U.C.; Lo, S.F.; Bennett, M.J. National academy of clinical biochemistry laboratory medicine practice guidelines: Follow-up testing for metabolic disease identified by expanded newborn screening using tandem mass spectrometry; executive summary. Clin. Chem. 2009, 55, 1615–1626. [Google Scholar] [CrossRef]
- Orsini, J.J.; Escolar, M.L.; Wasserstein, M.P.; Caggana, M. GeneReviews® [Internet] University of Washington: Washington, DC, USA, 1993–2025. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1238/ (accessed on 17 August 2025).
- OMIM. Glycine Encephalopathy 1. Available online: https://www.omim.org/entry/605899 (accessed on 5 July 2025).
- de Wert, G.A.-O.; Dondorp, W.A.-O.; Clarke, A.A.-O.; Dequeker, E.M.C.; Cordier, C.; Deans, Z.; van El, C.A.-O.; Fellmann, F.; Hastings, R.; Hentze, S.; et al. Opportunistic genomic screening. Recommendations of the European Society of Human Genetics. Eur. J. Hum. Genet. 2021, 29, 365–377. [Google Scholar] [CrossRef]
- Currier, R.J. Single-Gene Sequencing in Newborn Screening: Success, Challenge, Hope. Hastings Cent. Rep. 2018, 48, S37–S38. [Google Scholar] [CrossRef]
- Ries, M.; Gal, A. GeneReviews® [Internet]. Available online: https://www.ncbi.nlm.nih.gov/books/NBK11584/ (accessed on 17 August 2025).
- Mersch, J.; Brown, N.; Pirzadeh-Miller, S.; Mundt, E.; Cox, H.C.; Brown, K.; Aston, M.; Esterling, L.; Manley, S.; Ross, T. Prevalence of Variant Reclassification Following Hereditary Cancer Genetic Testing. JAMA 2018, 13, 106–115. [Google Scholar] [CrossRef]
- Grosse, S.; Rogowski, W.; Ross, L.; Cornel, M.; Dondorp, W.; Khoury, M. Population screening for genetic disorders in the 21st century: Evidence, economics, and ethics. Public Health Genom. 2010, 13, 105–115. [Google Scholar] [CrossRef]
- Carlson, J.J.; Henrikson, N.B.; Veenstra, D.L.; Ramsey, S.D. Economic analyses of human genetics services: A systematic review. Genet. Med. 2005, 7, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Stark, Z.A.-O.; Schofield, D.; Alam, K.; Wilson, W.; Mupfeki, N.; Macciocca, I.A.-O.; Shrestha, R.A.-O.; White, S.A.-O.X.; Gaff, C. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet. Med. 2017, 19, 867–874. [Google Scholar] [CrossRef]
- Tan, T.Y.; Dillon, O.J.; Stark, Z.; Schofield, D.; Alam, K.; Shrestha, R.; Chong, B.; Phelan, D.; Brett, G.R.; Creed, E.; et al. Diagnostic Impact and Cost-effectiveness of Whole-Exome Sequencing for Ambulant Children with Suspected Monogenic Conditions. JAMA Pediatr. 2017, 171, 855–862. [Google Scholar] [CrossRef]
- Incerti, D.; Xu, X.M.; Chou, J.W.; Gonzaludo, N.; Belmont, J.W.; Schroeder, B.E. Cost-effectiveness of genome sequencing for diagnosing patients with undiagnosed rare genetic diseases. Genet. Med. 2022, 24, 109–118. [Google Scholar] [CrossRef]
- Lunke, S.; Bouffler, S.A.-O.; Downie, L.; Caruana, J.; Amor, D.J.; Archibald, A.; Bombard, Y.A.-O.; Christodoulou, J.; Clausen, M.; De Fazio, P.; et al. Prospective cohort study of genomic newborn screening: BabyScreen+ pilot study protocol. BMJ Open 2024, 14, e081426. [Google Scholar] [CrossRef]
- Smith, H.S.; Zettler, B.; Genetti, C.A.; Hickingbotham, M.R.; Coleman, T.F.; Lebo, M.; Nagy, A.; Zouk, H.; Mahanta, L.; Christensen, K.D.; et al. The BabySeq Project: A clinical trial of genome sequencing in a diverse cohort of infants. Am. J. Hum. Genet. 2024, 111, 2094–2106. [Google Scholar] [CrossRef]
- Boemer, F.A.-O.; Hovhannesyan, K.A.-O.; Piazzon, F.; Minner, F.; Mni, M.; Jacquemin, V.A.-O.; Mashhadizadeh, D.A.-O.; Benmhammed, N.; Bours, V.; Jacquinet, A.; et al. Population-based, first-tier genomic newborn screening in the maternity ward. Nat. Med. 2025, 31, 1339–1350. [Google Scholar] [CrossRef]
- Ziegler, A.; Koval-Burt, C.; Kay, D.M.; Suchy, S.F.; Begtrup, A.; Langley, K.G.; Hernan, R.; Amendola, L.M.; Boyd, B.M.; Bradley, J.; et al. Expanded Newborn Screening Using Genome Sequencing for Early Actionable Conditions. JAMA 2025, 333, 232–240. [Google Scholar] [CrossRef]
| Technology | Types of Variants Detected | Limitation | Complementary DNA-Based Technology/Methods Used to Address the Limitation (If Available) |
|---|---|---|---|
| WGS [21] | SNVs, 1 small indels | Intrinsic
|
|
Data quality
| |||
Data analysis and interpretation
| |||
|
| ||
| WES [22,23,31,37] | SNVs, 1 small indels, indels, large genomic rearrangements | Intrinsic
|
|
|
| ||
| |||
Data quality
| |||
| |||
Data analysis and interpretation:
| |||
|
| ||
| |||
|
| ||
| TGS [24,25,26,27,28,29,30,32,33,34,35,36,38,39,40,41,42,43,44,45] | SNVs, 1 small indels, indels (CNVs) | Intrinsic
|
|
|
| ||
|
| ||
| |||
|
| ||
Data quality
|
| ||
Data analysis and interpretation
| |||
|
| ||
| qPCR [8] | SNVs, deletion, quantity of T-cell receptor excision circles | Intrinsic
|
|
| 3 MassARRAY [30] | SNVs, small indels | Intrinsic
|
|
| VUS | Low/Incomplete-Penetrance Variants | 1 Heterozygous Carrier Status | |
|---|---|---|---|
| WGS [21] | Yes | Yes | Yes |
| WES | Yes [23] | Yes [23] | Yes [23,31] |
| TGS | Yes [24,25,26,29,32,33,34,38,40,42,43,45] | Yes [29,36,38,42] | Yes [27,28,32,33,34,35,38,39,40,41,42,44] |
| qPCR [8] | No | No | Yes |
| MassARRAY [30] | No | NR | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Published by MDPI on behalf of the International Society for Neonatal Screening. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fabella, T.D.; den Hoed, J.; Henneman, L.; Rodenburg, W.; Ket, J.C.F.; Schouten, J.; Sistermans, E.A. Multiplexable, High-Throughput DNA-Based Technologies in Screening and Confirmatory Testing of Newborn Conditions: A Scoping Review. Int. J. Neonatal Screen. 2025, 11, 104. https://doi.org/10.3390/ijns11040104
Fabella TD, den Hoed J, Henneman L, Rodenburg W, Ket JCF, Schouten J, Sistermans EA. Multiplexable, High-Throughput DNA-Based Technologies in Screening and Confirmatory Testing of Newborn Conditions: A Scoping Review. International Journal of Neonatal Screening. 2025; 11(4):104. https://doi.org/10.3390/ijns11040104
Chicago/Turabian StyleFabella, Terence Diane, Joery den Hoed, Lidewij Henneman, Wendy Rodenburg, Johannes C. F. Ket, Jan Schouten, and Erik A. Sistermans. 2025. "Multiplexable, High-Throughput DNA-Based Technologies in Screening and Confirmatory Testing of Newborn Conditions: A Scoping Review" International Journal of Neonatal Screening 11, no. 4: 104. https://doi.org/10.3390/ijns11040104
APA StyleFabella, T. D., den Hoed, J., Henneman, L., Rodenburg, W., Ket, J. C. F., Schouten, J., & Sistermans, E. A. (2025). Multiplexable, High-Throughput DNA-Based Technologies in Screening and Confirmatory Testing of Newborn Conditions: A Scoping Review. International Journal of Neonatal Screening, 11(4), 104. https://doi.org/10.3390/ijns11040104






