Design and Implementation of the Pre-Clinical DICOM Standard in Multi-Cohort Murine Studies
Abstract
:1. Introduction
2. Methodology
2.1. Image Acquisition Context Structured Report
- extract the worksheet from the spreadsheet in tab-delimited text form (tabdelimitedtoxml),
- convert each row to Extensible Markup Language (XML) files, one for each subject, using an eXtensible Stylesheet Language (XSL-T),
- convert each of the XML files into an XML-representation of a DICOM Structured Report conforming to the Pre-clinical Small Animal Image Acquisition Context SR Information Object Definition (IOD) (TID 8101), using an XSL-T stylesheet,
- use the XML to DICOM SR conversion capabilities of the PixelMed Java DICOM toolkit [17] to create DICOM SR files,
- upload DICOM SR files to TCIA for curation and inclusion in their archive alongside the split images.
- CONTAINER: (127001,DCM,“Preclinical Small Animal Imaging Acquisition Context”) [SEPARATE] (DCMR,8101)
- HAS CONCEPT MOD: CODE: (121049,DCM,“Language of Content Item and Descendants”) = (eng,RFC5646,“English”)
- HAS CONCEPT MOD: CODE: (121046,DCM,“Country of Language”) = (US,ISO3166_1,“United States”)
- HAS OBS CONTEXT: PNAME: (121008,DCM,“Person Observer Name”) = “SAIP^Imager”
- CONTAINS: CONTAINER: (127400,DCM,“Exogenous substance”) [SEPARATE]
- CONTAINS: CODE: (127460,DCM,“Tumor Graft”) = (2092003,SCT,“Melanoma”)
- HAS PROPERTIES: DATETIME: (111526,DCM,“DateTime Started”) = “20190722”
- HAS PROPERTIES: DATETIME: (111527,DCM,“DateTime Ended”) = “20190904”
- HAS PROPERTIES: TEXT: (111529,DCM,“Brand Name”) = “425362-245-T”
- HAS PROPERTIES: CODE: (410675002,SCT,“Route of administration”) = (34206005,SCT,“Subcutaneous route”)
- HAS PROPERTIES: CODE: (272737002,SCT,“Site of”) = (58602004,SCT,“Flank”)
- HAS CONCEPT MOD: CODE: (272741003,SCT,“Laterality”) = (24028007,SCT,“Right”)
- HAS PROPERTIES: CODE: (127401,DCM,“Tissue of origin”) = (39937001,SCT,“Skin”)
- HAS PROPERTIES: CODE: (127402,DCM,“Taxonomic rank of origin”) = (337915000,SCT,“Homo sapiens”)
- Anatomical Orientation Type (0010,2210) = “QUADRUPED”
- Patient Species Description (0010,2201) = “Mus musculus”
- Patient Species Code Sequence (0010,2202):
- > Code Value = “447612001”
- > Coding Scheme Designator = “SCT”
- > Code Meaning = “Mus musculus”
- Strain Description (0010,0212) = “NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ”
- Strain Nomenclature (0010,0213) = “MGI_2013”
- Strain Code Sequence (0010,0219):
- > Code Value = “3577020”
- > Coding Scheme Designator = “MGI”
- > Code Meaning = “NOD.Cg-Prkdc<scid> Il2rg<tm1Wjl>/SzJ”
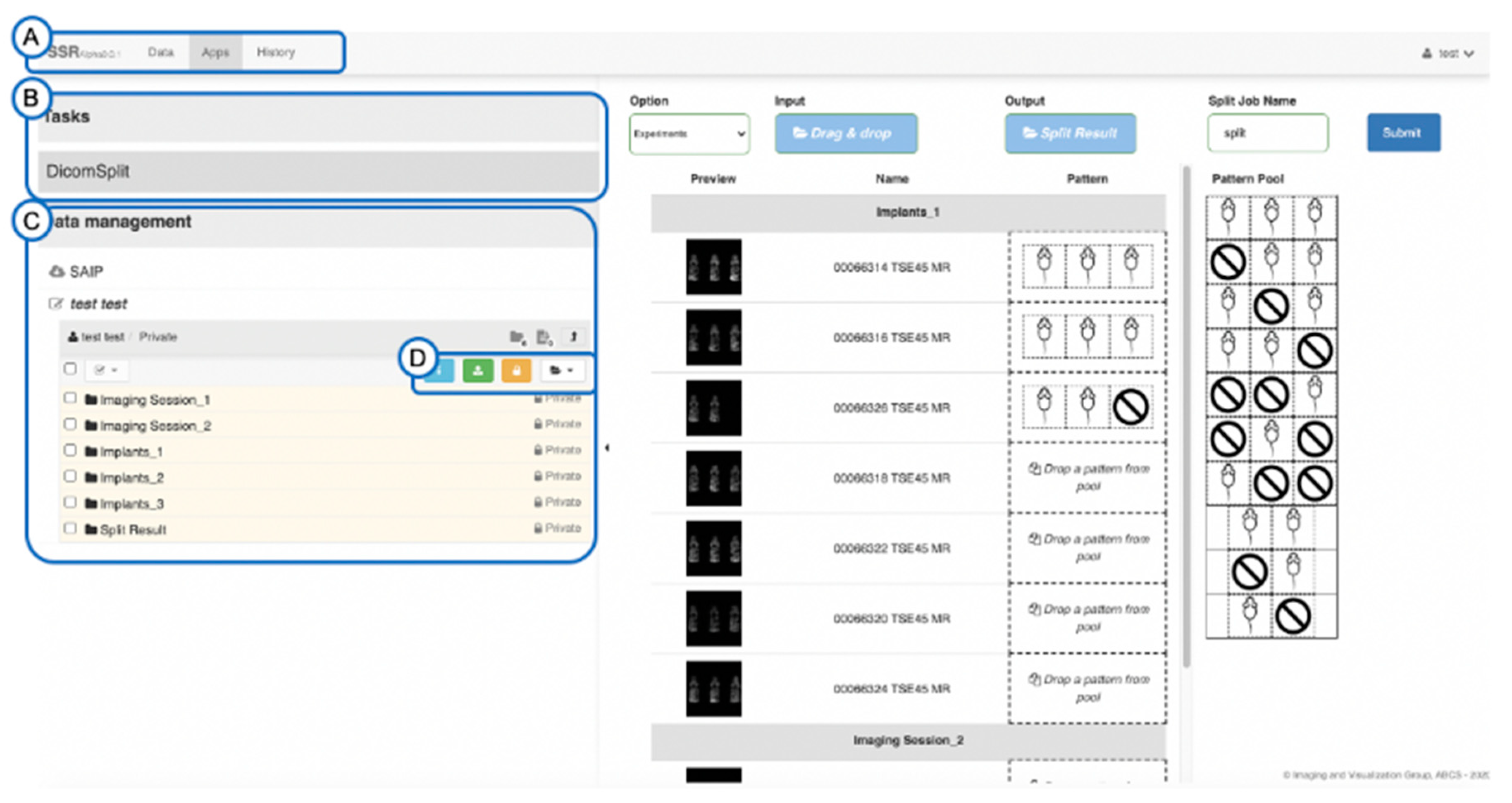
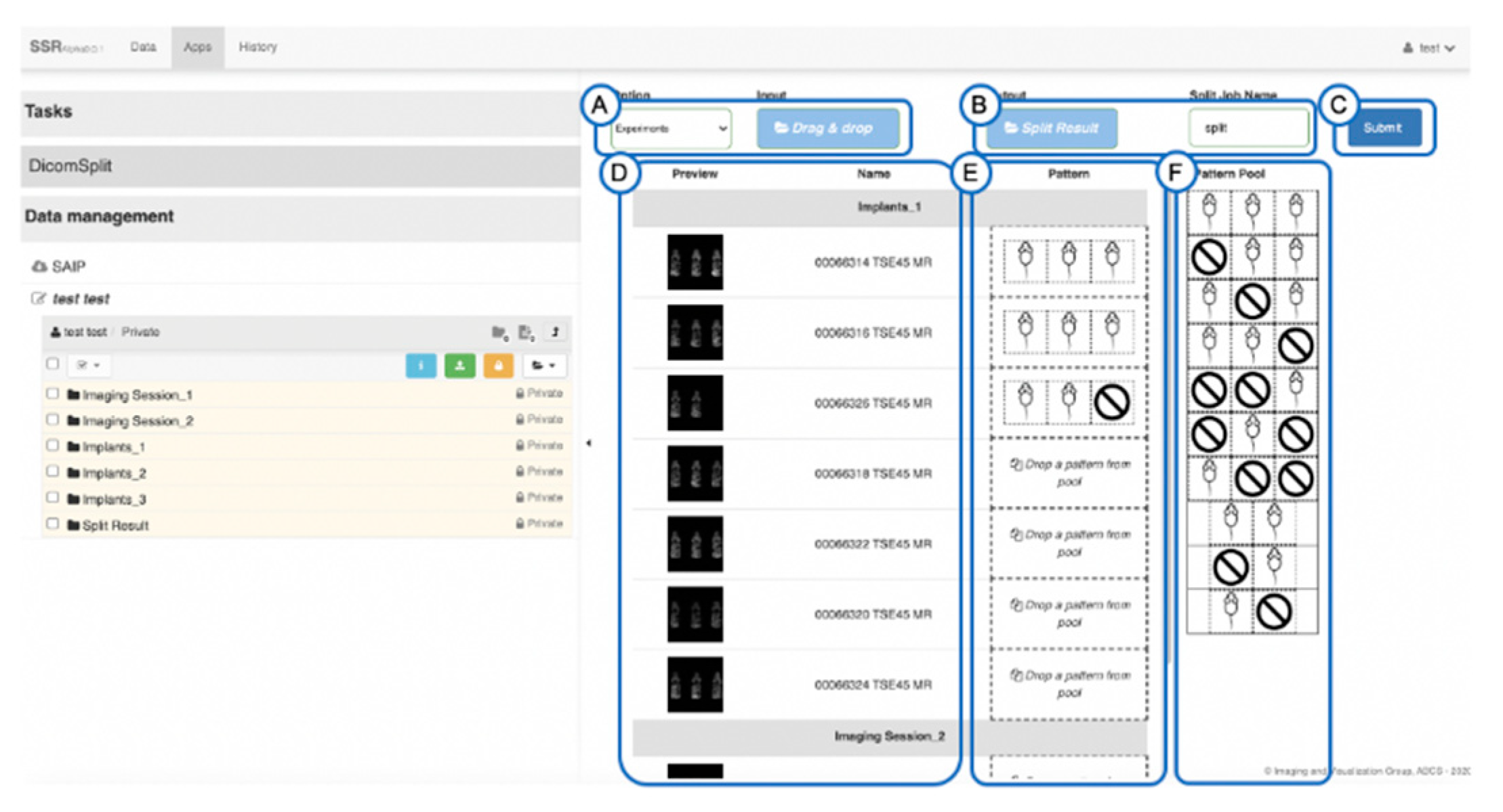
2.2. Splitting and Identification of Multi-Mouse Images
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DICOM | Digital Imaging and Communications in Medicine |
| MRI | Magnetic Resonance Imaging |
| TCIA | The Cancer Imaging Archive |
References
- Bidgood, W.D.; Bray, B.; Brown, N.; Mori, A.R.; Spackman, K.A.; Golichowski, A.; Jones, R.H.; Korman, L.; Dove, D.; Hildebrand, L.; et al. Image Acquisition Context: Procedure Description Attributes for Clinically Relevant Indexing and Selective Retrieval of Biomedical Images. JAMIA 1998, 6, 61–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- National Cancer Institute Patient-Derived Models Repository. Available online: https://pdmr.cancer.gov/ (accessed on 26 September 2020).
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meehan, T.F.; Conte, N.; Goldstein, T.; Inghirami, G.; Murakami, M.A.; Brabetz, S.; Gu, Z.; Wiser, J.A.; Dunn, P.; Begley, D.A.; et al. PDX-MI: Minimal Information for Patient-Derived Tumor Xenograft Models. Cancer Res. 2017, 77, e62–e66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, R.M. Patient-derived orthotopic xenografts: Better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer 2015, 15, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Stout, D.; Berr, S.S.; LeBlanc, A.; Kalen, J.D.; Osborne, D.; Price, J.; Schiffer, W.; Kuntnre, C.; Wall, J. Guidance for Methods Descriptions Used in Preclinical Imaging Papers. Mol. Imaging 2013, 12, 1–15. [Google Scholar] [CrossRef]
- Fueger, B.J.; Czernin, J.; Hildebrandt, I.; Tran, C.; Halpern, B.S.; Stout, D.; Phelps, M.E.; Weber, W.A. Impact of Animal Handling on the Results of 18F-FDG PET Studies in Mice. J. Nucl. Med. 2006, 47, 999–1006. [Google Scholar] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- NCI Metathesaurus. Available online: https://ncim.nci.nih.gov/ncimbrowser/ (accessed on 26 September 2020).
- National Library of Medicine (NLM) Unified Medical Language System (UMLS). Available online: https://www.nlm.nih.gov/research/umls/index.html (accessed on 26 September 2020).
- The Mouse Genome Informatics. Available online: www.informatics.jax.org (accessed on 26 September 2020).
- American Society of Anesthesiologists. Available online: www.asahq.org (accessed on 26 September 2020).
- The Systemized Nomenclature of Medicine Clinical Terms (SNOMED CT). Available online: http://www.snomed.org (accessed on 26 September 2020).
- Clunie, D.A. DICOM Structured Reporting; PixelMed Publishing: Bangor, ME, USA, 2000; ISBN 978-0-9701369-0-9. Available online: http://pixelmed.com/srbook.html (accessed on 26 September 2020).
- DICOM Standards Committee. DICOM Supplement 187—Preclinical Small Animal Imaging Acquisition Context. National Electrical Manufacturers Association (NEMA). 2016. Available online: http://medical.nema.org/medical/dicom/final/sup187_ft_preclinicalanimalacquisitioncontext.pdf (accessed on 26 September 2020).
- National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.16—Content Mapping Resource—TID 8101 Preclinical Small Animal Image Acquisition Context. Available online: http://dicom.nema.org/medical/dicom/current/output/chtml/part16/sect_AcquisitionContextSRIODTemplates.html#sect_TID_8101 (accessed on 26 September 2020).
- Clunie, D.A. PixelMed Publishing™ Java DICOM Toolkit. Available online: http://www.pixelmed.com/dicomtoolkit.html (accessed on 26 September 2020).
- Clunie, D.A. DICOM Validator—Dciodvfy. Available online: http://www.dclunie.com/dicom3tools/dciodvfy.html (accessed on 26 September 2020).
- Clunie, D.A. DicomSRValidator. Available online: http://www.dclunie.com/pixelmed/software/javadoc/com/pixelmed/validate/DicomSRValidator.html (accessed on 26 September 2020).
- DICOM Standards Committee. DICOM CP 1478—Identification of Species and Strain of Pre-Clinical Research Small Animal Subjects. National Electrical Manufacturers Association (NEMA). 2016. Available online: http://medical.nema.org/medical/dicom/final/cp1478_ft_speciesandstrain.pdf (accessed on 26 September 2020).
- DICOM Standards Committee. DICOM CP 1457—Identification of Groups of Pre-Clinical Research Small Animal Subjects. National Electrical Manufacturers Association (NEMA). 2015. Available online: http://medical.nema.org/medical/dicom/final/cp1457_ft_smallanimalidentification.pdf (accessed on 26 September 2020).
- National Electrical Manufacturers Association (NEMA). Digital Imaging and Communications in Medicine (DICOM) Standard PS3.3—C.7.1.4 Patient Group Macro; National Electrical Manufacturers Association (NEMA): Rosslyn, VA, USA. Available online: http://dicom.nema.org/medical/dicom/current/output/chtml/part03/sect_C.7.html#sect_C.7.1.4 (accessed on 26 September 2020).
- Pydicom Library. Available online: https://pydicom.github.io (accessed on 26 September 2020).
- Suloway, C.; Miao, T.; Liu, Y. Pydicom_Split. Available online: https://github.com/abcsFrederick/pydicom_split (accessed on 26 September 2020).
- Uuid4. Available online: https://docs.python.org/3/library/uuid.html (accessed on 26 September 2020).
- ITU-T Recommendation X667: Information technology—Procedures for the Operation of Object Identifier Registration Authorities: Generation of Universally Unique Identifiers and Their Use in Object Identifiers. Available online: https://www.itu.int/rec/T-REC-X.667/en (accessed on 8 October 2020).
- Suloway, C.; Miao, T.; Liu, Y. SSR—A Searchable, Sharable, and Reproducible Imaging Informatics Environment for Biomedical Image Storage, Annotation, and Analysis. In Proceedings of the Practice & Experience in Advanced Research Computing Conference Series (PEARC19), Chicago, IL, USA, 28 July–1 August 2019. [Google Scholar]
- Girder Data Management Platform; Kitware Inc.: Clifton Park, NY, USA. Available online: https://github.com/girder/girder (accessed on 26 September 2020).
- Tatum, J.L.; Kalen, J.D.; Jacobs, P.M.; Ileva, L.V.; Riffle, L.A.; Hollingshead, M.G.; Doroshow, J.H. A spontaneously metastatic model of bladder cancer: Imaging characterization. J. Transl. Med. 2019, 17, 425–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- The Cancer Imaging Archive. Available online: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=52757379 (accessed on 2 February 2021).
| DICOM Tag | DICOM Attribute | Action |
|---|---|---|
| (0008,0018) | SOPInstanceUID | Create new “2.25.” decimal encoded OID per X.667 |
| (0008,2111) | DerivationDescription | Added string attribute for reference purpose |
| (0008,9124) | DerivationImageSequence | Added SQ attribute for referencing back to original SOPClassUID and SOPInstanceUID |
| (0010,0010) | PatientName | Create new patient name based on the original patient name |
| (0010,0020) | PatientID | Create new patient ID based on the original patient ID |
| (0010,0026) | SourcePatientGroupIdentificationSequence | Added attribute stores the original patient name and ID |
| (0020,000d) | StudyInstanceUID | Create new “2.25.” decimal encoded OID per X.667 |
| (0020,000e) | SeriesInstanceUID | Create new “2.25.” decimal encoded ODI per X.667 |
| (0020,0011) | SeriesNumber | Update series number based on the original series number |
| (0088,0140) | StorageMediaFileSetUID | Create new “2.25.” decimal encoded OID per X.667 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalen, J.D.; Clunie, D.A.; Liu, Y.; Tatum, J.L.; Jacobs, P.M.; Kirby, J.; Freymann, J.B.; Wagner, U.; Smith, K.E.; Suloway, C.; et al. Design and Implementation of the Pre-Clinical DICOM Standard in Multi-Cohort Murine Studies. Tomography 2021, 7, 1-9. https://doi.org/10.3390/tomography7010001
Kalen JD, Clunie DA, Liu Y, Tatum JL, Jacobs PM, Kirby J, Freymann JB, Wagner U, Smith KE, Suloway C, et al. Design and Implementation of the Pre-Clinical DICOM Standard in Multi-Cohort Murine Studies. Tomography. 2021; 7(1):1-9. https://doi.org/10.3390/tomography7010001
Chicago/Turabian StyleKalen, Joseph D., David A. Clunie, Yanling Liu, James L. Tatum, Paula M. Jacobs, Justin Kirby, John B. Freymann, Ulrike Wagner, Kirk E. Smith, Christian Suloway, and et al. 2021. "Design and Implementation of the Pre-Clinical DICOM Standard in Multi-Cohort Murine Studies" Tomography 7, no. 1: 1-9. https://doi.org/10.3390/tomography7010001
APA StyleKalen, J. D., Clunie, D. A., Liu, Y., Tatum, J. L., Jacobs, P. M., Kirby, J., Freymann, J. B., Wagner, U., Smith, K. E., Suloway, C., & Doroshow, J. H. (2021). Design and Implementation of the Pre-Clinical DICOM Standard in Multi-Cohort Murine Studies. Tomography, 7(1), 1-9. https://doi.org/10.3390/tomography7010001





