A Flexible Multi-Channel Deep Network Leveraging Texture and Spatial Features for Diagnosing New COVID-19 Variants in Lung CT Scans
Abstract
1. Introduction
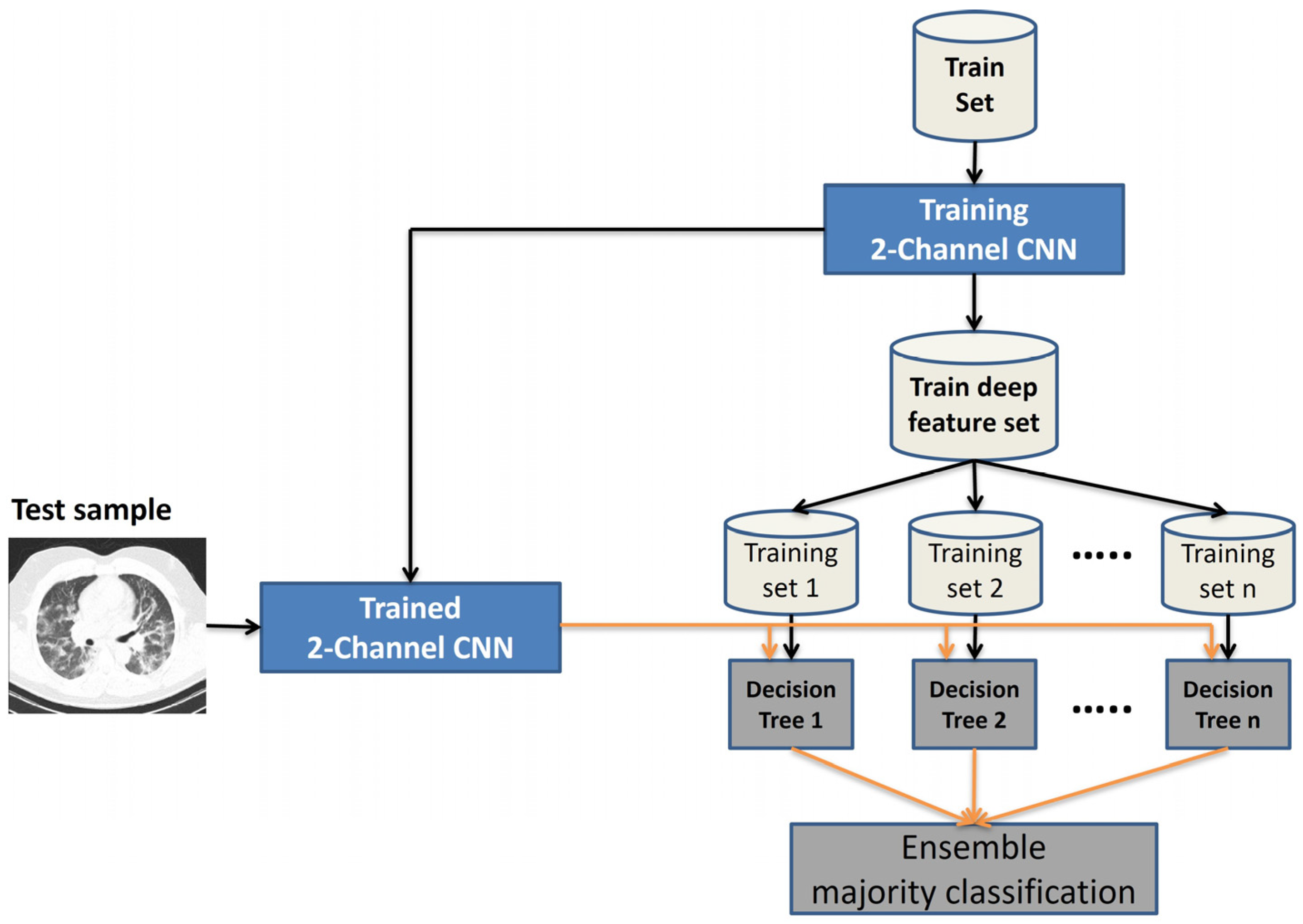
- Using a two-channel convolutional neural network with the same structure in channels and two different power sources can increase the accuracy of COVID-19 detection compared to existing methods.
- When using a consistent and tuned classifier, the use of deep features in a classical machine-learning pipeline in which feature extraction and classification are performed separately can achieve higher accuracy than many popular deep neural networks for COVID-19 diagnosis.
2. Related Works
3. Materials and Methods
- There is an imbalance between the number of features extracted using deep convolutional networks versus texture analysis operators and a much longer execution time of training and feature extraction in the deep network than using statistical texture analysis operators.
- The deep network needs to be trained, while in the other phase the texture operators are not trained.
3.1. Preprocessing Step
- Histogram equalization algorithm can be performed in the following three steps:
- Build the histogram of the input image;
- Compute the normalized summation of histogram bins;
- Transform the input image to an output image.
3.2. Feature Extraction Step
3.2.1. Modified Local Binary Patterns
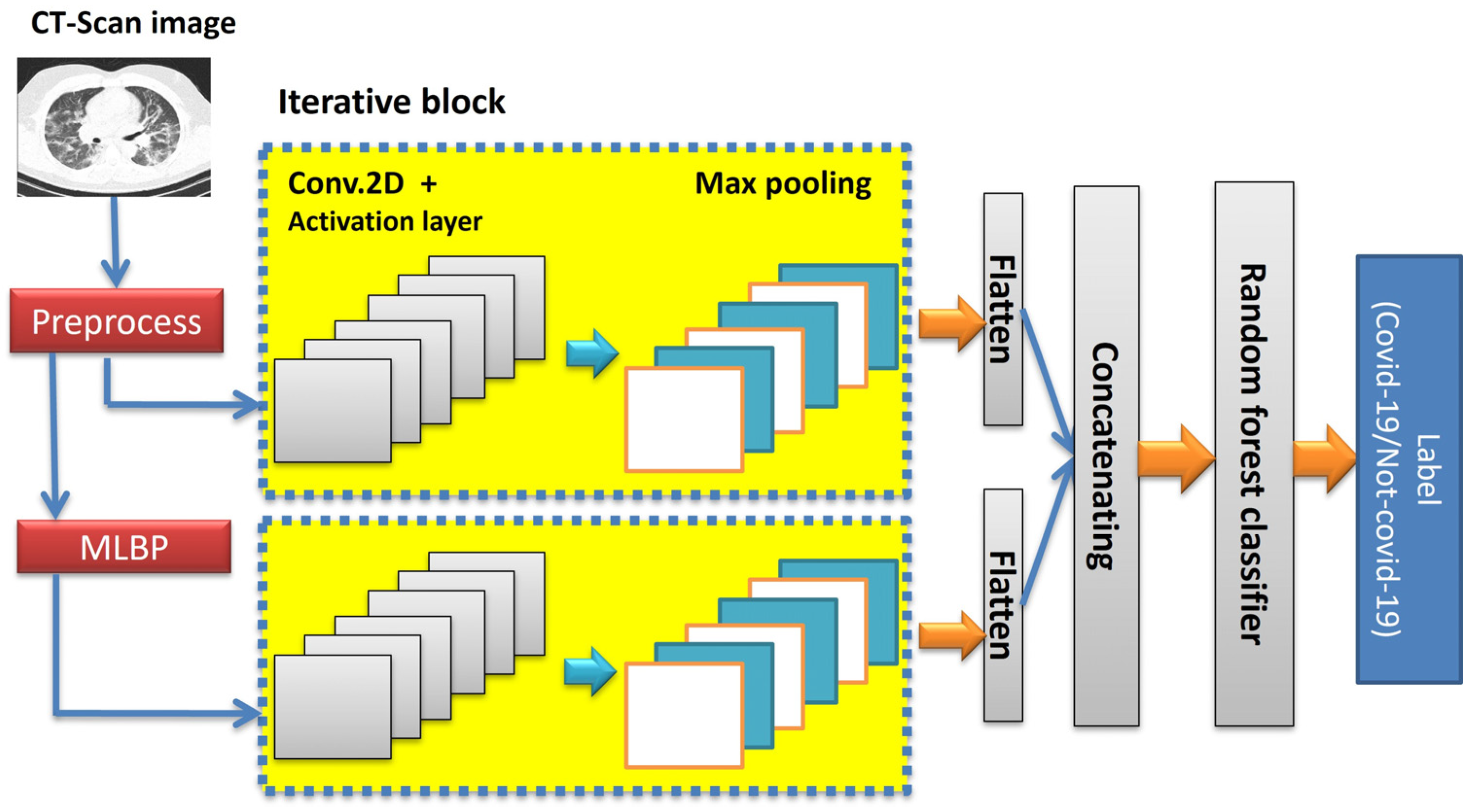
3.2.2. Proposed Deep CNN
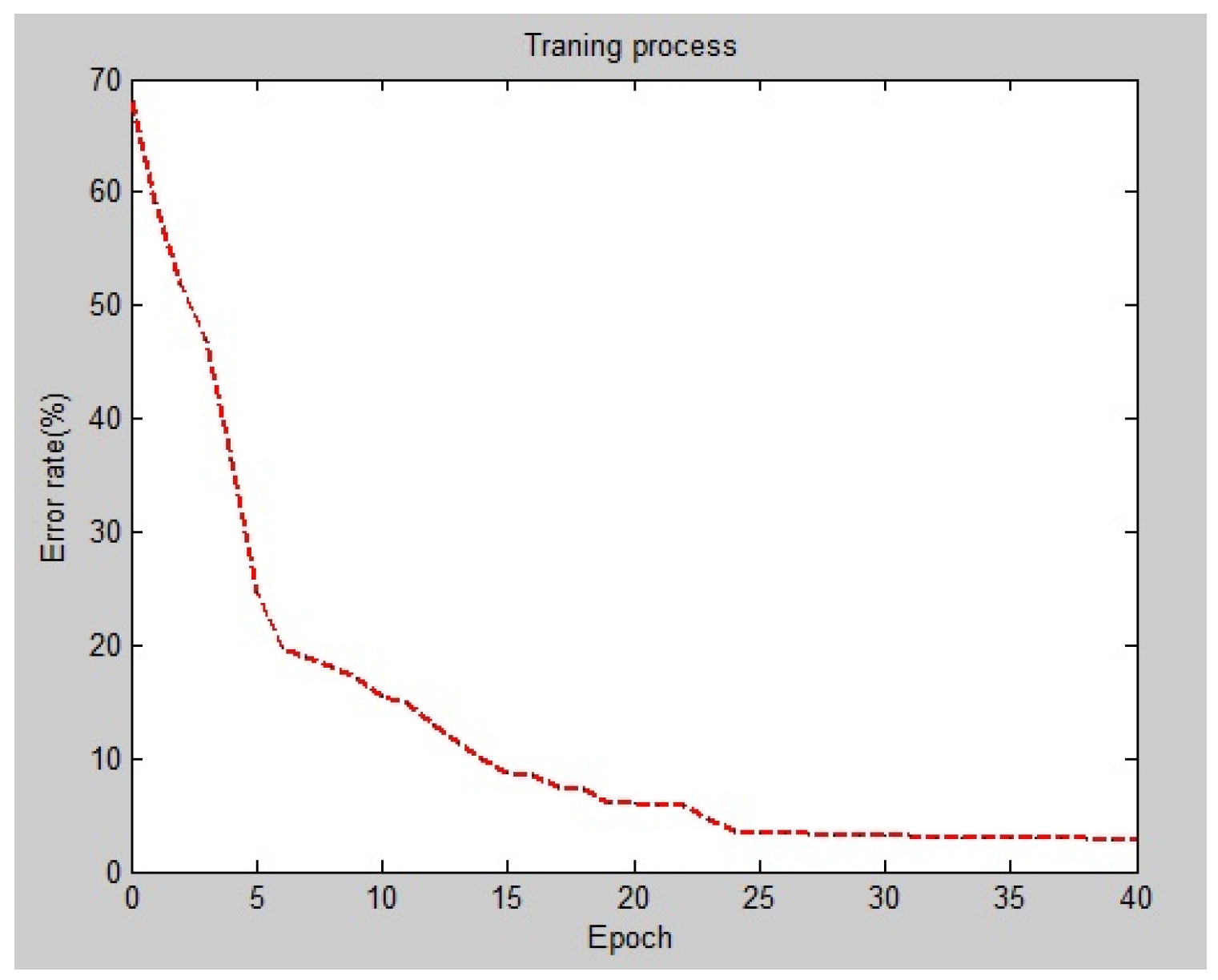
3.2.3. Hyperparameter Optimization
3.3. Classification Step
- In addition to classification, it can also be used for regression;
- Due to the concept of branches and trees, random forest predictions are more human-understandable than other classifiers;
- It can handle large datasets more efficiently than some other supervised classifiers;
- The random forest algorithm provides a higher level of accuracy in predicting outcomes than the decision tree algorithm.
4. Experimental Results
4.1. Datasets
4.2. Performance Evaluation Metrics
4.3. Performance Evaluation of the Proposed Approach in Terms of Different Classifiers
4.4. Random Forest Tuning
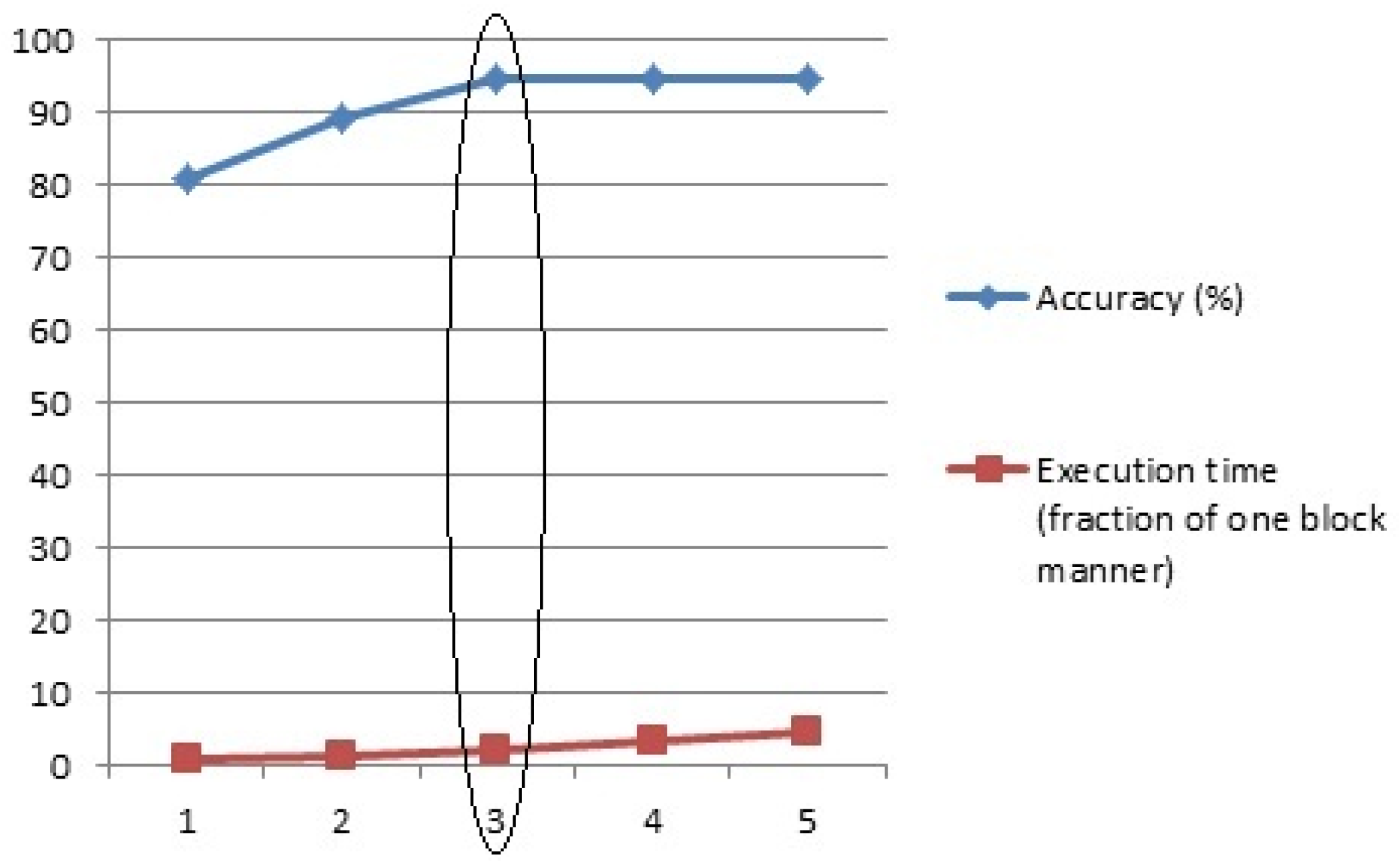
4.5. DCNN Optimization in Terms of Internal Blocks
4.6. DCNN Optimization in Terms of Learning Rate
4.7. Comparison Proposed Approach with Base-Line Methods
4.8. Comparison with State-of-the-Art Methods
4.9. Offline and Online Process
5. Discussion
- Using a two-channel CNN with the same architecture in channels and two different power sources provides better results for COVID-19 detection compared to existing one-channel methods.
- Using deep features in combination of supervised classifiers in which feature extraction and classification are performed separately achieves higher diagnostic accuracy than many popular deep CNNs for COVID-19 diagnosis.
6. Conclusions
- COVID-19 causes detectable lung texture changes. The proposed DCNN without the presence of a channel that feeds with image texture features provided a lower detection accuracy than the two-channel mode.
- Replacing deep CNN classification layers with supervised classifiers improved the COVID-19 diagnosis accuracy.
- Unlike methods that concatenate deep features with texture information, in this paper, the texture features were used to feed one of the channels of a deep convolutional neural network. The results showed that the presented two-channel CNN provides higher accuracy than many compared methods.
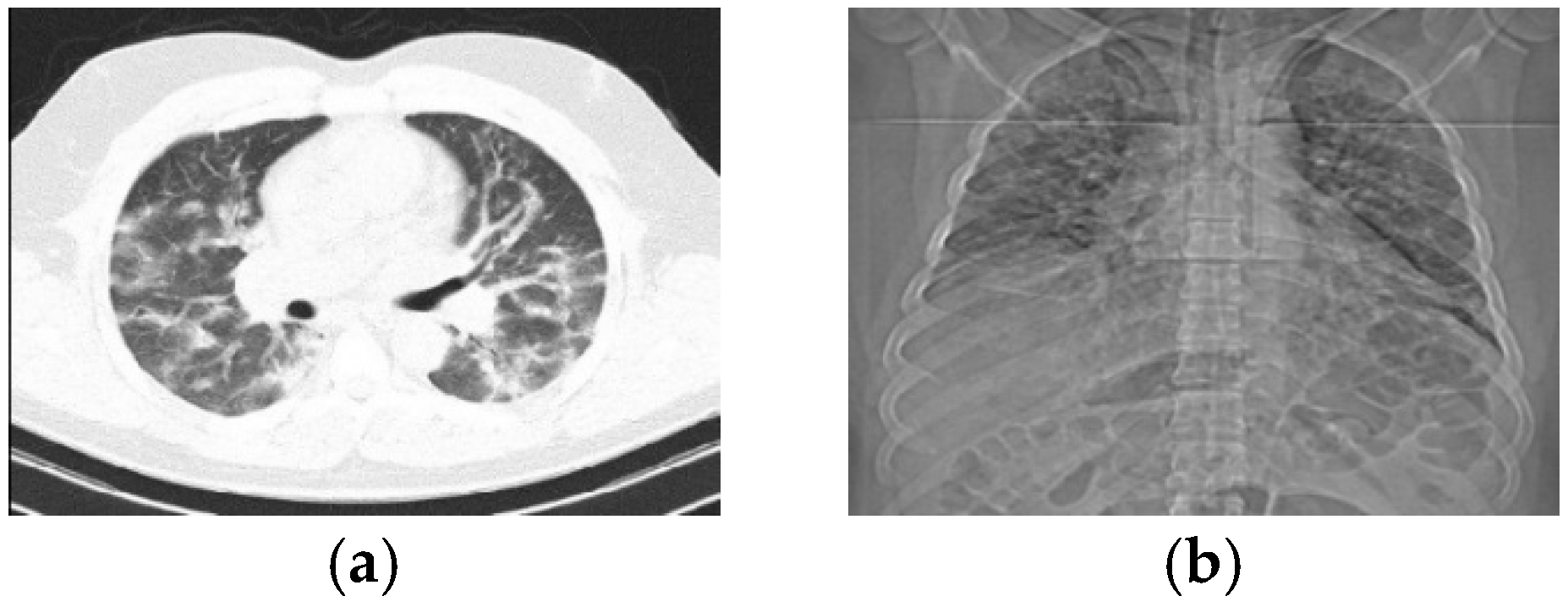
- Results show that COVID-19 disease, in addition to appearance properties such as color and shape, may disturb the texture of the patient’s lungs.
- The proposed method is a general approach where none of its steps are dependent on the type of input image. Therefore, an idea for future research is to apply the presented model in other visual pattern classification problems. The classification task in the proposed method originates from the trained two-channel deep neural network that is trained by spatial information and texture features of CT scan images. Therefore, the proposed two-channel deep neural network has a general architecture and can be used in similar classification problems such as diagnosing various lung diseases, diagnosing breast cancer, and diagnosing malignant tumors, in which texture variations play an important role.
- Like other viruses, COVID-19 continually evolves and undergoes mutations, resulting in new variants with distinct characteristics such as altered transmissibility and disease severity. Therefore, current detection methods, including ours, are inherently limited to recognizing variants present in their training data. However, our proposed method’s trainable architecture allows for adaptation to emerging variants by incorporating new CT scan images from patients infected with novel COVID-19 variants.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yi, J.; Zhang, H.; Mao, J.; Chen, Y.; Zhong, H.; Wang, Y. Review on the COVID-19 pandemic prevention and control system based on AI. Eng. Appl. Artif. Intell. 2022, 114, 105184. [Google Scholar] [CrossRef]
- Filchakova, O.; Dossym, D.; Ilyas, A.; Kuanysheva, T.; Abdizhamil, A.; Bukasov, R. Review of COVID-19 testing and diagnostic methods. Talanta 2022, 244, 123409. [Google Scholar] [CrossRef] [PubMed]
- Pu, R.; Liu, S.; Ren, X.; Shi, D.; Ba, Y.; Huo, Y.; Zhang, W.; Ma, L.; Liu, Y.; Yang, Y.; et al. The screening value of RT-LAMP and RT-PCR in the diagnosis of COVID-19: Systematic review and meta-analysis. J. Virol. Methods 2022, 300, 114392. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.L.; Mirza, A.; Gallagher, N.; Boudreau, A.; Jacinto, L.G.; Yu, T.; Norton, J.; Luo, C.H.; Conte, A.; Zhou, R.; et al. Limitations of molecular and antigen test performance for SARS-CoV-2 in symptomatic and asymptomatic COVID-19 contacts. J. Clin. Microbiol. 2022, 60, e0018722. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Tong, L.; Zhu, Y.; Wang, M.D. COVID-19 automatic diagnosis with radiographic imaging: Explainable attention transfer deep neural networks. IEEE J. Biomed. Health Inform. 2021, 25, 2376–2387. [Google Scholar] [CrossRef]
- Nascimento, E.D.; Fonseca, W.T.; de Oliveira, T.R.; de Correia, C.R.; Faça, V.M.; de Morais, B.P.; Silvestrini, V.C.; Pott-Junior, H.; Teixeira, F.R.; Faria, R.C. COVID-19 diagnosis by SARS-CoV-2 Spike protein detection in saliva using an ultrasensitive magneto-assay based on disposable electrochemical sensor. Sens. Actuators B Chem. 2022, 353, 131128. [Google Scholar] [CrossRef]
- Saslow, D.; Runowicz, C.D.; Solomon, D.; Moscicki, A.; Smith, R.A.; Eyre, H.J.; Cohen, C. American Cancer Society guideline for the early detection of cervical neoplasia and cancer. CA A Cancer J. Clin. 2002, 52, 342–362. [Google Scholar] [CrossRef]
- Hussain, E.; Hasan, M.; Rahman, A.; Lee, I.; Tamanna, T.; Parvez, M.Z. CoroDet: A deep learning based classification for COVID-19 detection using chest X-ray images. Chaos Solitons Fractals 2021, 142, 110495. [Google Scholar] [CrossRef]
- Satu, S.; Ahammed, K.; Abedin, M.Z.; Rahman, A.; Islam, S.M.S.; Azad, A.K.M.; Alyami, S.A.; Moni, M.A. Convolutional neural network model to detect COVID-19 patients utilizing lung X-ray images. medRxiv 2020. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef]
- Huang, M.-L.; Liao, Y.-C. A lightweight CNN-based network on COVID-19 detection using X-ray and CT images. Comput. Biol. Med. 2022, 146, 105604. [Google Scholar] [CrossRef] [PubMed]
- Kathamuthu, N.D.; Subramaniam, S.; Le, Q.H.; Muthusamy, S.; Panchal, H.; Sundararajan, S.C.M.; Alrubaie, A.J.; Zahra, M.M.A. A deep transfer learning-based convolution neural network model for COVID-19 detection using computed tomography scan images for medical applications. Adv. Eng. Softw. 2023, 175, 103317. [Google Scholar] [CrossRef] [PubMed]
- Jalali, S.M.J.; Ahmadian, M.; Ahmadian, S.; Hedjam, R.; Khosravi, A.; Nahavandi, S. X-ray image based COVID-19 detection using evolutionary deep learning approach. Expert Syst. Appl. 2022, 201, 116942. [Google Scholar] [CrossRef] [PubMed]
- Celik, G. Detection of COVID-19 and other pneumonia cases from CT and X-ray chest images using deep learning based on feature reuse residual block and depthwise dilated convolutions neural network. Appl. Soft Comput. 2023, 133, 109906. [Google Scholar] [CrossRef]
- Shah, V.; Keniya, R.; Shridharani, A.; Punjabi, M.; Shah, J.; Mehendale, N. Diagnosis of COVID-19 using CT scan images and deep learning techniques. Emerg. Radiol. 2021, 28, 497–505. [Google Scholar] [CrossRef]
- Alquran, H.; Alsleti, M.; Alsharif, R.; Abu Qasmieh, I.; Alqudah, A.M.; Harun, N.H.B. Employing texture features of chest x-ray images and machine learning in COVID-19 detection and classification. Mendel 2021, 27, 9–17. [Google Scholar] [CrossRef]
- Yasar, H.; Ceylan, M. A novel comparative study for detection of COVID-19 on CT lung images using texture analysis, machine learning, and deep learning methods. Multimed. Tools Appl. 2021, 80, 5423–5447. [Google Scholar] [CrossRef]
- Hussain, L.; Nguyen, T.; Li, H.; Abbasi, A.A.; Lone, K.J.; Zhao, Z.; Zaib, M.; Chen, A.; Duong, T.Q. Machine-learning classification of texture features of portable chest X-ray accurately classifies COVID-19 lung infection. BioMed. Eng. Online 2020, 19, 88. [Google Scholar] [CrossRef]
- Ravi, V.; Narasimhan, H.; Chakraborty, C.; Pham, T.D. Deep learning-based meta-classifier approach for COVID-19 classification using CT scan and chest X-ray images. Multimed. Syst. 2022, 28, 1401–1415. [Google Scholar] [CrossRef]
- Soni, M.; Singh, A.K.; Babu, K.S.; Kumar, S.; Kumar, A.; Singh, S. Convolutional neural network based CT scan classification method for COVID-19 test validation. Smart Health 2022, 25, 100296. [Google Scholar] [CrossRef]
- Perumal, V.; Narayanan, V.; Rajasekar, S.J.S. Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl. Intell. 2021, 51, 341–358. [Google Scholar] [CrossRef]
- Varela-Santos, S.; Melin, P. A new approach for classifying coronavirus COVID-19 based on its manifestation on chest X-rays using texture features and neural networks. Inf. Sci. 2021, 545, 403–414. [Google Scholar] [CrossRef]
- Öztürk, Ş.; Özkaya, U.; Barstuğan, M. Classification of Coronavirus (COVID-19) from X-ray and CT images using shrunken features. Int. J. Imaging Syst. Technol. 2021, 31, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Liu, X.; Chen, S.; Liu, S.; Liu, X.; Muhammad, K.; Bhattacharyya, S. A deep fuzzy model for diagnosis of COVID-19 from CT images. Appl. Soft Comput. 2022, 122, 108883. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.K.; Kashyap, M. Automated diagnosis of COVID stages from lung CT images using statistical features in 2-dimensional flexible analytic wavelet transform. Biocybern. Biomed. Eng. 2022, 42, 829–841. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. A computer-aided diagnostic framework for coronavirus diagnosis using texture-based radiomics images. Digit. Health 2022, 8, 20552076221092543. [Google Scholar] [CrossRef]
- Dabov, K.; Foi, A.; Katkovnik, V.; Egiazarian, K. Image denoising by sparse 3-D transform-domain collaborative filtering. IEEE Trans. Image Process. 2007, 16, 2080–2095. [Google Scholar] [CrossRef]
- Rubel, A.; Lukin, V.; Uss, M.; Vozel, B.; Pogrebnyak, O.; Egiazarian, K. Efficiency of texture image enhancement by DCT-based filtering. Neurocomputing 2016, 175, 948–965. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikäinen, M.; Mäenpää, T. Gray scale and rotation invariant texture classification with local binary patterns. In Proceedings of the European Conference on Computer Vision, Dublin, Ireland, 26 June–1 July 2000; pp. 404–420. [Google Scholar]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, Y.; He, X.; Xie, P. Covid-ct-dataset: A ct scan dataset about COVID-19. arXiv 2020, arXiv:2003.13865. [Google Scholar]
- Salama, W.M.; Aly, M.H. Framework for COVID-19 segmentation and classification based on deep learning of computed tomography lung images. J. Electron. Sci. Technol. 2022, 20, 100161. [Google Scholar] [CrossRef]
- Wang, Z.; Dong, J.; Zhang, J. Multi-model ensemble deep learning method to diagnose COVID-19 using chest computed tomography images. J. Shanghai Jiaotong Univ. (Sci.) 2022, 27, 70–80. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Zhao, H.; Jie, Y.; Wang, R. Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef]
- Imani, M. Automatic diagnosis of coronavirus (COVID-19) using shape and texture characteristics extracted from X-Ray and CT-Scan images. Biomed. Signal Process. Control. 2021, 68, 102602. [Google Scholar] [CrossRef]
- Chen, X.; Bai, Y.; Wang, P.; Luo, J. Data augmentation based semi-supervised method to improve COVID-19 CT classification. Math. Biosci. Eng. 2023, 20, 6838–6852. [Google Scholar] [CrossRef]
- COVID-19 CT Segmentation Dataset. Available online: http://medicalsegmentation.com/covid19/ (accessed on 10 October 2023).
- Kordnoori, S.; Sabeti, M.; Mostafaei, H.; Banihashemi, S.S.A. Analysis of lung scan imaging using deep multi-task learning structure for COVID-19 disease. IET Image Process. 2023, 17, 1534–1545. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of coronavirus Disease (COVID-19) based on deep features and support vector machine. Int. J. Math. Eng. Manag. Sci. 2020, 5, 643–651. [Google Scholar] [CrossRef]
- Amyar, A.; Modzelewski, R.; Li, H.; Ruan, S. Multi-task deep learning based CT imaging analysis for COVID-19 pneumonia: Classification and segmentation. Comput. Biol. Med. 2020, 126, 104037. [Google Scholar] [CrossRef]
- Daniel; Cenggoro, T.W.; Pardamean, B. A systematic literature review of machine learning application in COVID-19 medical image classification. Procedia Comput. Sci. 2023, 216, 749–756. [Google Scholar] [CrossRef]
- Lee, M.-H.; Shomanov, A.; Kudaibergenova, M.; Viderman, D. Deep learning methods for interpretation of pulmonary CT and X-ray images in patients with COVID-19-related lung involvement: A systematic review. J. Clin. Med. 2023, 12, 3446. [Google Scholar] [CrossRef]
- Awadelkarim, S.A.M.; Karrar, A.E. Utilization of machine learning algorithms in COVID-19 classification and prediction: A review. IJCSNS 2025, 25, 81. [Google Scholar]
- AhmedK, A.; Aljahdali, S.; Hussain, S.N. Comparative prediction performance with support vector machine and random forest classification techniques. Int. J. Comput. Appl. 2013, 69, 12–16. [Google Scholar] [CrossRef]
- Memos, V.A.; Minopoulos, G.; Stergiou, K.D.; Psannis, K.E. Internet-of-Things-Enabled infrastructure against infectious diseases. IEEE Internet Things Mag. 2021, 4, 20–25. [Google Scholar] [CrossRef]
- Yasar, H.; Ceylan, M. Deep learning–based approaches to improve classification parameters for diagnosing COVID-19 from CT Images. Cogn. Comput. 2024, 16, 1806–1833. [Google Scholar] [CrossRef]
- Ibrahim, A.U.; Ozsoz, M.; Serte, S.; Al-Turjman, F.; Yakoi, P.S. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cogn. Comput. 2024, 16, 1589–1601. [Google Scholar] [CrossRef]
- Kaushik, B.; Chadha, A.; Mahajan, A.; Ashok, M. A three layer stacked multimodel transfer learning approach for deep feature extraction from Chest Radiographic images for the classification of COVID-19. Eng. Appl. Artif. Intell. 2025, 147, 110241. [Google Scholar] [CrossRef]
- Liu, H.; Zhao, M.; She, C.; Peng, H.; Liu, M.; Li, B.; Zafar, B. Classification of CT scan and X-ray dataset based on deep learning and particle swarm optimization. PLoS ONE 2025, 20, e0317450. [Google Scholar] [CrossRef]
- Rashid, H.-A.; Sajadi, M.M.; Mohsenin, T. CoughNet-V2: A scalable multimodal DNN framework for point-of-care edge devices to detect symptomatic COVID-19 cough. In Proceedings of the 2022 IEEE Healthcare Innovations and Point of Care Technologies (HI-POCT), Houston, TX, USA, 10–11 March 2022; pp. 37–40. [Google Scholar]

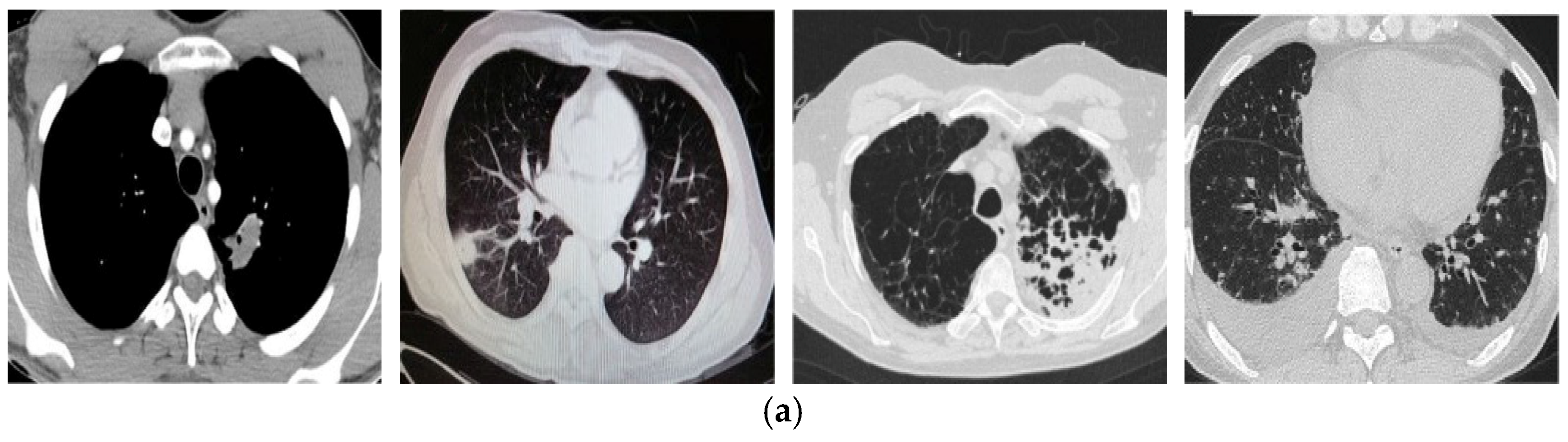
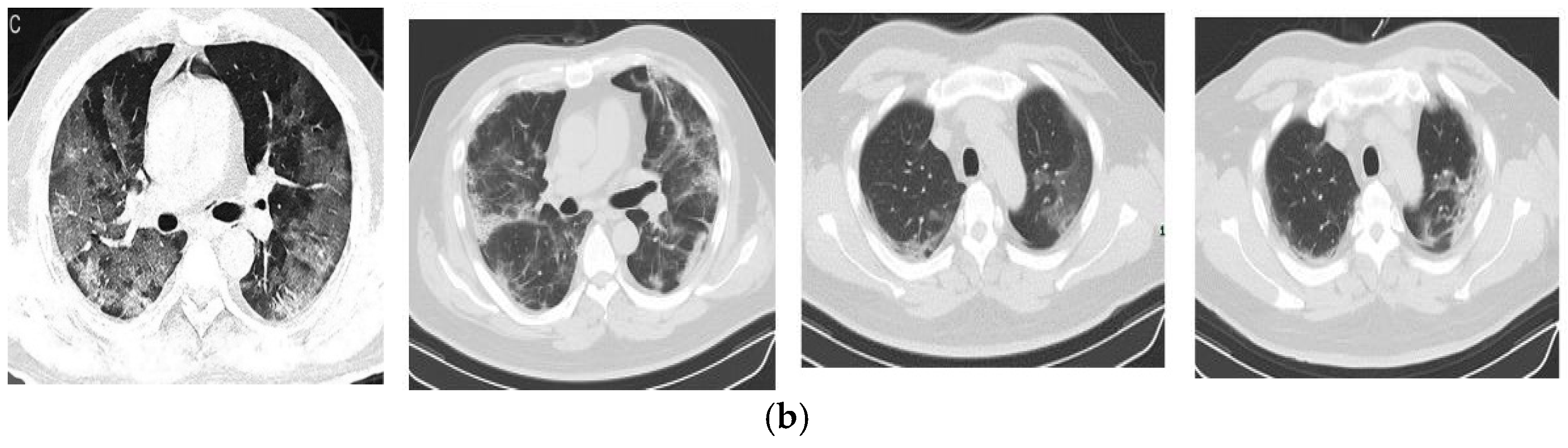

| Ref. | Year | Method | Classification | Advantages | Limitations |
|---|---|---|---|---|---|
| [24] | 2021 | Haralick features + pre-trained networks (VGG16, ResNet50, InceptionV3) | Deep learning | Lower execution time than trained deep networks | Lack of deep network training Lower performance than newer methods |
| [25] | 2021 | GLCM + LBP + MLP | Machine learning | Lower computational complexity than deep method | Lower performance than trained deep networks |
| [26] | 2021 | Handcrafted features (GLCM + LBP + SFTA) + PCA + SVM | Machine learning | Lower computational complexity than deep-based methods | Lower performance than deep learning-based methods |
| [22] | 2022 | EfficientNet + KPCA + Ensemble classifiers | Hybrid | Higher performance than compared methods in reference | Higher tunable parameters than single classifiers |
| [23] | 2022 | Fusion of Deep networks (Resnet50/Inception V3/Efficientb7) | Deep learning | Higher performance than compared machine-learning-based methods | Higher execution time than single deep neural network-based methods |
| [27] | 2022 | Deep features + Fuzzy classification | Deep learning | Higher performance than compared networks in the reference | Dependence of variants diagnosis on the redefinition of fuzzy rules |
| [28] | 2022 | Statistical features + DWT + PCA + SVM | Machine learning | Lower computational complexity than deep-based | Lower performance than trained deep networks |
| [29] | 2022 | Texture (GLCM/Wavelet transform) + ResNet | Hybrid | Higher performance than compared deep methods in reference | Lower detection accuracy than newer methods |
| [34] | 2024 | Pipeline (LBP + DWT + CNN) | Hybrid | Higher performance than compared networks in the reference | High ratio of COVID-19 samples to non-COVID-19 sample Lower detection accuracy than newer methods |
| [35] | 2024 | Fine-tuned AlexNet | Deep learning | Higher performance than pre-trained Alex-Net and compared networks in the reference | Very high ratio of non-COVID-19 samples to COVID-19 samples More parameters than some benchmark deep networks |
| [30] | 2025 | Fusion of pre-trained deep networks | Deep learning | Lower complexity than trained deep network-based methods | Lack of deep network training |
| [31] | 2025 | Residual neural network + Evolutionary algorithm | Deep learning | Higher performance than classical deep learning-based methods | High execution computation time due to using genetic algorithm |
| Block | Layer Type | Input | Output | Details |
|---|---|---|---|---|
| 1 | Conv.2D | 128 × 128 × 3 | 128 × 128 × 3 | 3 Filters 5 × 5, Stride 1, Pad 2 |
| Activation | 128 × 128 × 3 | 128 × 128 × 3 | Swish | |
| Pooling.2D | 128 × 128 × 3 | 64 × 64 × 3 | Max pooling, Stride 1, Spatial extent 1 | |
| 2 | Conv.2D | 64 × 64 × 3 | 64 × 64 × 20 | 20 Filters 5 × 5, Stride 1, Pad 2 |
| Activation | 64 × 64 × 20 | 64 × 64 × 20 | Swish | |
| Pooling.2D | 64 × 64 × 20 | 32 × 32 × 20 | Max pooling, Stride 1, Spatial extent 1 | |
| 3 | Conv.2D | 32 × 32 × 20 | 32 × 32 × 25 | 25 Filters 5 × 5, Stride 1, Pad 2 |
| Activation | 32 × 32 × 25 | 32 × 32 × 25 | Swish | |
| Pooling.2D | 32 × 32 × 25 | 16 × 16 × 25 | Max pooling, Stride 1, Spatial extent 1 | |
| 4 | Flatten | 16 × 16 × 25 | 6400 | Flatten |
| Approach | Accuracy | Precision | Recall | F-Score |
|---|---|---|---|---|
| Naïve bayes | 90.79 | 88.63 | 91.56 | 90.07 |
| C4.5 tree | 90.87 | 89.52 | 90.29 | 89.90 |
| SVM | 94.25 | 91.89 | 95.78 | 93.79 |
| KNN, K = 3 | 91.45 | 90.76 | 90.71 | 90.73 |
| KNN, K = 5 | 93.37 | 92.79 | 92.40 | 92.59 |
| KNN, K = 7 | 93.08 | 92.27 | 92.40 | 92.33 |
| Random forest | 94.63 | 91.63 | 97.04 | 94.25 |
| Number of Trees | Maximum Depth | Accuracy |
|---|---|---|
| 10 | 1 | 87.53 |
| 3 | 89.28 | |
| 5 | 91.87 | |
| 7 | 90.36 | |
| 20 | 1 | 87.69 |
| 3 | 90.45 | |
| 5 | 94.01 | |
| 7 | 92.95 | |
| 30 | 1 | 88.02 |
| 3 | 92.76 | |
| 5 | 94.63 | |
| 7 | 92.61 | |
| 40 | 1 | 87.91 |
| 3 | 92.37 | |
| 5 | 93.35 | |
| 7 | 92.07 |
| Label | COVID-19 | Non COVID-19 | |
|---|---|---|---|
| Predicted | |||
| COVID-19 | 230 | 21 | |
| Non COVID-19 | 7 | 264 | |
| Learning Rate (First 20 Epochs) | Learning Rate (Second 20 Epochs) | Weight Decay Value | Accuracy |
|---|---|---|---|
| 10−4 | 10−5 | 10−3 | 92.35 |
| 10−4 | 10−5 | 10−2 | 92.28 |
| 10−4 | 10−6 | 10−3 | 91.38 |
| 10−4 | 10−6 | 10−2 | 92.54 |
| 10−5 | 10−5 | 10−3 | 93.61 |
| 10−5 | 10−5 | 10−2 | 93.76 |
| 10−5 | 10−6 | 10−3 | 94.31 |
| 10−5 | 10−6 | 10−2 | 94.63 |
| 10−6 | 10−6 | 10−3 | 94.01 |
| 10−6 | 10−6 | 10−2 | 94.21 |
| 10−6 | 10−7 | 10−3 | 92.61 |
| 10−6 | 10−7 | 10−2 | 92.82 |
| Approach | Accuracy | Precision | Recall | F-Score |
|---|---|---|---|---|
| MLBP + RF | 89.76 | 89.23 | 86.36 | 87.77 |
| Proposed DCNN (first channel without RF) | 91.61 | 90.65 | 91.79 | 91.21 |
| Proposed DCNN (second channel without RF) | 90.72 | 89.36 | 91.45 | 90.39 |
| Proposed DCNN (first channel with RF as classifier) | 92.74 | 91.88 | 91.74 | 91.80 |
| Proposed DCNN (second channel with RF as classifier) | 91.86 | 90.41 | 92.08 | 91.23 |
| Proposed approach | 94.63 | 91.63 | 97.04 | 94.25 |
| Approach | Accuracy | Precision | Recall | F-Score |
|---|---|---|---|---|
| ResNet-50 [27] | 77.4 | NR | NR | 74.6 |
| VGG-16 [27] | 76 | NR | NR | 76 |
| EfficientNet-B1 [27] | 79 | NR | NR | 79 |
| DenseNet-169 [27] | 79.5 | NR | NR | 76 |
| Fine-tuned DenseNet-169 [43] | 87.1 | NR | NR | 88.1 |
| Deep fuzzy model [27] | 94.20 | NR | NR | 93.8 |
| ResNet-18 [44] | 90.42 | 88.24 | 91.71 | 89.43 |
| CovidNet-CT [44] | 90.48 | NR | NR | NR |
| DRE-Net [45] | 84.74 | NR | NR | 84 |
| CFRCF + Gabor + RF [46] | 76.68 | NR | NR | 74.26 |
| CFRCF + EMAP + SVM [46] | 66.37 | NR | NR | 61.14 |
| LBP + RF [46] | 69.96 | NR | NR | 65.64 |
| Conventional CNN [46] | 77.03 | NR | NR | 67.92 |
| Teacher–student framework + data augmentation [47] | 79.56 | 84.59 | 74.93 | 79.47 |
| Residual neural network + evolutionary algorithm [31] | 83.04 | 88.18 | 79.50 | 83.62 |
| Proposed approach | 94.63 | 91.63 | 97.04 | 94.25 |
| Approach | Accuracy | Precision | Recall | F-Score |
|---|---|---|---|---|
| Deep encoder–decoder + MLP [48] | 95.40 | NR | 93.1 | NR |
| COVID-Net [49] | 92.60 | NR | NR | NR |
| ShuffleNet [50] | 70.66 | 53.48 | 65.26 | 58.79 |
| Multitask [51] | 94.67 | NR | 96 | NR |
| Proposed approach | 95.42 | 98.17 | 93.36 | 95.70 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fekri-Ershad, S.; Dehkordi, K.B. A Flexible Multi-Channel Deep Network Leveraging Texture and Spatial Features for Diagnosing New COVID-19 Variants in Lung CT Scans. Tomography 2025, 11, 99. https://doi.org/10.3390/tomography11090099
Fekri-Ershad S, Dehkordi KB. A Flexible Multi-Channel Deep Network Leveraging Texture and Spatial Features for Diagnosing New COVID-19 Variants in Lung CT Scans. Tomography. 2025; 11(9):99. https://doi.org/10.3390/tomography11090099
Chicago/Turabian StyleFekri-Ershad, Shervan, and Khalegh Behrouz Dehkordi. 2025. "A Flexible Multi-Channel Deep Network Leveraging Texture and Spatial Features for Diagnosing New COVID-19 Variants in Lung CT Scans" Tomography 11, no. 9: 99. https://doi.org/10.3390/tomography11090099
APA StyleFekri-Ershad, S., & Dehkordi, K. B. (2025). A Flexible Multi-Channel Deep Network Leveraging Texture and Spatial Features for Diagnosing New COVID-19 Variants in Lung CT Scans. Tomography, 11(9), 99. https://doi.org/10.3390/tomography11090099






