Discussion of a Simple Method to Generate Descriptive Images Using Predictive ResNet Model Weights and Feature Maps for Recurrent Cervix Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
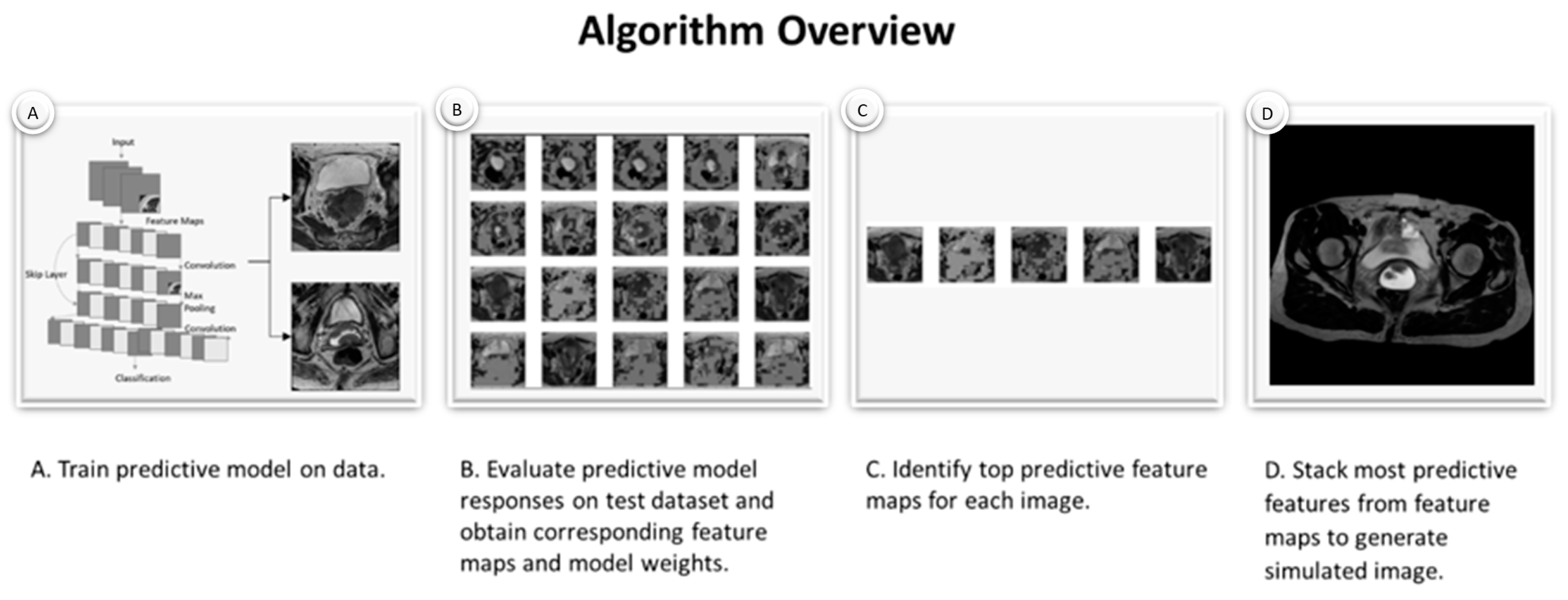
2.2. Simulated Images Generated by Custom Algorithm
2.3. Image Classification
2.4. Statistical Significance
3. Results
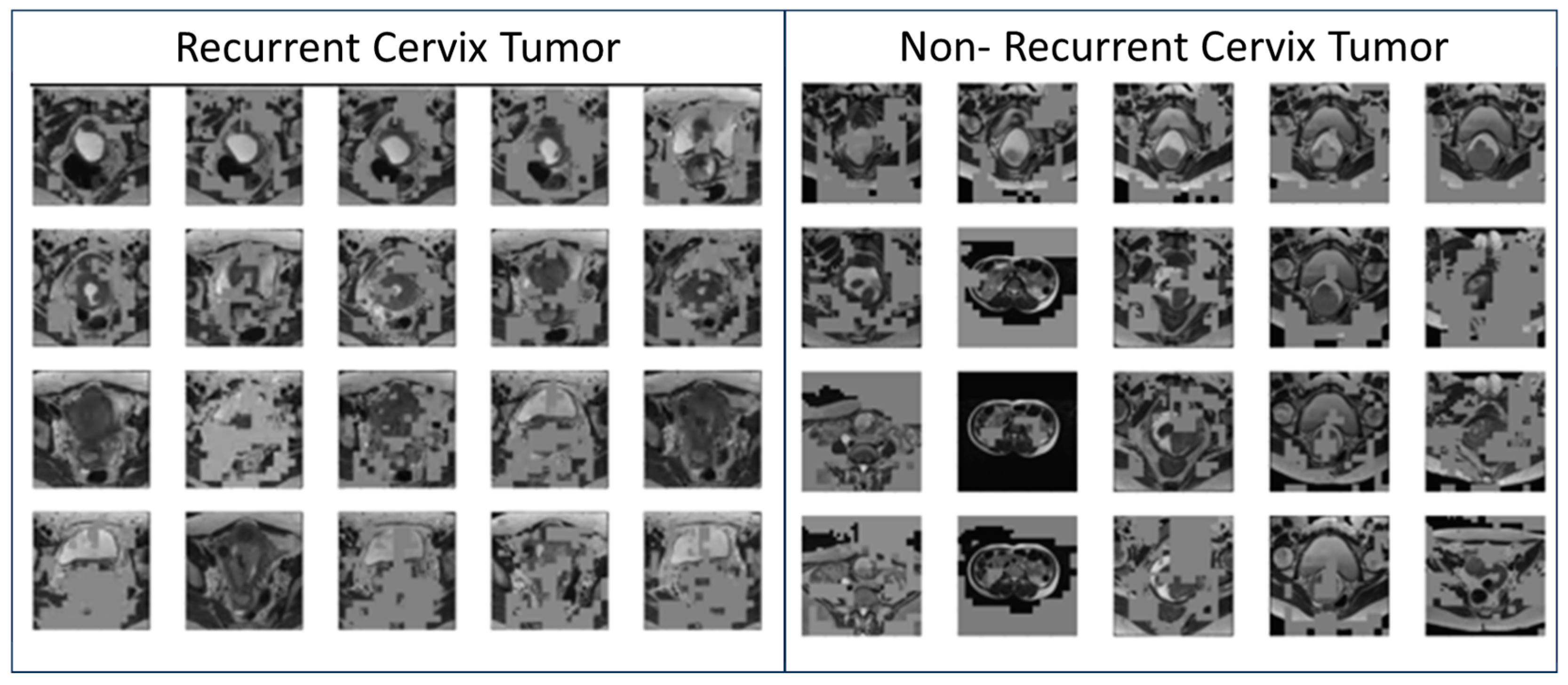
3.1. Model Accuracy and Sample Feature Maps
3.2. Generated Simulated Cervix Tumor Images
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RT | Radiation Therapy |
| ResNet | Residual Neural Network |
| ML | Machine Learning |
| AI | Artificial Intelligence |
| XAI | Explainable AI |
| MRI | Magnetic Resonance Imaging |
References
- Buskwofie, A.; David-West, G.; Clare, C.A. A Review of Cervical Cancer: Incidence and Disparities. J. Natl. Med. Assoc. 2020, 112, 229–232. [Google Scholar] [CrossRef] [PubMed]
- Quinn, M.; Benedet, J.; Odicino, F.; Maisonneuve, P.; Beller, U.; Creasman, W.; Heintz, A.; Nan, H.; Pecorelli, S. Carcinoma of the Cervix Uteri. Int. J. Gynecol. Obstet. 2006, 95, S43–S103. [Google Scholar]
- Chino, J.; Annunziata, C.M.; Beriwal, S.; Bradfield, L.; Erickson, B.A.; Fields, E.C.; Fitch, K.; Harkenrider, M.M.; Holschneider, C.H.; Kamrava, M.; et al. Radiation Therapy for Cervical Cancer: Executive Summary of an ASTRO Clinical Practice Guideline. Pract. Radiat. Oncol. 2020, 10, 220–234. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, S.; Chen, Y.; Yu, L.; Hu, X. YTHDC1 inhibits cell proliferation and angiogenesis in cervical cancer by regulating m6 A modification of SOCS4 mRNA. Mol. Cell. Toxicol. 2024, 20, 533–540. [Google Scholar] [CrossRef]
- Wen, Y.; Liang, H.; Zhang, H. Clinical utility of HPV typing and quantification combined with PAX1/ZNF582 methylation detection in accurate cervical cancer screening. CytoJournal 2023, 20, 26. [Google Scholar] [PubMed]
- Wang, Y.; Xu, Y.; Song, J.; Liu, X.; Liu, S.; Yang, N.; Wang, L.; Liu, Y.; Zhao, Y.; Zhou, W.; et al. Tumor Cell-Targeting and Tumor Microenvironment-Responsive Nanoplatforms for the Multimodal Imaging-Guided Photodynamic/Photothermal/Chemodynamic Treatment of Cervical Cancer. Int. J. Nanomed. 2024, 19, 5837–5858. [Google Scholar] [CrossRef]
- Bendifallah, S.; de Foucher, T.; Bricou, A.; Ouldamer, L.; Lavoue, V.; Varinot, J.; Canlorbe, G.; Carcopino, X.; Raimond, E.; Huguet, F.; et al. Cervical Cancer Recurrence: Proposal for a Classification Based on Anatomical Dissemination Pathways and Prognosis. Surg. Oncol. 2019, 30, 40–46. [Google Scholar]
- Choi, J.I.; Kim, S.H.; Seong, C.K.; Sim, J.S.; Lee, H.J.; Do, K.H. Recurrent uterine cervical carcinoma: Spectrum of imaging findings. Korean J. Radiol. 2000, 1, 198–207. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chiappa, V.; Bogani, G.; Interlenghi, M.; Vittori Antisari, G.; Salvatore, C.; Zanchi, L.; Ludovisi, M.; Leone Roberti Maggiore, U.; Calareso, G.; Haeusler, E.; et al. Using Radiomics and Machine Learning Applied to MRI to Predict Response to Neoadjuvant Chemotherapy in Locally Advanced Cervical Cancer. Diagnostics 2023, 13, 3139. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mathivanan, S.; Francis, D.; Srinivasan, S.; Khatavkar, V.; P, K.; Shah, M.A. Enhancing cervical cancer detection and robust classification through a fusion of deep learning models. Sci. Rep. 2014, 14, 10812. [Google Scholar] [CrossRef]
- Shanthi, P.B.; Faruqi, F.; Hareesha, K.S.; Kudva, R. Deep Convolution Neural Network for Malignancy Detection and Classification in Microscopic Uterine Cervix Cell Images. Asian Pac. J. Cancer Prev. 2019, 20, 3447–3456. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wang, J.; Yu, Y.; Tan, Y.; Wan, H.; Zheng, N.; He, Z.; Mao, L.; Ren, W.; Chen, K.; Lin, Z.; et al. Artificial intelligence enables precision diagnosis of cervical cytology grades and cervical cancer. Nat. Commun. 2024, 15, 4369. [Google Scholar] [CrossRef]
- Egemen, D.; Perkins, R.B.; Cheung, L.C.; Befano, B.; Rodriguez, A.C.; Desai, K.; Lemay, A.; Ahmed, S.R.; Antani, S.; Jeronimo, J.; et al. Artificial intelligence–based image analysis in clinical testing: Lessons from cervical cancer screening. JNCI J. Natl. Cancer Inst. 2024, 116, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, D.; Wang, J.; Haji-Momenian, S.; Shin, B.; Riess, J.; Khati, N.; Bauman, J.; Goyal, S.; Loew, M.; Chappell, N.; et al. Prediction of Progression After Cervix Cancer Radiotherapy Using a Machine-Learning Model on Pre-Treatment MRI. Int. J. Radiat. Oncol. Biol. Phys. 2023, 117, S132. [Google Scholar] [CrossRef]
- Ghassemi, M.; Naumann, T.; Schulam, P.; Beam, A.L.; Chen, I.Y.; Ranganath, R. A Review of Challenges and Opportunities in Machine Learning for Health. AMIA Summits Transl. Sci. Proc. 2020, 2020, 191–200. [Google Scholar] [PubMed] [PubMed Central]
- Gupta, V.; Erdal, B.; Ramirez, C.; Floca, R.; Genereaux, B.; Bryson, S.; Bridge, C.; Kleesiek, J.; Nensa, F.; Braren, R.; et al. Current State of Community-Driven Radiological AI Deployment in Medical Imaging. JMIR AI 2024, 3, e55833. [Google Scholar] [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Alsuradi, H.; Park, W.; Eid, M. Explainable classification of EEG data for an active touch task using Shapley values. In International Conference on Human-Computer Interaction, Copenhagen, Denmark, 19–24 July 2020; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Zafar, M.R.; Khan, N. Deterministic local interpretable model-agnostic explanations for stable explainability. Mach. Learn. Knowl. Extr. 2021, 3, 525–541. [Google Scholar] [CrossRef]
- Ghassemi, M.; Oakden-Rayner, L.; Beam, A.L. The false hope of current approaches to explainable artificial intelligence in health care. Lancet Digit. Health 2021, 3, e745–e750. [Google Scholar] [CrossRef]
- Slack, D.; Hilgard, S.; Jia, E.; Singh, S.; Lakkaraju, H. Fooling lime and shap: Adversarial attacks on post hoc explanation methods. In Proceedings of the AAAI/ACM Conference on AI, Ethics, and Society, New York, NY, USA, 7–9 February 2020. [Google Scholar]
- Islam, A.; Mridha, M.F.; Safran, M.; Alfarhood, S.; Kabir, M. Revolutionizing Brain Tumor Detection Using Explainable AI in MRI Images. NMR Biomed. 2025, 38, e70001. [Google Scholar] [CrossRef]
- Li, W.; Rao, Q.; Dong, S.; Zhu, M.; Yang, Z.; Huang, X.; Liu, G. PIDGN: An explainable multimodal deep learning framework for early prediction of Parkinson’s disease. J. Neurosci. Methods 2025, 415, 110363. [Google Scholar] [CrossRef]
- Hossain, M.M.; Ahmed, M.M.; Nafi, A.A.N.; Islam, M.R.; Ali, M.S.; Haque, J.; Miah, M.S.; Rahman, M.M.; Islam, M.K. A novel hybrid ViT-LSTM model with explainable AI for brain stroke detection and classification in CT images: A case study of Rajshahi region. Comput. Biol. Med. 2025, 186, 109711. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable deep learning models in medical image analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef]
- Cheng, J.; Tian, S.; Yu, L.; Gao, C.; Kang, X.; Ma, X.; Wu, W.; Liu, S.; Lu, H. ResGANet: Residual group attention network for medical image classification and segmentation. Med. Image. Anal. 2022, 76, 102313. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.D.; Gorre, N.; Hu, Q.; Li, H.; El Naqa, I.; Giger, M.L. A review of explainable and interpretable AI with applications in COVID-19 imaging. Med. Phys. 2022, 49, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Marinescu, R.; Dalca, A.V.; Bonkhoff, A.K.; Bretzner, M.; Rost, N.S.; Golland, P. 3D-StyleGAN: A style-based generative adversarial network for generative modeling of three-dimensional medical images. In DGM4MICCAI DALI 2021 Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Hasenstab, K.A.; Hahn, L.; Chao, N.; Hsiao, A. Simulating clinical features on chest radiographs for medical image exploration and CNN explainability using a style-based generative adversarial autoencoder. Sci. Rep. 2024, 14, 24427. [Google Scholar] [CrossRef]
- Provenzano, D.; Haji-Momenian, S.; Batheja, V.; Loew, M. Exploring the Explainability of a Machine Learning Model for Prostate Cancer: Do Lesions Localize with the Most Important Feature Maps? medRxiv 2024. [Google Scholar] [CrossRef]
- Provenzano, D.; Loew, M.; Goyal, S.; Rao, Y. A Simple Method to Generate Descriptive Images for Recurrent Cervix Cancer Using Predictive ResNet Model Weights and Feature Maps. Int. J. Radiat. Oncol. Biol. Phys. 2024, 120, e709. [Google Scholar]
- Lucchesi, F.R.; Aredes, N.D. The Cancer Genome Atlas Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma Collection (TCGA-CESC) (Version 3) [Data Set]; The Cancer Imaging Archive: Bethesda, MD, USA, 2016. [Google Scholar] [CrossRef]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Martín, A.; Ashish, A.; Paul, B.; Eugene, B.; Zhifeng, C.; Craig, C.; Greg, S.C.; Andy, D.; Jeffrey, D.; Matthieu, D.; et al. TensorFlow: Large-scale machine learning on heterogeneous systems. arXiv 2015, arXiv:1603.04467. Software available from tensorflow.org (Access Date 03 03 2025. [Google Scholar]
- Ng, A.; Jordan, M. On discriminative vs. generative classifiers: A comparison of logistic regression and naive bayes. Adv. Neural Inf. Process. Syst. 2021, 14. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. Adv. Neural Inf. Process. Syst. 2014, 27. [Google Scholar]
- Nazir, S.; Dickson, D.M.; Akram, M.U. Survey of explainable artificial intelligence techniques for biomedical imaging with deep neural networks. Comput. Biol. Med. 2023, 156, 106668. [Google Scholar] [CrossRef]
- Muhammad, D.; Bendechache, M. Unveiling the black box: A systematic review of Explainable Artificial Intelligence in medical image analysis. Comput. Struct. Biotechnol. J. 2024, 24, 542–560. [Google Scholar] [CrossRef]
- De-Giorgio, F.; Benedetti, B.; Mancino, M.; Sala, E.; Pascali, V.L. The need for balancing ’black box’ systems and explainable artificial intelligence: A necessary implementation in radiology. Eur. J. Radiol. 2025, 185, 112014. [Google Scholar] [CrossRef]
- Hafeez, Y.; Memon, K.; Al-Quraishi, M.S.; Yahya, N.; Elferik, S.; Ali, S.S.A. Explainable AI in Diagnostic Radiology for Neurological Disorders: A Systematic Review, and What Doctors Think About It. Diagnostics 2025, 15, 168. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, C.; Hu, W.; Luo, L.; Shi, D.; Zhang, J.; Yin, Q.; Zhang, L.; Han, X.; He, M. Economic evaluation for medical artificial intelligence: Accuracy vs. cost-effectiveness in a diabetic retinopathy screening case. npj Digit. Med. 2024, 7, 43. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Provenzano, D.; Wang, J.; Goyal, S.; Rao, Y.J. Discussion of a Simple Method to Generate Descriptive Images Using Predictive ResNet Model Weights and Feature Maps for Recurrent Cervix Cancer. Tomography 2025, 11, 38. https://doi.org/10.3390/tomography11030038
Provenzano D, Wang J, Goyal S, Rao YJ. Discussion of a Simple Method to Generate Descriptive Images Using Predictive ResNet Model Weights and Feature Maps for Recurrent Cervix Cancer. Tomography. 2025; 11(3):38. https://doi.org/10.3390/tomography11030038
Chicago/Turabian StyleProvenzano, Destie, Jeffrey Wang, Sharad Goyal, and Yuan James Rao. 2025. "Discussion of a Simple Method to Generate Descriptive Images Using Predictive ResNet Model Weights and Feature Maps for Recurrent Cervix Cancer" Tomography 11, no. 3: 38. https://doi.org/10.3390/tomography11030038
APA StyleProvenzano, D., Wang, J., Goyal, S., & Rao, Y. J. (2025). Discussion of a Simple Method to Generate Descriptive Images Using Predictive ResNet Model Weights and Feature Maps for Recurrent Cervix Cancer. Tomography, 11(3), 38. https://doi.org/10.3390/tomography11030038






