Advancements in Neurosurgical Intraoperative Histology
Abstract
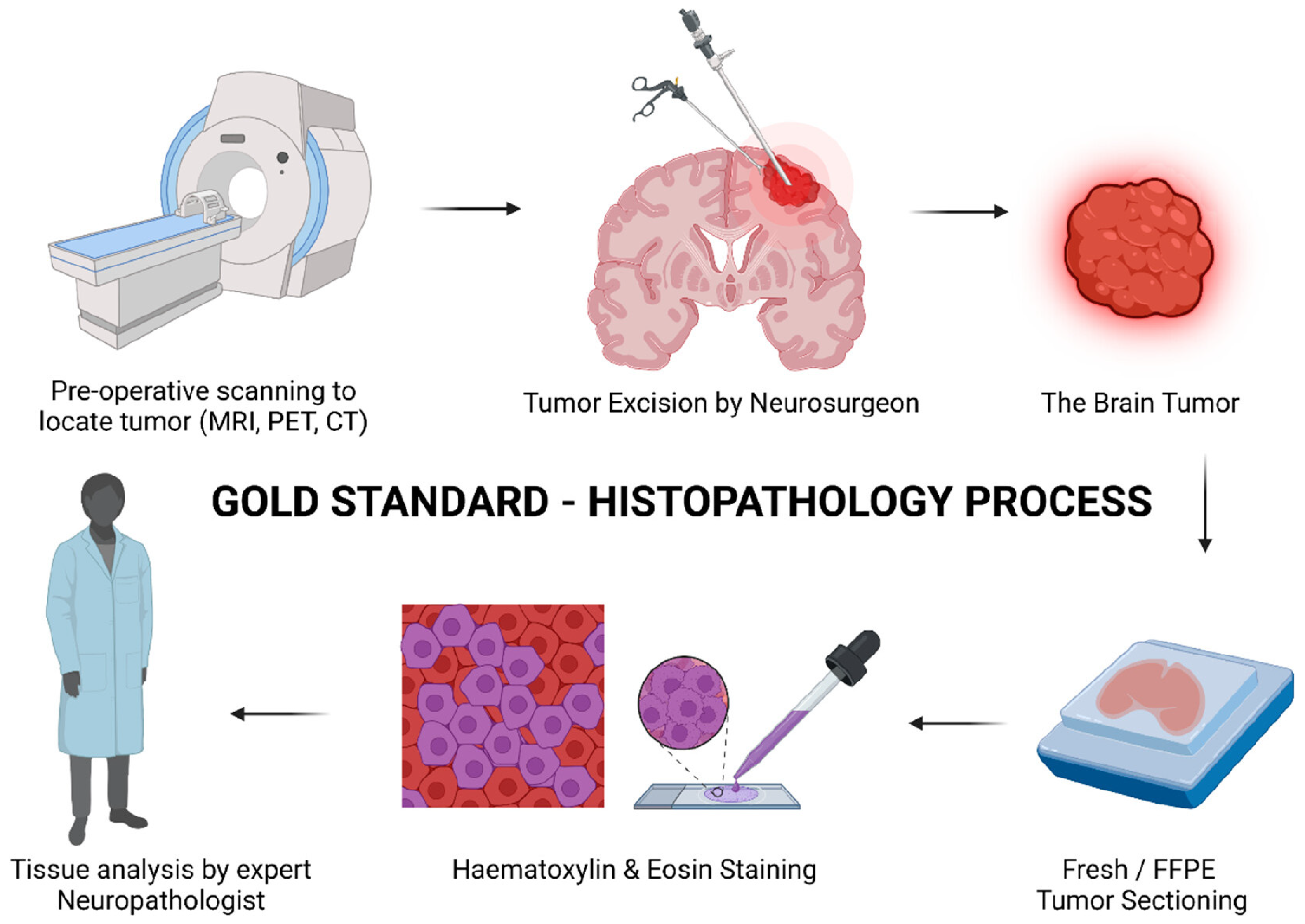
1. Introduction
2. Frozen Sectioning
3. Other Cytological Preparations
4. Raman Histology
4.1. Indications
4.2. Techniques
4.2.1. Raman Spectroscopy
4.2.2. Stimulated Raman Scattering (SRS) Microscopy
4.2.3. Coherent Anti-Strokes Raman Scattering (CARS)
4.2.4. Surface-Enhanced Raman Scattering (SERS) Microscopy
4.2.5. Tip-Enhanced Raman Scattering (TERS) Microscopy
4.3. Outcomes
5. Confocal Laser Endomicroscopy
5.1. Indications
5.2. Techniques
5.3. Outcomes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Mattiuzzi, C.; Lippi, G. Current cancer epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Santucci, C.; Carioli, G.; Bertuccio, P.; Malvezzi, M.; Pastorino, U.; Boffetta, P.; Negri, E.; Bosetti, C.; La Vecchia, C. Progress in cancer mortality, incidence, and survival: A global overview. Eur. J. Cancer Prev. 2020, 29, 367–381. [Google Scholar] [CrossRef] [PubMed]
- Ilic, I.; Ilic, M. International patterns and trends in the brain cancer incidence and mortality: An observational study based on the global burden of disease. Heliyon 2023, 9, e18222. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, G.; Waite, K.A.; Edelson, J.L.; Kruchko, C.; Ostrom, Q.T.; Barnholtz-Sloan, J.S. Changes in survival over time for primary brain and other CNS tumors in the United States, 2004–2017. J. Neuro-Oncol. 2022, 160, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Mannen, H. Historical review of development of neuro-historogical techniques and three-dimensional reconstruction of individual neurons. Clin. Neurol. 2006, 46, 751–759. [Google Scholar]
- Orringer, D.A.; Pandian, B.; Niknafs, Y.S.; Hollon, T.C.; Boyle, J.; Lewis, S.; Garrard, M.; Hervey-Jumper, S.L.; Garton, H.J.L.; Maher, C.O.; et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat. Biomed. Eng. 2017, 1, 0027. [Google Scholar] [CrossRef]
- Nikova, A.; Birbilis, T. The Basic Steps of Evolution of Brain Surgery. Maedica 2017, 12, 297. [Google Scholar] [PubMed]
- Gal, A.A. The centennial anniversary of the frozen section technique at the Mayo Clinic. Arch. Pathol. Lab. Med. 2005, 129, 1532–1535. [Google Scholar] [CrossRef]
- Wheeler, T.M. Origin and Development of American Surgical Pathology. Trans. Am. Clin. Climatol. Assoc. 2020, 131, 326–334. [Google Scholar]
- Jaafar, H. Intra-operative frozen section consultation: Concepts, applications and limitations. Malays. J. Med. Sci. 2006, 13, 4–12. [Google Scholar] [PubMed]
- Novis, D.A.; Zarbo, R.J. Interinstitutional comparison of frozen section turnaround time. A College of American Pathologists Q-Probes study of 32868 frozen sections in 700 hospitals. Arch. Pathol. Lab. Med. 1997, 121, 559–567. [Google Scholar]
- Murugappan S, Tofail SAM, Thorat ND: Raman Spectroscopy: A Tool for Molecular Fingerprinting of Brain Cancer. ACS Omega 2023, 8, 27845–27861. [CrossRef] [PubMed]
- Khoddami, M.; Akbarzadeh, A.; Mordai, A.; Bidari-Zerehpoush, F.; Alipour, H.; Samadzadeh, S.; Alipour, B. Diagnostic accuracy of frozen section of central nervous system lesions: A 10-year study. Iran. J. Child Neurol. 2015, 9, 25–30. [Google Scholar]
- Kang, M.; Chung, D.H.; Kim, N.R.; Cho, H.Y.; Ha, S.Y.; Lee, S.; An, J.; Seok, J.Y.; Yie, G.-T.; Yoo, C.J.; et al. Intraoperative Frozen Cytology of Central Nervous System Neoplasms: An Ancillary Tool for Frozen Diagnosis. J. Pathol. Transl. Med. 2019, 53, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Uematsu, Y.; Owai, Y.; Okita, R.; Tanaka, Y.; Itakura, T. The usefulness and problem of intraoperative rapid diagnosis in surgical neuropathology. Brain Tumor Pathol. 2007, 24, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Chand, P.; Amit, S.; Gupta, R.; Agarwal, A. Errors, limitations, and pitfalls in the diagnosis of central and peripheral nervous system lesions in intraoperative cytology and frozen sections. J. Cytol. 2016, 33, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Plesec, T.P.; Prayson, R.A. Frozen section discrepancy in the evaluation of central nervous system tumors. Arch. Pathol. Lab. Med. 2007, 131, 1532–1540. [Google Scholar] [CrossRef]
- Mitra, S.; Kumar, M.; Sharma, V.; Mukhopadhyay, D. Squash preparation: A reliable diagnostic tool in the intraoperative diagnosis of central nervous system tumors. J. Cytol. 2010, 27, 81–85. [Google Scholar] [CrossRef]
- Gandour-Edwards, R.F.; Donald, P.J.; Boggan, J.E. Intraoperative frozen section diagnosis in skull base surgery. Skull Base Surg. 1993, 3, 159–163. [Google Scholar] [CrossRef][Green Version]
- Savargaonkar, P.; Farmer, P.M. Utility of intra-operative consultations for the diagnosis of central nervous system lesions. Ann. Clin. Lab. Sci. 2001, 31, 133–139. [Google Scholar]
- Rao, S.; Rajkumar, A.; Ehtesham, M.D.; Duvuru, P. Challenges in neurosurgical intraoperative consultation. Neurol. India 2009, 57, 464–468. [Google Scholar] [CrossRef]
- Shakir, M.; Altaf, A.; Hussain, H.; Abidi, S.M.A.; Petitt, Z.; Tariq, M.; Gilani, A.; Enam, S.A. Unveiling the potential application of intraoperative brain smear for brain tumor diagnosis in low-middle-income countries: A comprehensive systematic review. Surg. Neurol. Int. 2023, 14, 325. [Google Scholar] [CrossRef]
- Jindal, A.; Kaur, K.; Mathur, K.; Kumari, V.; Diwan, H. Intraoperative Squash Smear Cytology in CNS Lesions: A Study of 150 Pediatric Cases. J. Cytol. 2017, 34, 217–220. [Google Scholar] [CrossRef] [PubMed]
- Govindaraman, P.; Arumugam, N.; Ramasamy, C.; Prakasam, G. Role of squash smear in intraoperative consultation of central nervous system tumors. J. Sci. Soc. 2017, 44, 7. [Google Scholar] [CrossRef]
- Koyuncuer, A. Intraoperative Touch Imprint Cytology of Brain Neoplasms: A Useful High-Diagnostic Tool in 93 Consecutive Cases; Differential Diagnoses, Pitfalls, and Traps. Anal. Cell. Pathol. 2024, 2024, 2346092. [Google Scholar] [CrossRef]
- Tanabe, N.; Inoshita, N.; Ishida, A.; Kato, M.; Yoshimoto, H.; Shiramizu, H.; Suga, H.; Tateno, T.; Ohashi, K.; Yamada, S. Touch imprint cytology is useful for the intraoperative pathological diagnosis of PitNETs’ surgical margins. Brain Tumor Pathol. 2023, 40, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Saletnik, A.; Saletnik, B.; Puchalski, C. Overview of Popular Techniques of Raman Spectroscopy and Their Potential in the Study of Plant Tissues. Molecules 2021, 26, 1537. [Google Scholar] [CrossRef]
- Terrones, O.; Olazar-Intxausti, J.; Anso, I.; Lorizate, M.; Nieto-Garai, J.A.; Contreras, F.X. Raman Spectroscopy as a Tool to Study the Pathophysiology of Brain Diseases. Int. J. Mol. Sci. 2023, 24, 2384. [Google Scholar] [CrossRef]
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; He, C.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 2008, 322, 1857–1861. [Google Scholar] [CrossRef]
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman Techniques: Fundamentals and Frontiers. Nanoscale Res. Lett. 2019, 14, 231. [Google Scholar] [CrossRef] [PubMed]
- Wadiura, L.I.; Kiesel, B.; Roetzer-Pejrimovsky, T.; Mischkulnig, M.; Vogel, C.C.; Hainfellner, J.A.; Matula, C.; Freudiger, C.W.; Orringer, D.A.; Wöhrer, A.; et al. Toward digital histopathological assessment in surgery for central nervous system tumors using stimulated Raman histology. Neurosurg. Focus 2022, 53, E12. [Google Scholar] [CrossRef] [PubMed]
- Hollon, T.; Lewis, S.; Freudiger, C.W.; Sunney Xie, X.; Orringer, D.A. Improving the accuracy of brain tumor surgery via Raman-based technology. Neurosurg. Focus 2016, 40, E9. [Google Scholar] [CrossRef]
- Hollon, T.; Orringer, D.A. Label-free brain tumor imaging using Raman-based methods. J. Neurooncol. 2021, 151, 393–402. [Google Scholar] [CrossRef]
- Ranasinghe, J.C.; Wang, Z.; Huang, S. Raman Spectroscopy on Brain Disorders: Transition from Fundamental Research to Clinical Applications. Biosensors 2022, 13, 27. [Google Scholar] [CrossRef]
- Ryzhikova, E.; Ralbovsky, N.M.; Sikirzhytski, V.; Kazakov, O.; Halamkova, L.; Quinn, J.; Zimmerman, E.A.; Lednev, I.K. Raman spectroscopy and machine learning for biomedical applications: Alzheimer’s disease diagnosis based on the analysis of cerebrospinal fluid. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 248, 119188. [Google Scholar] [CrossRef]
- Lochocki, B.; Morrema, T.H.J.; Ariese, F.; Hoozemans, J.J.M.; de Boer, J.F. The search for a unique Raman signature of amyloid-beta plaques in human brain tissue from Alzheimer’s disease patients. Analyst 2020, 145, 1724–1736. [Google Scholar] [CrossRef] [PubMed]
- Paraskevaidi, M.; Martin-Hirsch, P.L.; Martin, F.L. Progress and Challenges in the Diagnosis of Dementia: A Critical Review. ACS Chem. Neurosci. 2018, 9, 446–461. [Google Scholar] [CrossRef]
- Stern, D. Corneal Ablation by Nanosecond, Picosecond, and Femtosecond Lasers at 532 and 625 nm. Arch. Ophthalmol. 1989, 107, 587. [Google Scholar] [CrossRef]
- Orillac, C.; Hollon, T.; Orringer, D.A. Clinical Translation of Stimulated Raman Histology. Biomed. Eng. Technol. 2022, 1, 225–236. [Google Scholar] [CrossRef]
- Brzozowski, K.; Matuszyk, E.; Pieczara, A.; Firlej, J.; Nowakowska, A.M.; Baranska, M. Stimulated Raman scattering microscopy in chemistry and life science—Development, innovation, perspectives. Biotechnol. Adv. 2022, 60, 108003. [Google Scholar] [CrossRef] [PubMed]
- Neidert, N.; Straehle, J.; Erny, D.; Sacalean, V.; El Rahal, A.; Steybe, D.; Schmelzeisen, R.; Vlachos, A.; Reinacher, P.C.; Coenen, V.A.; et al. Stimulated Raman histology in the neurosurgical workflow of a major European neurosurgical center—Part A. Neurosurg. Rev. 2022, 45, 1731–1739. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Yi, R.; Liu, L.; Qu, J. Coherent Anti-Stokes Raman Scattering Microscopy and Its Applications. Front. Phys. 2020, 8. [Google Scholar] [CrossRef]
- Evans, C.L.; Xie, X.S. Coherent anti-Stokes Raman scattering microscopy: Chemical imaging for biology and medicine. Annu. Rev. Anal. Chem. 2008, 1, 883–909. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R. Surface-Enhanced Vibrational Spectroscopy. Wiley: Hoboken, NJ, USA, 2006. [CrossRef]
- Pérez-Jiménez, A.I.; Lyu, D.; Lu, Z.; Liu, G.; Ren, B. Surface-enhanced Raman spectroscopy: Benefits, trade-offs and future developments. Chem. Sci. 2020, 11, 4563–4577. [Google Scholar] [CrossRef]
- Han, X.X.; Zhao, B.; Ozaki, Y. Surface-enhanced Raman scattering for protein detection. Anal. Bioanal. Chem. 2009, 394, 1719–1727. [Google Scholar] [CrossRef]
- Kumar, N.; Mignuzzi, S.; Su, W.; Roy, D. Tip-enhanced Raman spectroscopy: Principles and applications. EPJ Tech. Instrum. 2015, 2, 9. [Google Scholar] [CrossRef]
- Li, Z.; Persits, N.; Gray, D.J.; Ram, R.J. Computational polarized Raman microscopy on sub-surface nanostructures with sub-diffraction-limit resolution. Opt. Express 2021, 29, 38027. [Google Scholar] [CrossRef]
- Doran, C.E.; Frank, C.B.; McGrath, S.; Packer, R.A. Use of Handheld Raman Spectroscopy for Intraoperative Differentiation of Normal Brain Tissue From Intracranial Neoplasms in Dogs. Front. Vet. Sci. 2022, 8, 819200. [Google Scholar] [CrossRef]
- Zhang, R.R.; Kuo, J.S. Detection of Human Brain Tumor Infiltration With Quantitative Stimulated Raman Scattering Microscopy. Neurosurgery 2016, 78, N9–N11. [Google Scholar] [CrossRef][Green Version]
- Aguiar, R.P.; Silveira, L.; Falcão, E.T.; Pacheco, M.T.T.; Zângaro, R.A.; Pasqualucci, C.A. Discriminating Neoplastic and Normal Brain Tissues in Vitro Through Raman Spectroscopy: A Principal Components Analysis Classification Model. Photomed. Laser Surg. 2013, 31, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Jermyn, M.; Desroches, J.; Mercier, J.; St-Arnaud, K.; Guiot, M.-C.; Leblond, F.; Petrecca, K. Raman spectroscopy detects distant invasive brain cancer cells centimeters beyond MRI capability in humans. Biomed. Opt. Express 2016, 7, 5129. [Google Scholar] [CrossRef] [PubMed]
- Freudiger, C.W.; Pfannl, R.; Orringer, D.A.; Saar, B.G.; Ji, M.; Zeng, Q.; Ottoboni, L.; Ying, W.; Waeber, C.; Sims, J.R.; et al. Multicolored stain-free histopathology with coherent Raman imaging. Lab. Investig. 2012, 92, 1492–1502. [Google Scholar] [CrossRef]
- Evans, C.L.; Potma, E.O.; Puoris’haag, M.; Côté, D.; Lin, C.P.; Xie, X.S. Chemical imaging of tissue in vivo with video-rate coherent anti-Stokes Raman scattering microscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 16807–16812. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Yue, Q.; Duan, W.; Sui, A.; Zhao, B.; Deng, Y.; Zhai, Y.; Zhang, Y.; Sun, T.; Zhang, G.; et al. Intelligent SERS Navigation System Guiding Brain Tumor Surgery by Intraoperatively Delineating the Metabolic Acidosis. Adv. Sci. 2022, 9, 2104935. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sheng, S.; Wang, R.; Sun, M. Tip-Enhanced Raman Spectroscopy. Anal. Chem. 2016, 88, 9328–9346. [Google Scholar] [CrossRef] [PubMed]
- Abramov, I.; Park, M.T.; Gooldy, T.C.; Xu, Y.; Lawton, M.T.; Little, A.S.; Porter, R.W.; Smith, K.A.; Eschbacher, J.M.; Preul, M.C. Real-time intraoperative surgical telepathology using confocal laser endomicroscopy. Neurosurg. Focus 2022, 52, E9. [Google Scholar] [CrossRef]
- Shahid, M.W.; Buchner, A.M.; Coron, E.; Woodward, T.A.; Raimondo, M.; Dekker, E.; Fockens, P.; Wallace, M.B. Diagnostic accuracy of probe-based confocal laser endomicroscopy in detecting residual colorectal neoplasia after EMR: A prospective study. Gastrointest. Endosc. 2012, 75, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Restelli, F.; Pollo, B.; Vetrano, I.G.; Cabras, S.; Broggi, M.; Schiariti, M.; Falco, J.; de Laurentis, C.; Raccuia, G.; Ferroli, P.; et al. Confocal laser microscopy in neurosurgery: State of the art of actual clinical applications. J. Clin. Med. 2021, 10, 2035. [Google Scholar] [CrossRef]
- Aisenberg, J. Gastrointestinal endoscopy nears “the molecular era”. Gastrointest. Endosc. 2008, 68, 528–530. [Google Scholar] [CrossRef]
- Wang, T.D.; Van Dam, J. Optical biopsy: A new frontier in endoscopic detection and diagnosis. Clin. Gastroenterol. Hepatol. 2004, 2, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Abramov, I.; Belykh, E.; Mignucci-Jiménez, G.; Park, M.T.; Eschbacher, J.M.; Preul, M.C. Characterization of ex vivo and in vivo intraoperative neurosurgical confocal laser endomicroscopy imaging. Front. Oncol. 2022, 12, 979748. [Google Scholar] [CrossRef] [PubMed]
- Restelli, F.; Mathis, A.M.; Höhne, J.; Mazzapicchi, E.; Acerbi, F.; Pollo, B.; Quint, K. Confocal laser imaging in neurosurgery: A comprehensive review of sodium fluorescein-based CONVIVO preclinical and clinical applications. Front. Oncol. 2022, 12, 998384. [Google Scholar] [CrossRef]
- Areias, L.R.P.; Mariz, I.; Maçôas, E.; Farinha, J.P.S. Reflectance Confocal Microscopy: A Powerful Tool for Large Scale Characterization of Ordered/Disordered Morphology in Colloidal Photonic Structures. ACS Nano 2021, 15, 11779–11788. [Google Scholar] [CrossRef] [PubMed]
- Wirth, D.; Snuderl, M.; Sheth, S.; Kwon, C.-S.; Frosch, M.P.; Curry, W.; Yaroslavsky, A.N. Identifying brain neoplasms using dye-enhanced multimodal confocal imaging. J. Biomed. Opt. 2012, 17, 026012. [Google Scholar] [CrossRef] [PubMed]
- Kiesslich, R.; Neurath, M.F. Chromoendoscopy and Other Novel Imaging Techniques. Gastroenterol. Clin. N. Am. 2006, 35, 605–619. [Google Scholar] [CrossRef] [PubMed]
- Martirosyan, N.L.; Eschbacher, J.M.; Kalani, M.Y.S.; Turner, J.D.; Belykh, E.; Spetzler, R.F.; Nakaji, P.; Preul, M.C. Prospective evaluation of the utility of intraoperative confocal laser endomicroscopy in patients with brain neoplasms using fluorescein sodium: Experience with 74 cases. Neurosurg. Focus 2016, 40, E11. [Google Scholar] [CrossRef]
- Breuskin, D.; Szczygielski, J.; Urbschat, S.; Kim, Y.-J.; Oertel, J. Confocal Laser Endomicroscopy in Neurosurgery—An Alternative to Instantaneous Sections? World Neurosurg. 2017, 100, 180–185. [Google Scholar] [CrossRef]
- Charalampaki, P.; Javed, M.; Daali, S.; Heiroth, H.-J.; Igressa, A.; Weber, F. Confocal Laser Endomicroscopy for Real-time Histomorphological Diagnosis. Neurosurgery 2015, 62, 171–176. [Google Scholar] [CrossRef]
- Pavlov, V.; Meyronet, D.; Meyer-Bisch, V.; Armoiry, X.; Pikul, B.; Dumot, C.; Beuriat, P.-A.; Signorelli, F.; Guyotat, J. Intraoperative Probe-Based Confocal Laser Endomicroscopy in Surgery and Stereotactic Biopsy of Low-Grade and High-Grade Gliomas. Neurosurgery 2016, 79, 604–612. [Google Scholar] [CrossRef]
- Acerbi, F.; Pollo, B.; De Laurentis, C.; Restelli, F.; Falco, J.; Vetrano, I.G.; Broggi, M.; Schiariti, M.; Tramacere, I.; Ferroli, P.; et al. Ex Vivo Fluorescein-Assisted Confocal Laser Endomicroscopy (CONVIVO® System) in Patients With Glioblastoma: Results From a Prospective Study. Front. Oncol. 2020, 10, 606574. [Google Scholar] [CrossRef] [PubMed]
- Höhne, J.; Schebesch, K.-M.; Zoubaa, S.; Proescholdt, M.; Riemenschneider, M.J.; Schmidt, N.O. Intraoperative imaging of brain tumors with fluorescein: Confocal laser endomicroscopy in neurosurgery. Clinical and user experience. Neurosurg. Focus 2021, 50, E19. [Google Scholar] [CrossRef] [PubMed]
- Abramov, I.; Park, M.T.; Belykh, E.; Dru, A.B.; Xu, Y.; Gooldy, T.C.; Scherschinski, L.; Farber, S.H.; Little, A.S.; Porter, R.W.; et al. Intraoperative confocal laser endomicroscopy: Prospective in vivo feasibility study of a clinical-grade system for brain tumors. J. Neurosurg. 2023, 138, 587–597. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohamed, A.A.; Sargent, E.; Williams, C.; Karve, Z.; Nair, K.; Lucke-Wold, B. Advancements in Neurosurgical Intraoperative Histology. Tomography 2024, 10, 693-704. https://doi.org/10.3390/tomography10050054
Mohamed AA, Sargent E, Williams C, Karve Z, Nair K, Lucke-Wold B. Advancements in Neurosurgical Intraoperative Histology. Tomography. 2024; 10(5):693-704. https://doi.org/10.3390/tomography10050054
Chicago/Turabian StyleMohamed, Ali A., Emma Sargent, Cooper Williams, Zev Karve, Karthik Nair, and Brandon Lucke-Wold. 2024. "Advancements in Neurosurgical Intraoperative Histology" Tomography 10, no. 5: 693-704. https://doi.org/10.3390/tomography10050054
APA StyleMohamed, A. A., Sargent, E., Williams, C., Karve, Z., Nair, K., & Lucke-Wold, B. (2024). Advancements in Neurosurgical Intraoperative Histology. Tomography, 10(5), 693-704. https://doi.org/10.3390/tomography10050054








