On Effect of Chloroform on Electrical Activity of Proteinoids
Abstract
1. Introduction
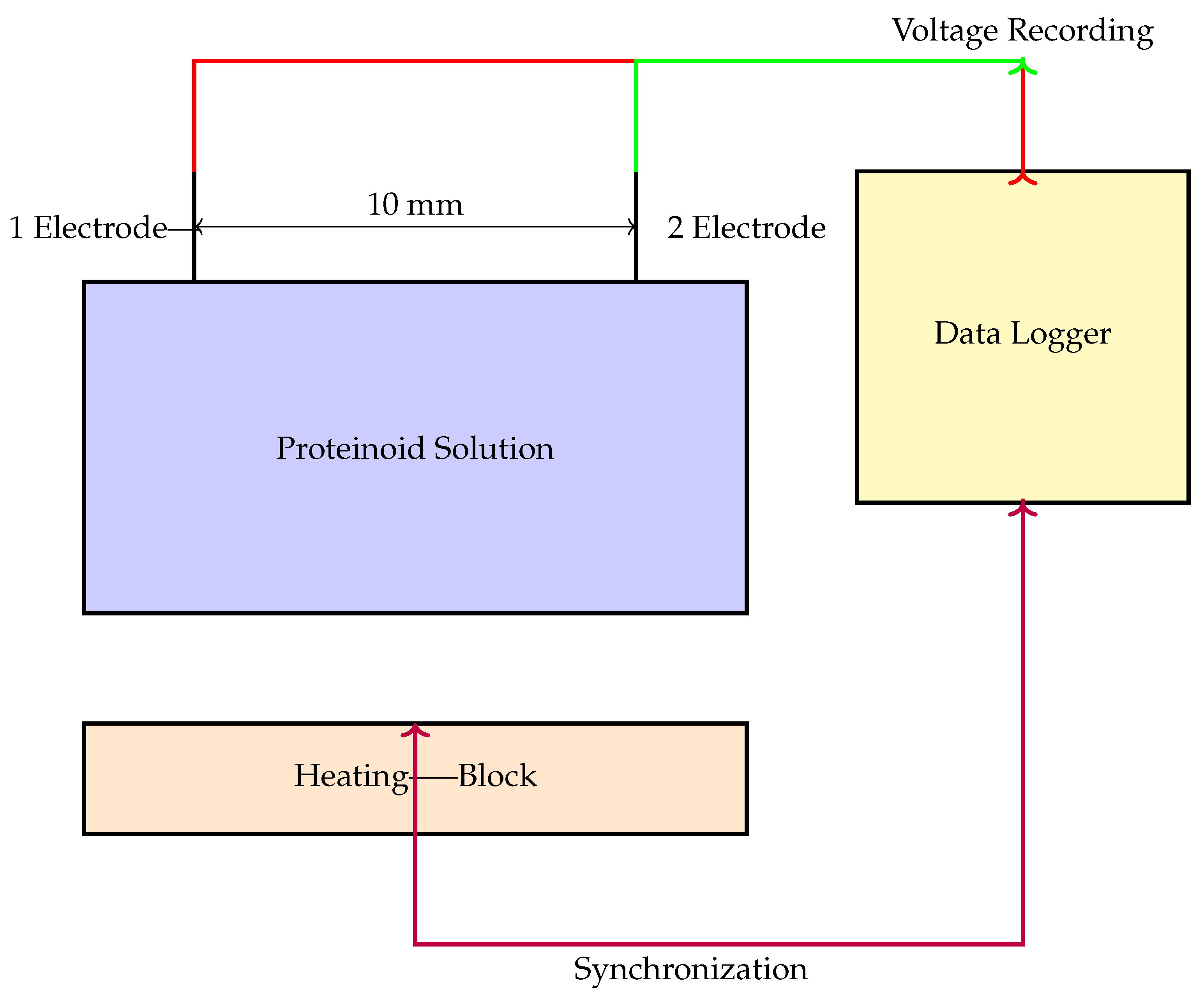
2. Methods and Materials
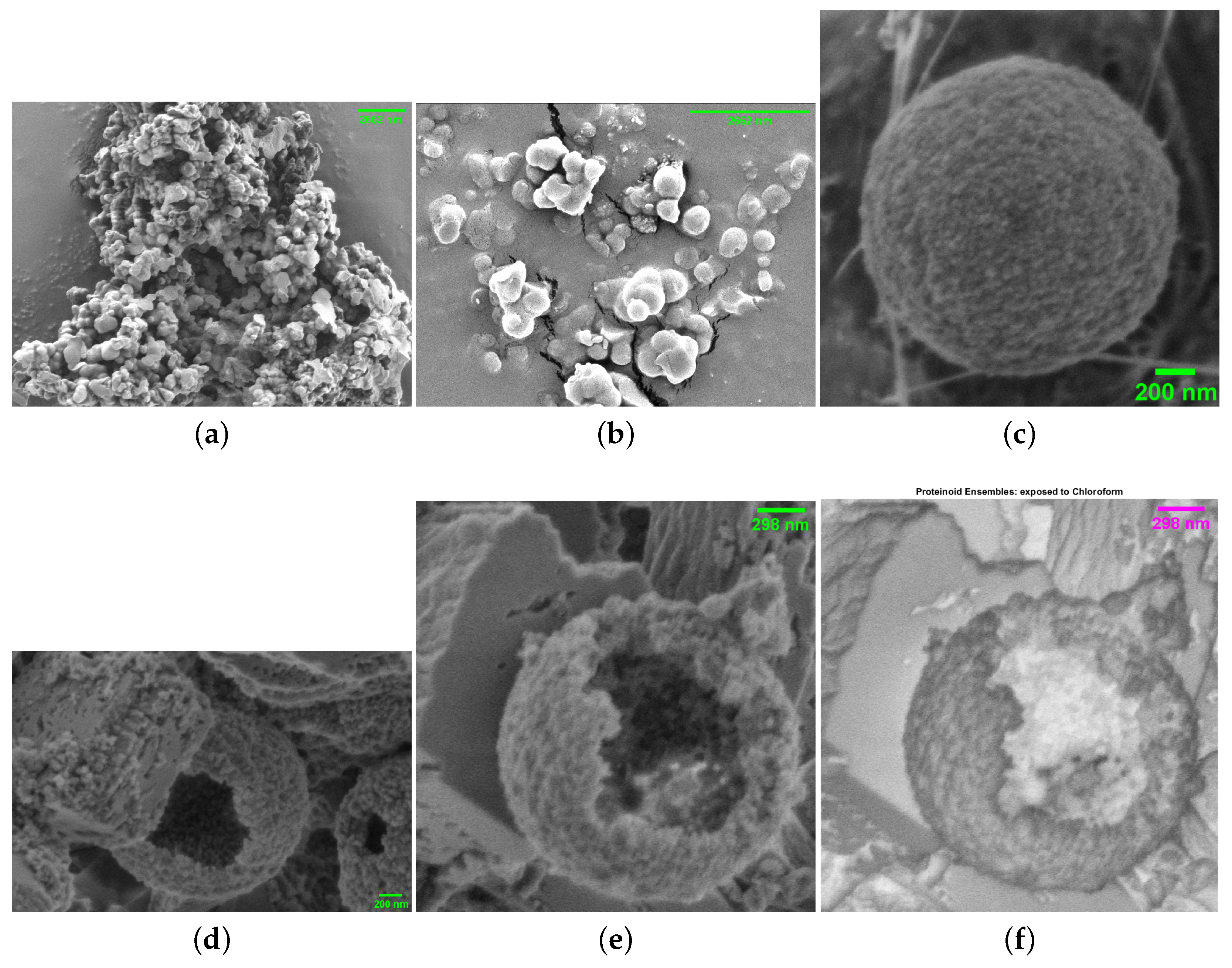
3. Results
3.1. Computational Analysis: Molecular Mechanics Simulation of Chloroform and Dipeptide Systems
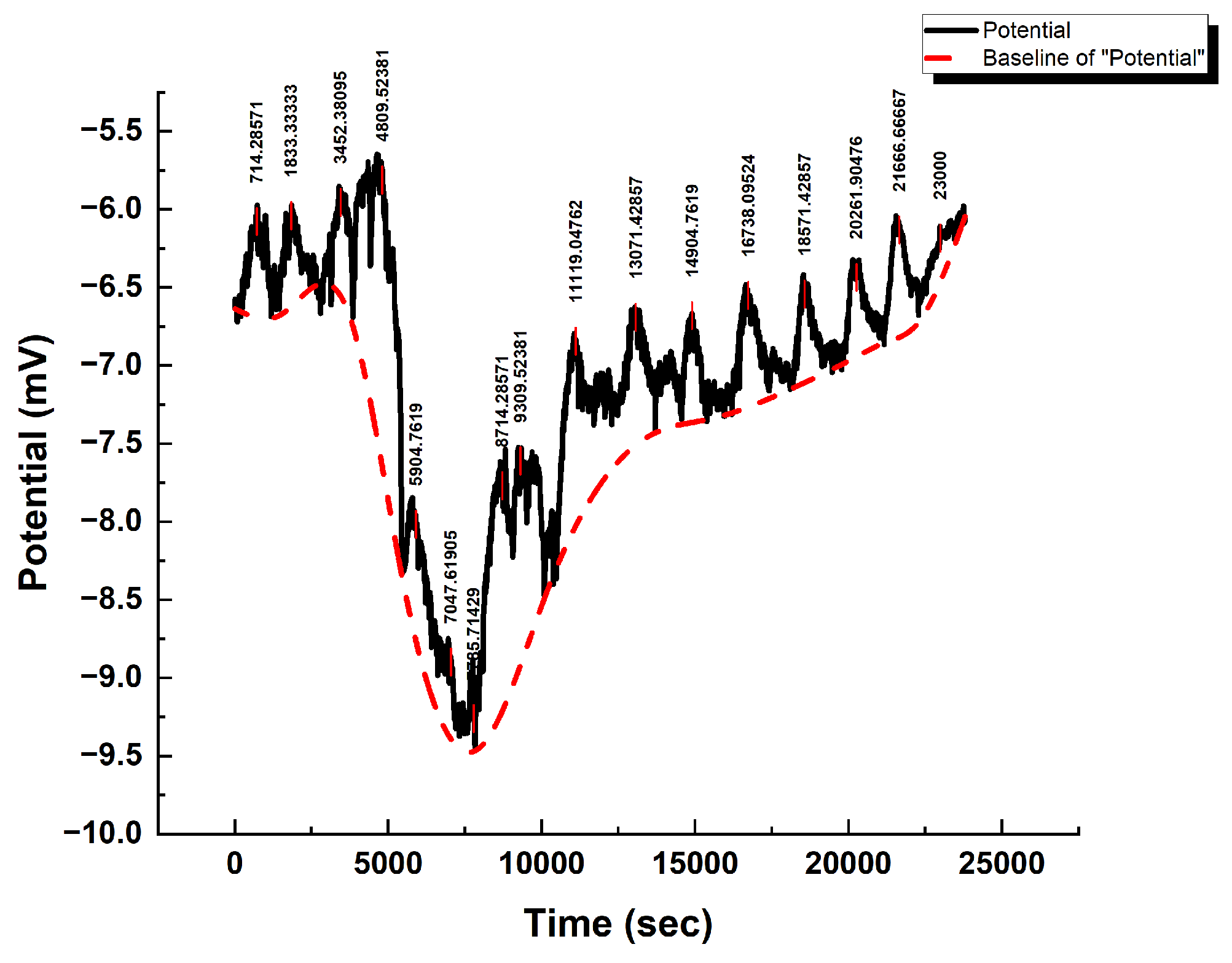
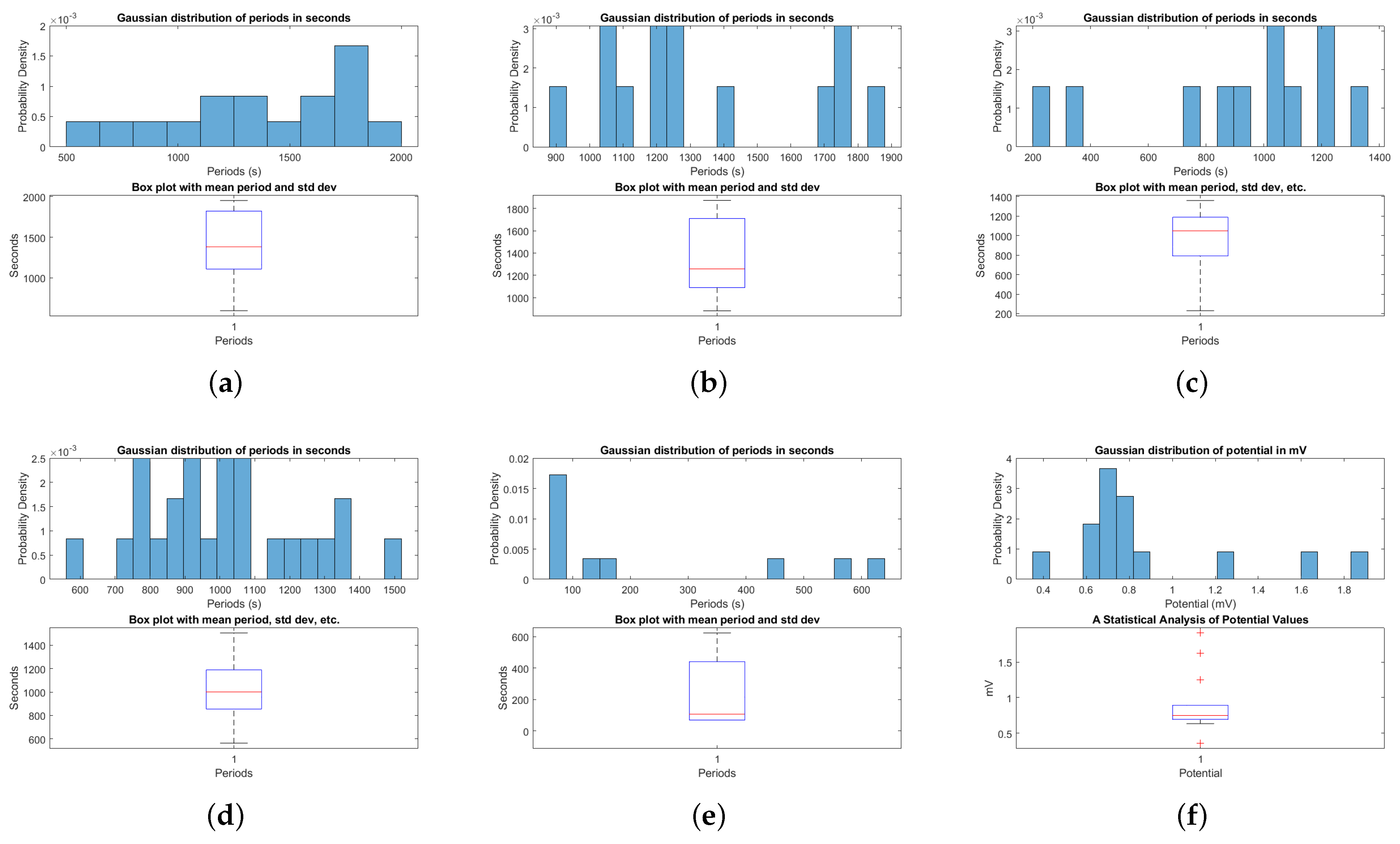
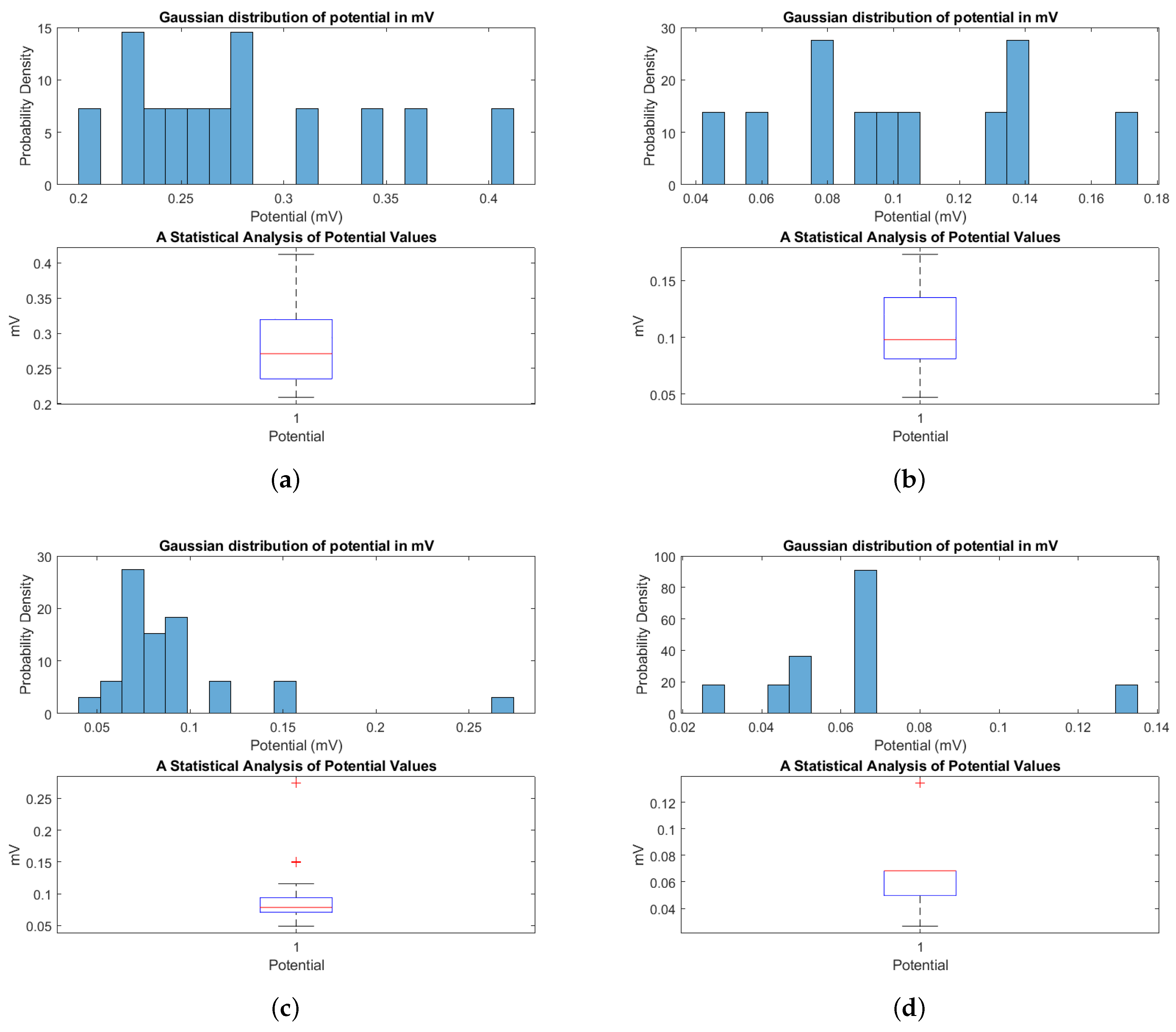
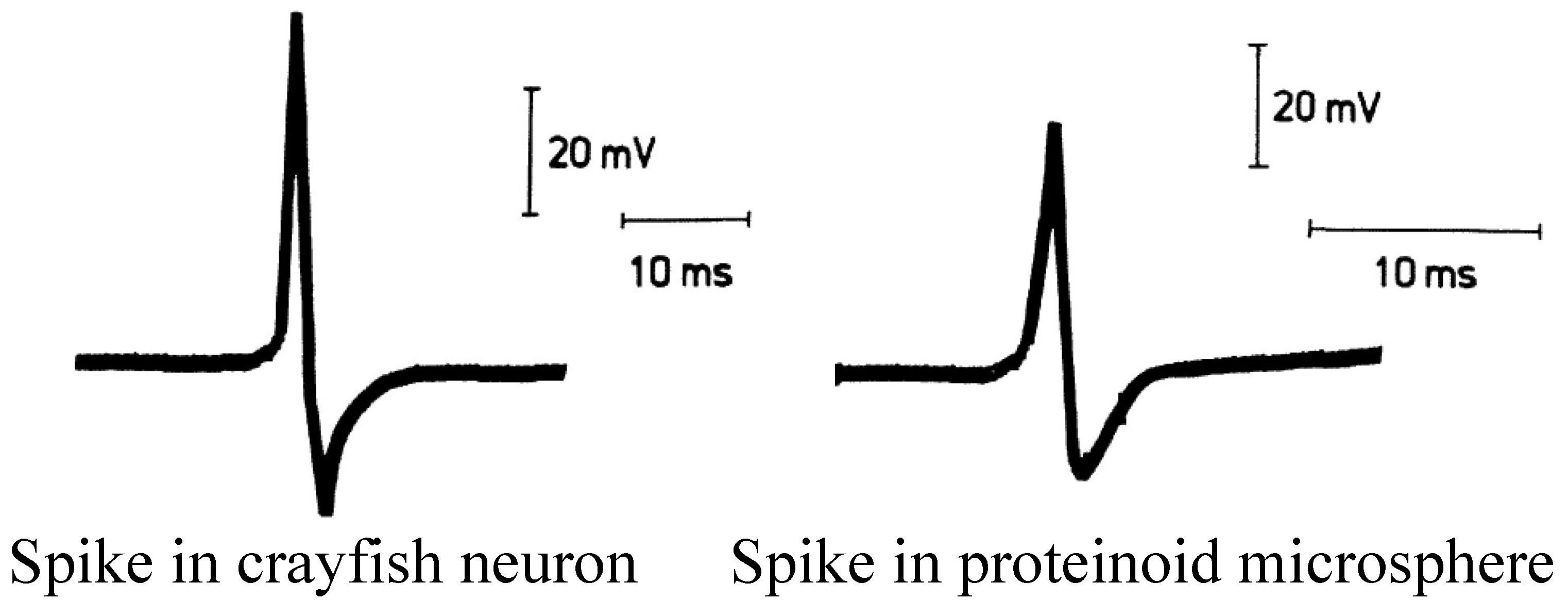
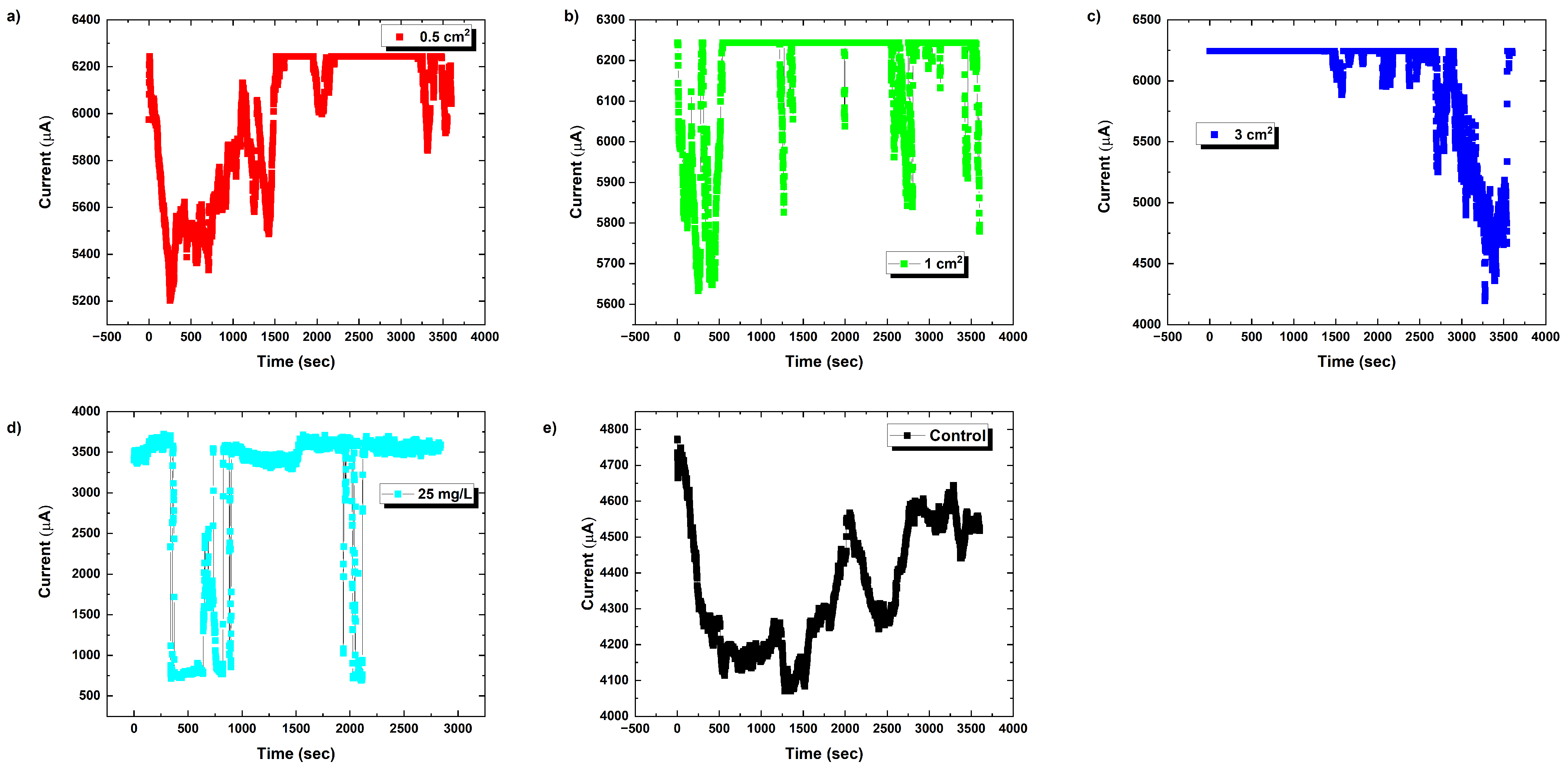
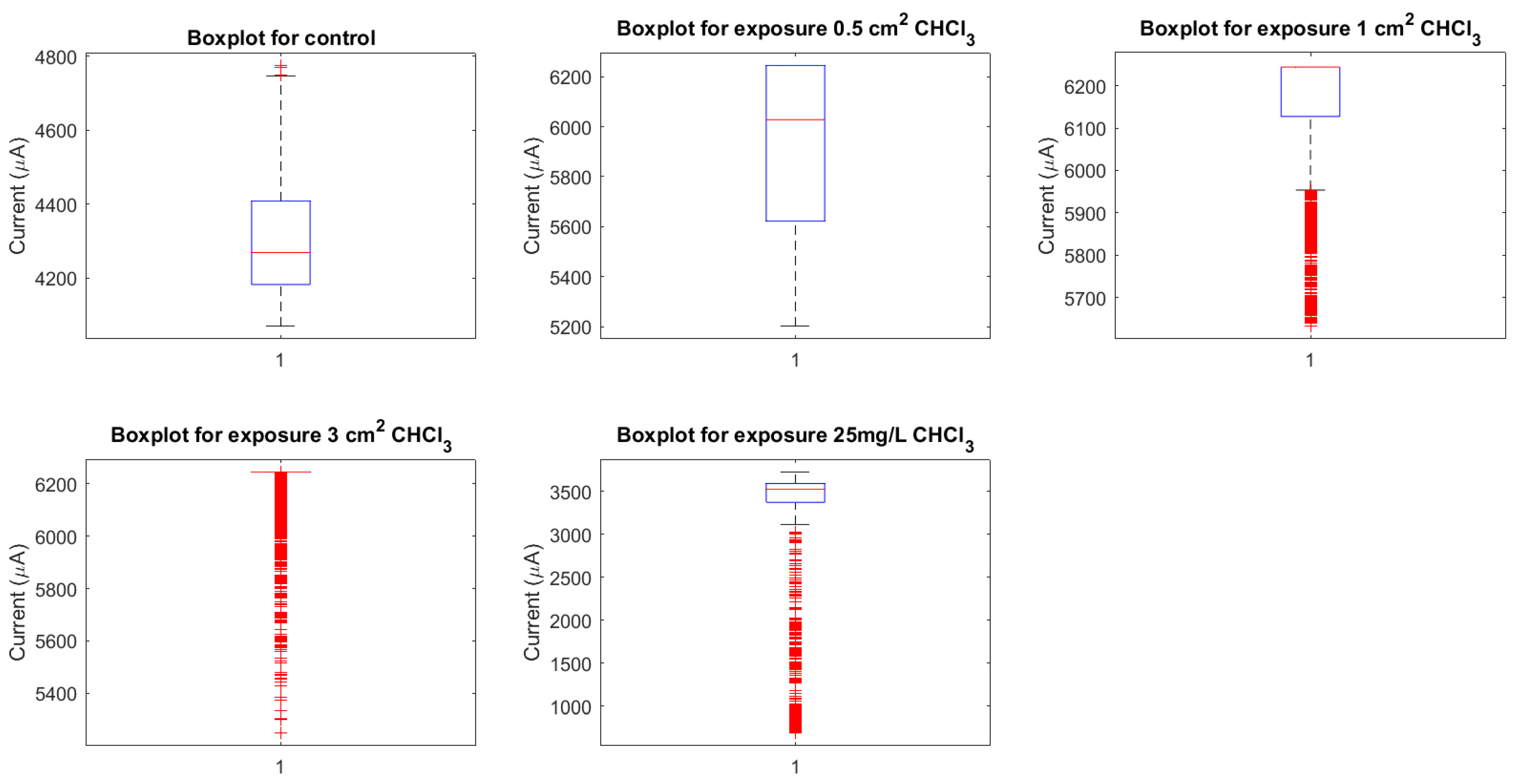
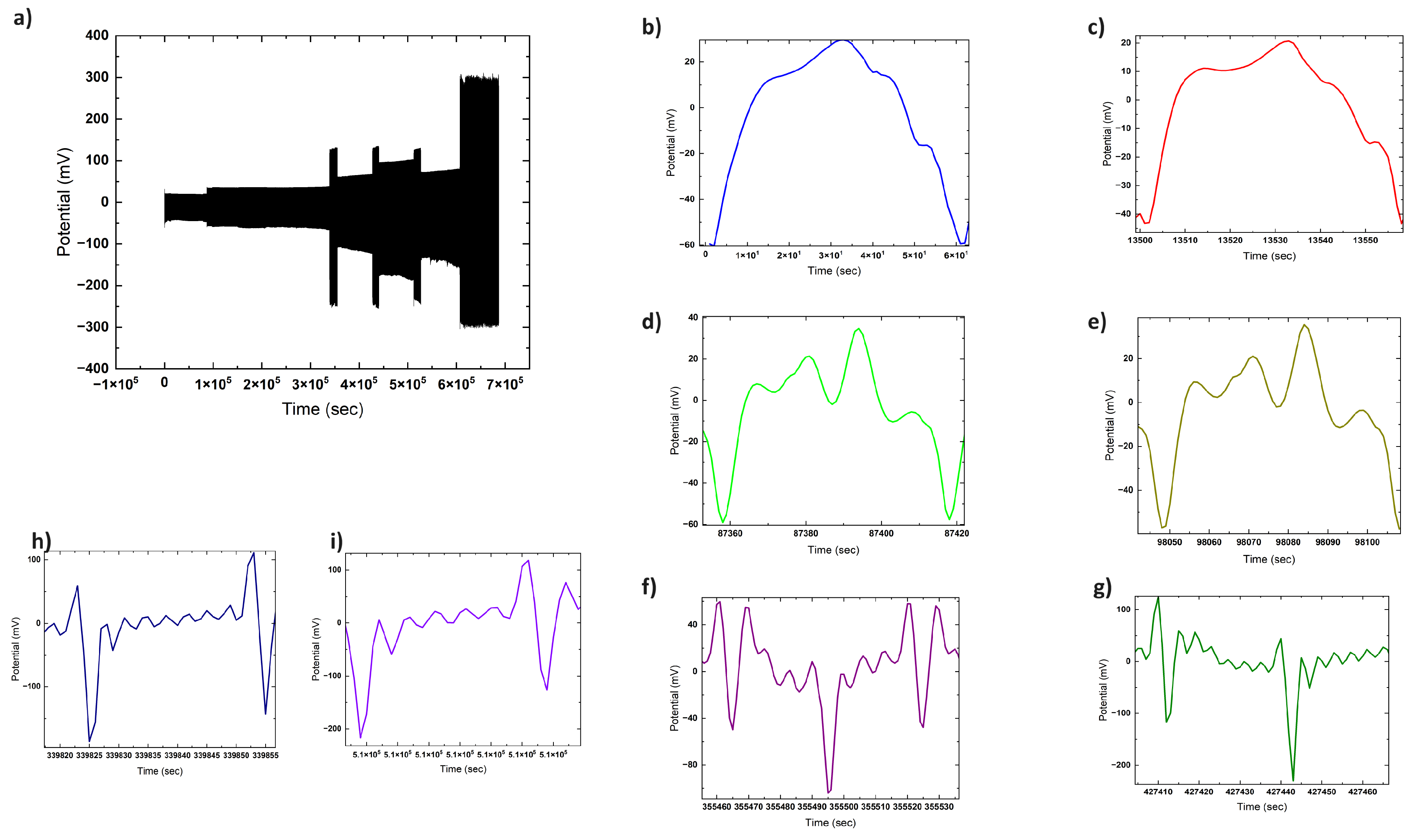
3.2. Baseline Electrical Dynamics
3.3. Vapour-Phase Chloroform Exposures
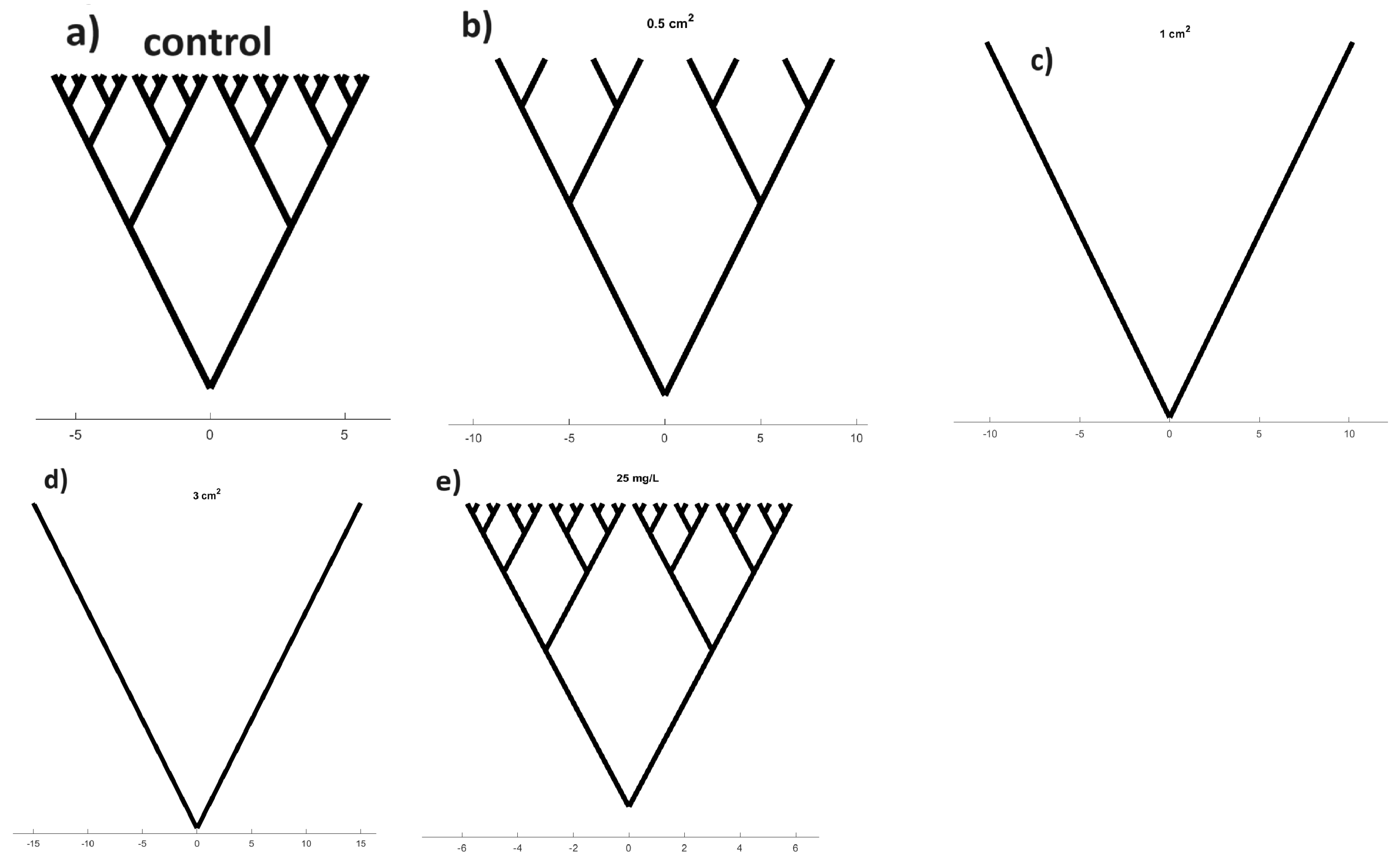
3.3.1. Modulation of Proteinoid Spiking upon Exposure to 0.5 cm × 0.5 cm Chloroform-Embedded Filters
3.3.2. Modulation of Proteinoid Spiking upon Exposure to 1.0 cm × 1.0 cm Chloroform-Embedded Filters
3.3.3. Modulation of Proteinoid Spiking upon Exposure to 3.0 cm × 3.0 cm Chloroform-Embedded Filters
3.4. Solvated Chloroform Response Dynamics
3.5. Concentration-Dependent Effects
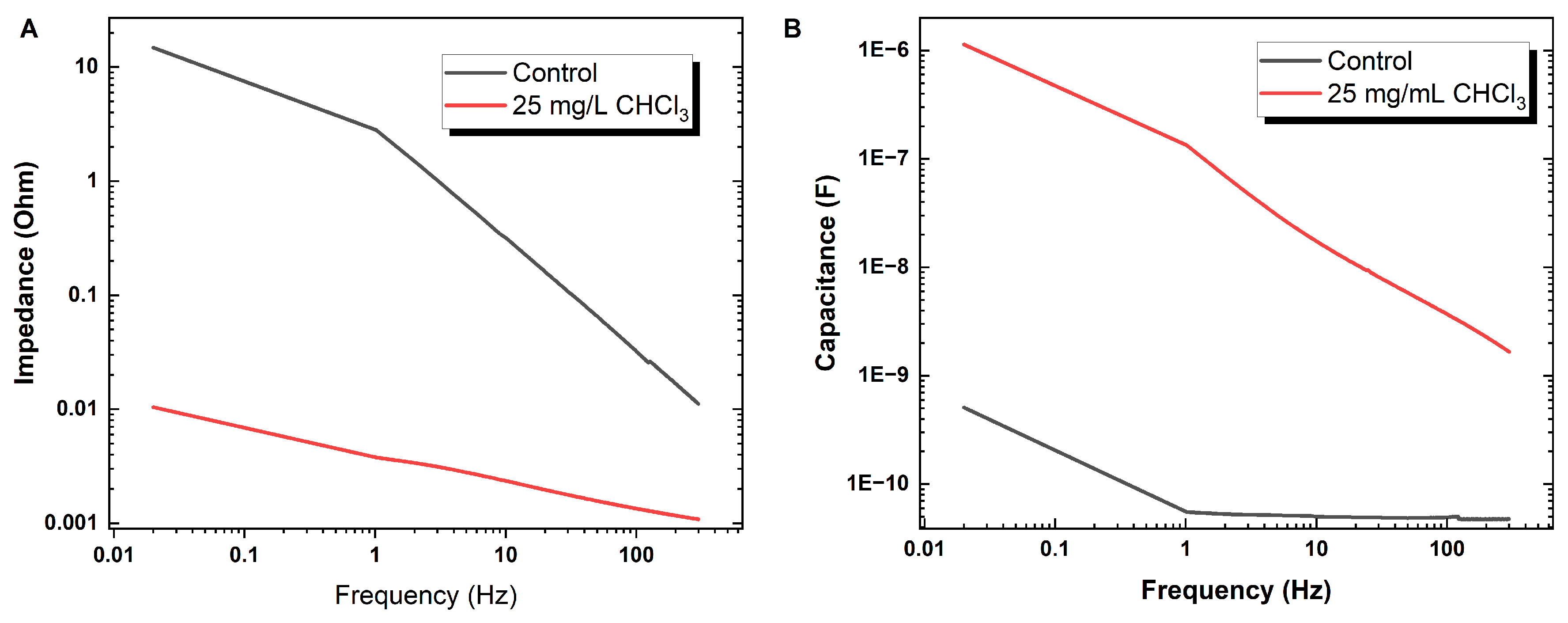
3.5.1. Tuning Resistance and Reactance in Proteinoid Assemblies through Chloroform Incorporation
3.5.2. Concentration-Dependent Modulation of Proteinoids–Chloroform Memristive Dynamics Characterised by Chronoamperometry
3.6. Elucidating Input–Output Relationships of Chloroform-Tuned Proteinoid Network Biointerfaces under Periodic Voltage Harmonic Forcing
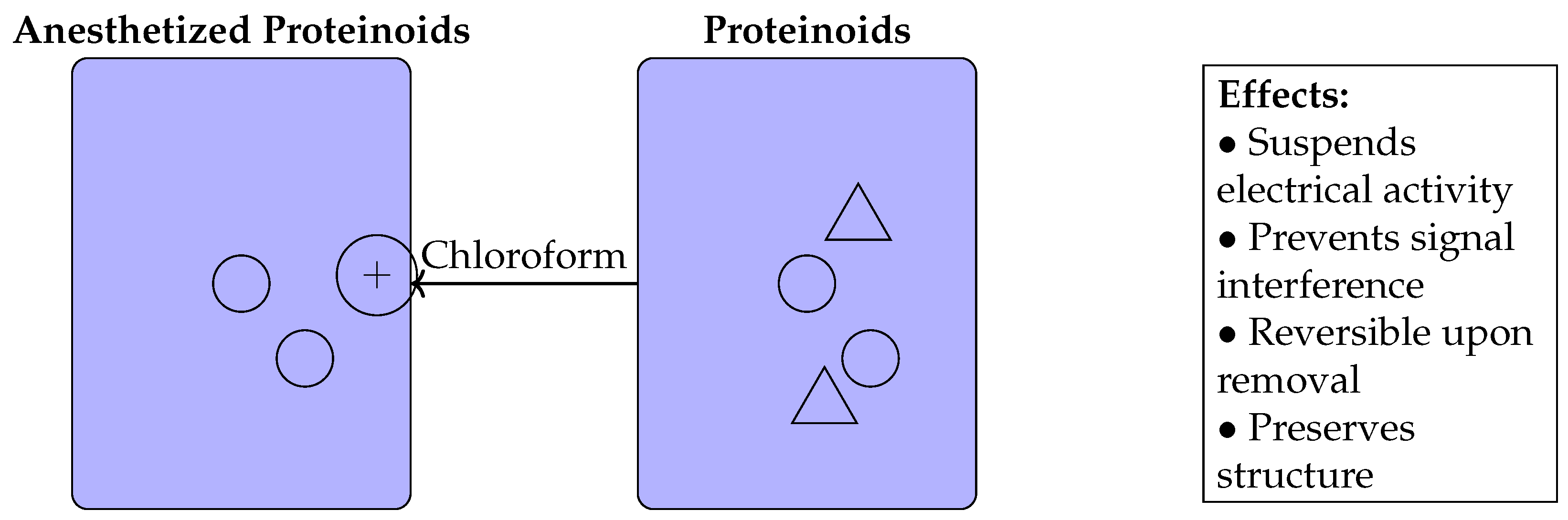
4. Potential Molecular Mechanisms
- Transient pore formation.Small chloroform molecules infiltrate and cause temporary defects in cross-linked peptide matrix.
- Swelling strain effects.Proteinoid microsphere wall dimensions increase or decrease with solvent attack.
- Modulated molecular clustering.Disrupts interior packing dynamics and charge mobility pathways.
- Lack of ion channel disruption.Unlike cells, no insulation from the impedance of selective ion flow.
- Bulk matrix effects rather than surface phenomena.Deep permeation into peptide structure underpins observed dynamics.
5. Discussion
- The direct binding of chloroform molecules within hydrophobic pockets on proteinoid surfaces, structurally distorting excitable domains.
- Changes in the ion permeability and transport kinetics of the membrane, altering the spike propensity.
- Conformational shifts of intrinsic pore-forming subunits, temporarily blocking conductive states.
- The insertion of chloroform into lipid bilayers, modifying membrane dielectric properties and capacitance.
- Combinatorial effects on proteinoid morphology and physiology, disrupting synchronised oscillations.
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A


References
- Fox, S.W.; Nakashima, T.; Przybylski, A.; Syren, R.M. The updated experimental proteinoid model. Int. J. Quantum Chem. 1982, 22, 195–204. [Google Scholar] [CrossRef]
- Nakashima, T. Metabolism of proteinoid microspheres. In Organic Geo- and Cosmochemistry; Springer: Berlin/Heidelberg, Germany, 2005; pp. 57–81. [Google Scholar]
- Nakashima, T.; Fox, S. Synthesis of peptides from amino acids and atp with lysine-rich proteinoid. J. Mol. Evol. 1980, 15, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Mougkogiannis, P.; Phillips, N.; Adamatzky, A. Transfer functions of proteinoid microspheres. Biosystems 2023, 227, 104892. [Google Scholar] [CrossRef] [PubMed]
- Mougkogiannis, P.; Adamatzky, A. Low frequency electrical waves in ensembles of proteinoid microspheres. Sci. Rep. 2023, 13, 1992. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.W. Metabolic microspheres: Origins and evolution. Naturwissenschaften 1980, 67, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.M.; Rao, K.P. Preparation and characterization of ph-sensitive proteinoid microspheres for the oral delivery of methotrexate. Biomaterials 1998, 19, 725–732. [Google Scholar] [CrossRef]
- Matsuno, K. Electrical excitability of proteinoid microspheres composed of basic and acidic proteinoids. BioSystems 1984, 17, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Nikolaidou, A.; Mougkogiannis, P.; Adamatzky, A. Electroactive composite biofilms integrating kombucha, chlorella and synthetic proteinoid proto–brains. R. Soc. Open Sci. 2023, 11, 240238. [Google Scholar] [CrossRef]
- Mougkogiannis, P.; Adamatzky, A. Proto–neurons from abiotic polypeptides. Encyclopedia 2023, 4, 512–543. [Google Scholar] [CrossRef]
- Mougkogiannis, P.; Adamatzky, A. Spiking frequency modulation of proteinoids with light and realisation of boolean gates. arXiv 2023, arXiv:2305.02433. [Google Scholar]
- Talwar, G. Textbook of Biochemistry, Biotechnology, Allied and Molecular Medicine; PHI Learning Pvt. Ltd.: Delhi, India, 2015. [Google Scholar]
- Rohlfing, D.L. The development of the proteinoid model for the origin of life. In Molecular Evolution and Protobiology; Springer: Berlin/Heidelberg, Germany, 1984; pp. 29–43. [Google Scholar]
- Fox, S.W. From Inanimate Matter to Living Systems; Technical Report; NASA: Washington, DC, USA, 1980.
- Schiffmann, Y. Self-organization in biology and development. Prog. Biophys. Mol. Biol. 1997, 68, 145–206. [Google Scholar] [CrossRef]
- Torday, J.S.; Miller, W.B., Jr. On the evolution of the mammalian brain. Front. Syst. Neurosci. 2016, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.H. Emergence of life. Zygon® 2007, 42, 837–856. [Google Scholar] [CrossRef]
- Tallawi, M. Proteinoid/hydroxyapatite hybrid microsphere composites. J. Biomed. Mater. Res. Part B Appl. Biomater. 2011, 96, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.W.; Yuyama, S. Dynamic phenomena in microspheres from thermal proteinoid. Comp. Biochem. Physiol. 1964, 11, 317–321. [Google Scholar] [CrossRef] [PubMed]
- Brooke, S.; Fox, S.W. Compartmentalization in proteinoid microspheres. BioSystems 1977, 9, 1–22. [Google Scholar] [PubMed]
- Bucher, E.S.; Wightman, R.M. Electrochemical analysis of neurotransmitters. Annu. Rev. Anal. Chem. 2015, 8, 239–261. [Google Scholar]
- Fox, S.W.; Bahn, P.R.; Dose, K.; Harada, K.; Hsu, L.; Ishima, Y.; Jungck, J.; Kendrick, J.; Krampitz, G.; Lacey, J.C.; et al. Experimental retracement of the origins of a protocell. J. Biol. Phys. 1995, 20, 17–36. [Google Scholar]
- Przybylski, A.T.; Fox, S.W. Excitable artificial cells of proteinoid. Appl. Biochem. Biotechnol. 1984, 10, 301–307. [Google Scholar] [PubMed]
- Ishima, Y.; Przybylski, A.T.; Fox, S.W. Electrical membrane phenomena in spherules from proteinoid and lecithin. BioSystems 1981, 13, 243–251. [Google Scholar] [PubMed]
- PMougkogiannis; Adamatzky, A. On interaction of proteinoids with simulated neural networks. bioRxiv 2023. [Google Scholar] [CrossRef]
- Kenyon, D. A comparison of proteinoid and aldocyanoin microsystems as models of the primordial protocell. In Molecular Evolution and Protobiology; Springer: Berlin/Heidelberg, Germany, 1984; pp. 163–188. [Google Scholar]
- Kim, J.-D.; Yang, S.R.; Cho, Y.W.; Park, K. 18 fast responsive nanoparticles of hydrophobically modified poly (amino acid) s and proteinoids. In Reflexive Polymers and Hydrogels: Understanding and Designing Fast Responsive Polymeric Systems; CRC Press: Boca Raton, FL, USA, 2004; Volume 373. [Google Scholar]
- Gombotz, W.R.; Wee, S.F. Protein release from alginate matrices. Adv. Drug Deliv. Rev. 2012, 64, 194–205. [Google Scholar] [CrossRef]
- Eisenman, G.; Dani, J.A. An introduction to molecular architecture and permeability of ion channels. Annu. Rev. Biophys. Biophys. Chem. 1987, 16, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Pang, X. Electrostatic interactions in protein structure, folding, binding, and condensation. Chem. Rev. 2018, 118, 1691–1741. [Google Scholar] [CrossRef] [PubMed]
- Przybylski, A.T. Physical background of excitability: Synthetic membranes and excitable cells. In Molecular Evolution and Protobiology; Springer: Berlin/Heidelberg, Germany, 1984; pp. 253–266. [Google Scholar]
- Kimizuka, H.; Koketsu, K. Ion transport through cell membrane. J. Theor. Biol. 1964, 6, 290–305. [Google Scholar] [CrossRef] [PubMed]
- Tamagawa, H. Membrane potential generation without ion transport. Ionics 2015, 21, 1631–1648. [Google Scholar] [CrossRef]
- Perouansky, M. The quest for a unified model of anesthetic action: A century in claude bernard’s shadow. J. Am. Soc. Anesthesiol. 2012, 117, 465–474. [Google Scholar] [CrossRef]
- Kennedy, R.; Galindo, A. Comparative site of action of various anaesthetic agents at the mammalian myoneural junction. BJA Br. J. Anaesth. 1975, 47, 533–540. [Google Scholar] [CrossRef][Green Version]
- Snow, J. On Chloroform and Other Anaesthetics: Their Action and Administration; Good Press: Glasgow, UK, 2022. [Google Scholar]
- Nunn, J.; Sharp, J.; Kimball, K. Reversible effect of an inhalational anaesthetic on lymphocyte motility. Nature 1970, 226, 85–86. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.H. Studies on the sensitivity of mimosa pudica ii. The effect of animal anesthetics and certain other compounds upon seismonic sensitivity. Am. J. Bot. 1931, 18, 215–235. [Google Scholar] [CrossRef]
- Bancroft, W.D.; Rutzler, J., Jr. Irritability and anesthesia in plants. J. Phys. Chem. 2002, 36, 273–285. [Google Scholar] [CrossRef]
- Grémiaux, A.; Yokawa, K.; Mancuso, S.; Baluška, F. Plant anesthesia supports similarities between animals and plants: Claude bernard’s forgotten studies. Plant Signal. Behav. 2014, 9, e27886. [Google Scholar] [CrossRef] [PubMed]
- Yokawa, K.; Kagenishi, T.; Baluška, F. Anesthetics, anesthesia, and plants. Trends Plant Sci. 2019, 24, 12–14. [Google Scholar] [CrossRef] [PubMed]
- Adamatzky, A.; Gandia, A. Fungi anaesthesia. Sci. Rep. 2022, 12, 340. [Google Scholar] [CrossRef] [PubMed]
- Seifriz, W. The swelling and shrinking of protoplasm. Trans. Faraday Soc. 1946, 42, B259–B266. [Google Scholar] [CrossRef]
- Halsey, M.J.; Smith, E. Effects of anaesthetics on luminous bacteria. Nature 1970, 227, 1363–1365. [Google Scholar] [CrossRef] [PubMed]
- Diamond, J.M.; Wright, E.M. Molecular forces governing non-electrolyte permeation through cell membranes. Proc. R. Soc. Lond. Ser. Biol. Sci. 1969, 172, 273–316. [Google Scholar]
- Cristani, M.; D’Arrigo, M.; Mandalari, G.; Castelli, F.; Sarpietro, M.G.; Micieli, D.; Venuti, V.; Bisignano, G.; Saija, A.; Trombetta, D. Interaction of four monoterpenes contained in essential oils with model membranes: Implications for their antibacterial activity. J. Agric. Food Chem. 2007, 55, 6300–6308. [Google Scholar] [CrossRef]
- Kelkar, D.A.; Chattopadhyay, A. The gramicidin ion channel: A model membrane protein. Biochim. Biophys. Acta BBA Biomembr. 2007, 1768, 2011–2025. [Google Scholar] [CrossRef]
- Fields, R.D.; Ni, Y. Nonsynaptic communication through atp release from volume-activated anion channels in axons. Sci. Signal. 2010, 3, ra73. [Google Scholar] [CrossRef] [PubMed]
- van Dongen, S.F.; de Hoog, H.-P.M.; Peters, R.J.; Nallani, M.; Nolte, R.J.; van Hest, J.C. Biohybrid polymer capsules. Chem. Rev. 2009, 109, 6212–6274. [Google Scholar] [CrossRef] [PubMed]
- Gundersen, V.; Storm-Mathisen, J.; Bergersen, L.H. Neuroglial transmission. Physiol. Rev. 2015, 95, 695–726. [Google Scholar] [CrossRef] [PubMed]
- Monnard, P.-A.; Deamer, D.W. Membrane self-assembly processes: Steps toward the first cellular life. Anat. Rec. 2002, 268, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Erokhin, V.; Berzina, T.; Camorani, P.; Smerieri, A.; Vavoulis, D.; Feng, J.; Fontana, M.P. Material memristive device circuits with synaptic plasticity: Learning and memory. BioNanoScience 2011, 1, 24–30. [Google Scholar] [CrossRef]
- Mougkogiannis, P.; Adamatzky, A. Learning in ensembles of proteinoid microspheres. R. Soc. Open Sci. 2023, 10, 230936. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, G.; Przybylski, A.T.; Fox, S.W. Thermal proteinoids as excitability-inducing materials. BioSystems 1987, 20, 219–223. [Google Scholar] [CrossRef] [PubMed]
- Ranke, J.; Stolte, S.; Störmann, R.; Arning, J.; Jastorff, B. Design of sustainable chemical products the example of ionic liquids. Chem. Rev. 2007, 107, 2183–2206. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, I.; Steinberg, S.; Tor, Y.; Shanzer, A.; Sagiv, J. Ionic recognition and selective response in self-assembling monolayer membranes on electrodes. Nature 1988, 332, 426–429. [Google Scholar] [CrossRef]
- Magin, C.M.; Alge, D.L.; Anseth, K.S. Bio-inspired 3D microenvironments: A new dimension in tissue engineering. Biomed. Mater. 2016, 11, 022001. [Google Scholar] [CrossRef] [PubMed]
- Biggins, P.; Kusterbeck, A.; Hiltz, J.A. Bio-Inspired Materials and Sensing Systems; Royal Society of Chemistry: London, UK, 2011. [Google Scholar]
- Hao, N.; Budnik, B.A.; Gunawardena, J.; O’Shea, E.K. Tunable signal processing through modular control of transcription factor translocation. Science 2013, 339, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Ryabko, B.; Reznikova, Z. Using shannon entropy and kolmogorov complexity to study the communicative system and cognitive capacities in ants. Complexity 1996, 2, 37–42. [Google Scholar] [CrossRef]
- Candès, E.J. Harmonic analysis of neural networks. Appl. Comput. Harmon. Anal. 1999, 6, 197–218. [Google Scholar] [CrossRef]
- Lewis, A.; Khatchatouriants, A.; Treinin, M.; Chen, Z.; Peleg, G.; Friedman, N.; Bouevitch, O.; Rothman, Z.; Loew, L.; Sheres, M. Second-harmonic generation of biological interfaces: Probing the membrane protein bacteriorhodopsin and imaging membrane potential around gfp molecules at specific sites in neuronal cells of C. elegans. Chem. Phys. 1999, 245, 133–144. [Google Scholar] [CrossRef]
- Pavel, M.A.; Petersen, E.N.; Wang, H.; Lerner, R.A.; Hansen, S.B. Studies on the mechanism of general anesthesia. Proc. Natl. Acad. Sci. USA 2020, 117, 13757–13766. [Google Scholar] [CrossRef] [PubMed]
- Sinha, M.; Mishra, S.; Joshi, P.G. Liquid-ordered microdomains in lipid rafts and plasma membrane of u-87 mg cells: A time-resolved fluorescence study. Eur. Biophys. J. 2003, 32, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Parton, R.G.; Hancock, J.F. Lipid rafts and plasma membrane microorganization: Insights from ras. Trends Cell Biol. 2004, 14, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Agbaga, M.-P.; McClellan, M.E.; Elliott, M.H. Analysis of lipids, fatty acid, and cholesterol in membrane microdomains. In Lipidomics: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2023; pp. 129–139. [Google Scholar]
- Adamatzky, A. Towards proteinoid computers. hypothesis paper. Biosystems 2021, 208, 104480. [Google Scholar] [CrossRef] [PubMed]
- Mougkogiannis, P.; Adamatzky, A. Logical gates in ensembles of proteinoid microspheres. PLoS ONE 2023, 18, e0289433. [Google Scholar] [CrossRef] [PubMed]
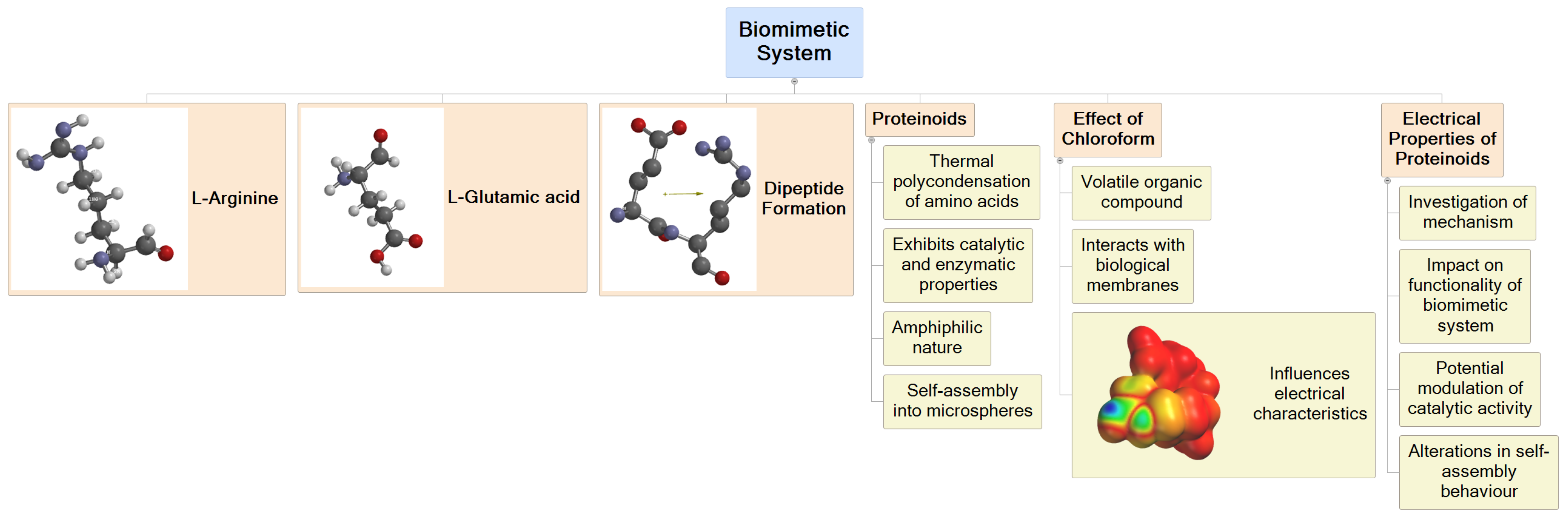
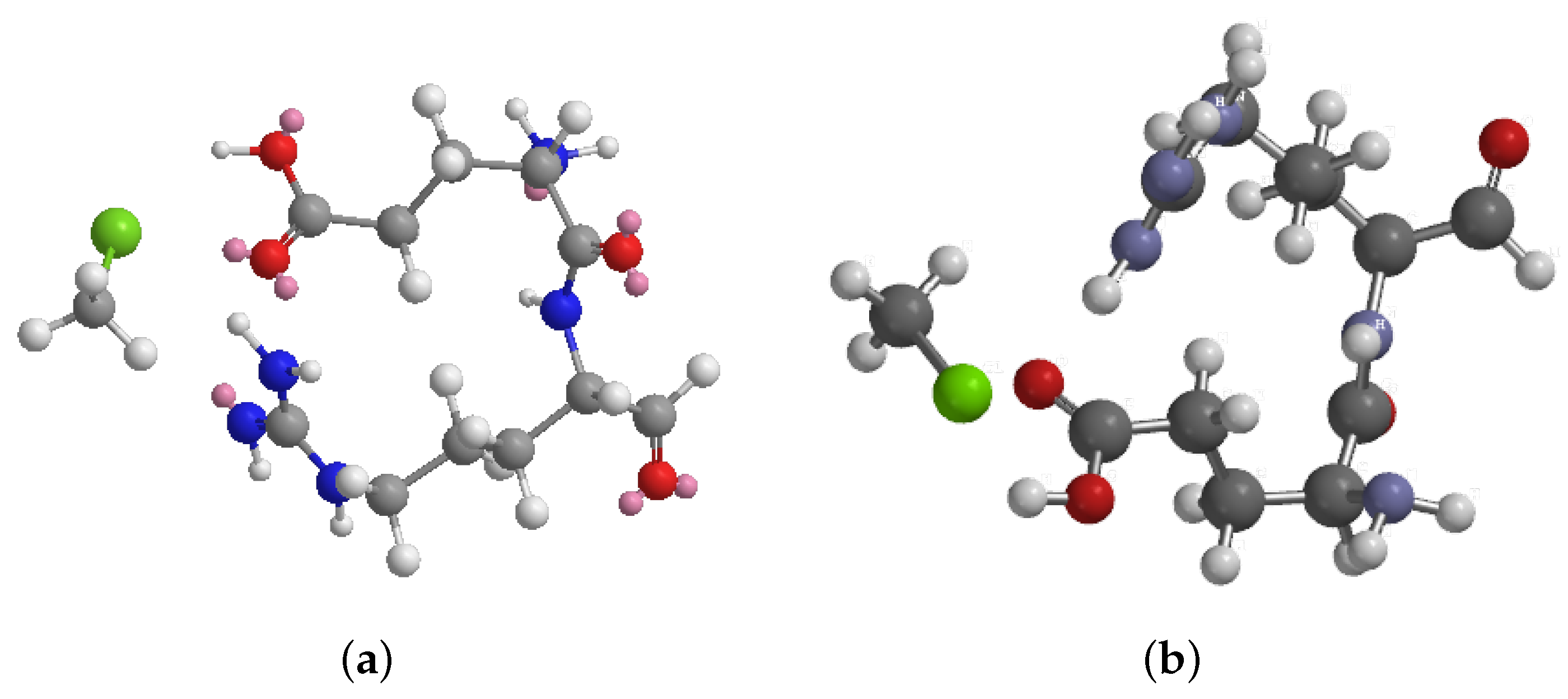



| Energy Component | Energy (kcal/mol) |
|---|---|
| Stretch | 0.6971 |
| Bend | 5.7351 |
| Stretch–bend | 0.2346 |
| Torsion | −0.5590 |
| Non-1,4 VDW | −9.7724 |
| 1,4 VDW | 7.9509 |
| Dipole/Dipole | −14.7107 |
| Total energy | −10.4245 |
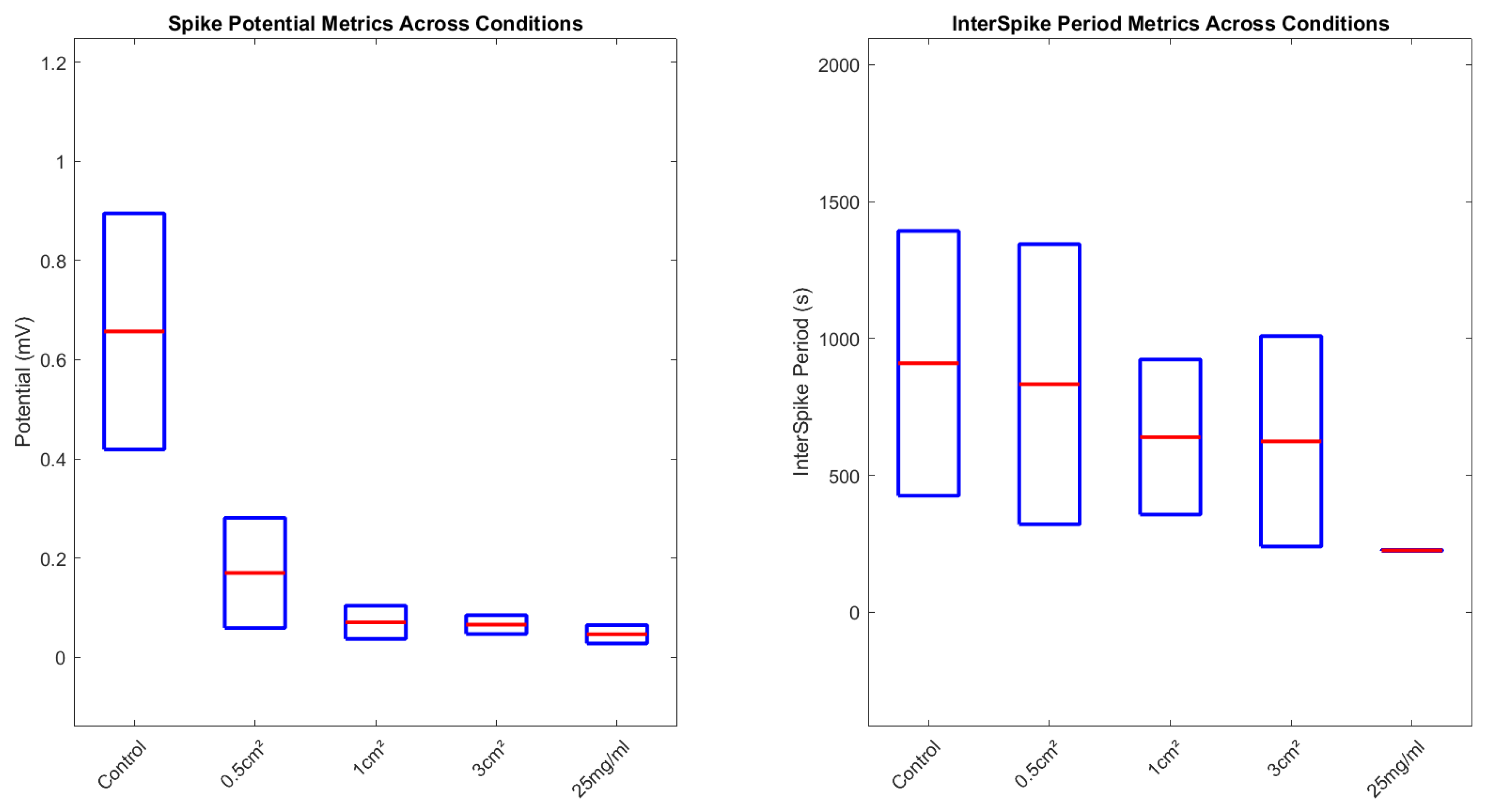
| Condition | Mean Pot (mV) | Std Dev | Max | Min | Kurtosis | Skewness |
|---|---|---|---|---|---|---|
| Control | 0.895 | 0.419 | 1.909 | 0.361 | 3.91 | 1.37 |
| 0.5 cm2 | 0.281 | 0.059 | 0.412 | 0.209 | 2.83 | 0.87 |
| 1 cm2 | 0.104 | 0.037 | 0.173 | 0.047 | 2.26 | 0.31 |
| 3 cm2 | 0.085 | 0.047 | 0.274 | 0.049 | 14.97 | 3.58 |
| 25 mg/mL | 0.065 | 0.028 | 0.134 | 0.027 | 5.05 | 1.39 |
| Condition | Mean Period (s) | Std Dev | Max | Min | Kurtosis | Skewness |
|---|---|---|---|---|---|---|
| Control | 1392.86 | 425.83 | 1952.38 | 595.24 | 1.95 | −0.36 |
| 0.5 cm2 | 1344.85 | 321.53 | 1871 | 882 | 1.82 | 0.41 |
| 1 cm2 | 923 | 356.92 | 1360 | 230 | 2.67 | −0.87 |
| 3 cm2 | 1008.75 | 240.08 | 1505 | 718 | 3.46 | 0.95 |
| 25 mg/mL | 228.2 | 223.91 | 623 | 69.8 | 2.08 | 0.92 |
| Condition | LZ Complexity |
|---|---|
| Control | 4.739922 |
| 0.5 cm2 | 2.790569 |
| 1 cm2 | 1.383151 |
| 3 cm2 | 1.002987 |
| 25 mg/L | 4.578150 |
| Property | Proteinoids | Anesthetised Proteinoids |
|---|---|---|
| Electrical Activity | Spontaneous spikes and oscillations | Suspended spiking, muted signals |
| Network Connectivity | Variable interproteinoid communication | Disrupted connections |
| Information Processing | Complex parallel logic and learning | Restricted processing capability |
| Handling | Requires insulation and interference avoidance | Permits direct manipulation and patterning |
| Reversibility | Inherent metastable state | Effects reversed upon chloroform removal |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mougkogiannis, P.; Adamatzky, A. On Effect of Chloroform on Electrical Activity of Proteinoids. Biomimetics 2024, 9, 380. https://doi.org/10.3390/biomimetics9070380
Mougkogiannis P, Adamatzky A. On Effect of Chloroform on Electrical Activity of Proteinoids. Biomimetics. 2024; 9(7):380. https://doi.org/10.3390/biomimetics9070380
Chicago/Turabian StyleMougkogiannis, Panagiotis, and Andrew Adamatzky. 2024. "On Effect of Chloroform on Electrical Activity of Proteinoids" Biomimetics 9, no. 7: 380. https://doi.org/10.3390/biomimetics9070380
APA StyleMougkogiannis, P., & Adamatzky, A. (2024). On Effect of Chloroform on Electrical Activity of Proteinoids. Biomimetics, 9(7), 380. https://doi.org/10.3390/biomimetics9070380







