Feature Selection Problem and Metaheuristics: A Systematic Literature Review about Its Formulation, Evaluation and Applications
Abstract
:1. Introduction
- Filter methods identify the optimal set of features by focusing on the specificities of the problem within the dataset without considering the classification algorithm to be used. These methods use statistical analysis to explore the connection between each input and target variable, assigning a relevance value to each feature. They stand out for their speed and computational efficiency. Examples of these methods include the correlation coefficient, the chi-squared test, and the Fisher score.
- Wrapper methods approach the feature selection iteratively, continuously adjusting the subset of features based on the training phase of the machine learning model. These methods offer a set of features ideally suited to the needs of the model and often performance improvement. Among its most well-known categories are forward selection, backward elimination, exhaustive selection, and metaheuristics.
- Embedded methods were introduced to overcome the difficulties filter and wrapper methods face. The purpose is to obtain quick results and with greater accuracy. Examples include lasso regression, decision trees, and random forest algorithms.
- An updated review of the literature analyzing and discussing objective functions proposed for the feature selection problem as well as metrics, classifiers, and metaheuristics used to solve the problem and benchmarks or real-world applications to validate the results obtained;
- A detailed classification of the objective functions and evaluation metrics provides a better understanding of the status of several aspects.
- A deep analysis of the metaheuristics used by researchers, following different criteria.
2. Methodology
- RQ1. How is the objective function of the feature selection problem formulated?
- RQ2. What metrics are used to analyze the performance of the feature selection problem?
- RQ3. What machine learning techniques have been used to calculate fitness in the feature selection problem?
- RQ4. What metaheuristics have been used to solve the feature selection problem?
- RQ5. Which datasets are commonly used as benchmarks, and which are derived from real-world applications?
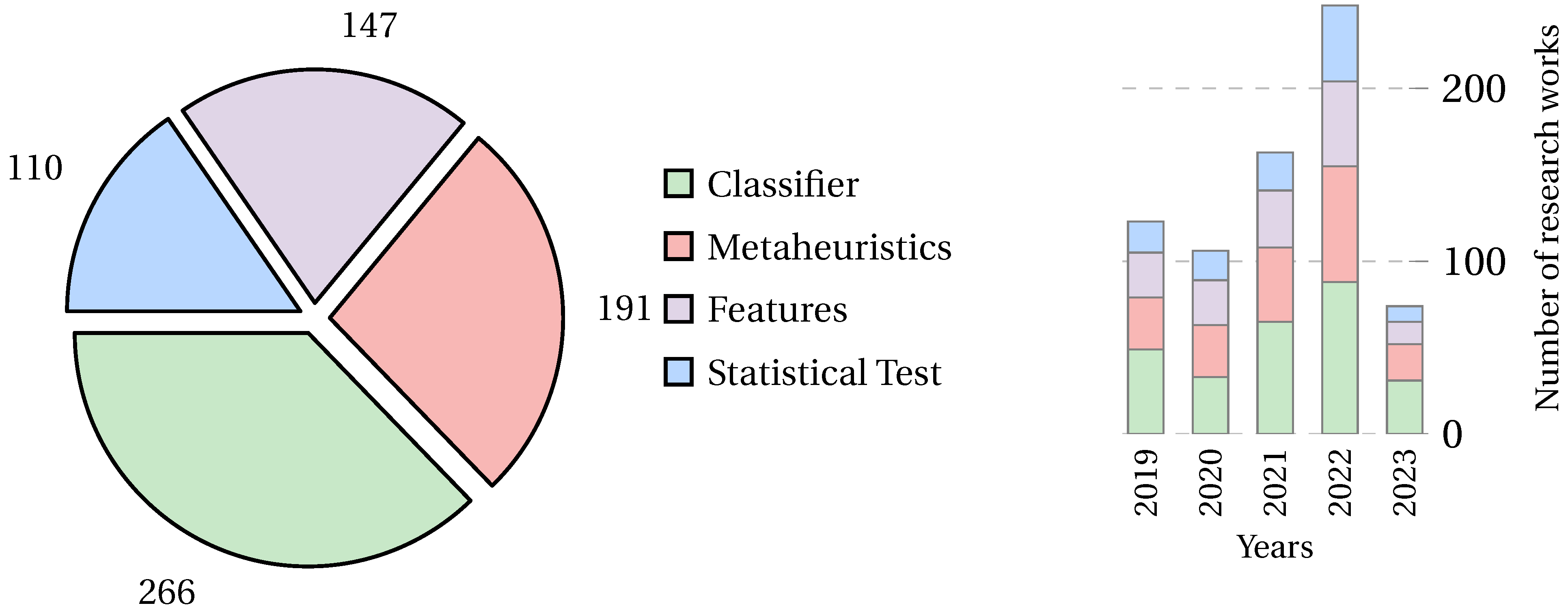
3. Bibliometric Analysis
4. Discussion
4.1. How Is the Objective Function of the Feature Selection Problem Formulated?
4.1.1. Single-Objective Functions
4.1.2. Pure Multi-Objective Functions
- = Number of features selected;
- = Accuracy;
- = Relevance;
- = Redundancy;
- = Interclass Distance;
- = Intraclass Distance.
4.1.3. Weighted Multi-Objective Functions
4.2. What Metrics Are Used to Analyze the Performance of the Feature Selection Problem?
4.2.1. Classifier Metrics
4.2.2. Metaheuristic Metrics
4.2.3. Feature Metrics
4.2.4. Statistical Tests
4.3. What Machine Learning Techniques Have Been Used to Calculate Fitness in the Feature Selection Problem?
4.3.1. Classifier Trends over Time
4.3.2. Classifier Usage by Year
- 2022: A significant leap is observed in the total number of articles, accompanied by a proportional increase in the use of diverse classifiers. The rise in articles employing multiple classifiers, including five classifiers [117], underscores a dynamic approach to optimization challenges.
4.3.3. Classifier Descriptions
4.3.4. Most Common Classifiers
4.3.5. Classifier Categories
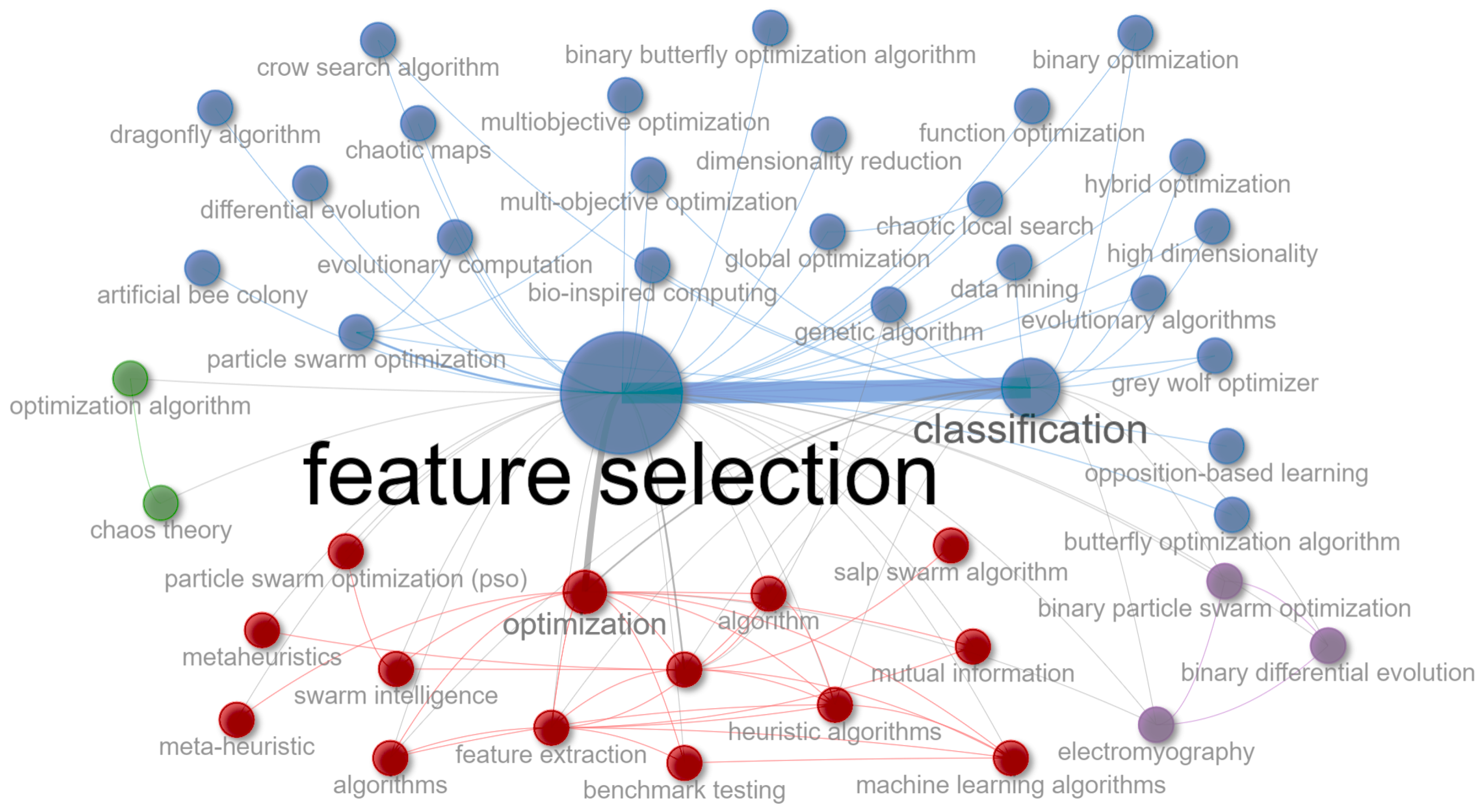
4.4. What Metaheuristics Have Been Used to Solve the Feature Selection Problem?
4.4.1. Frequency of Source Metaheuristics Utilization
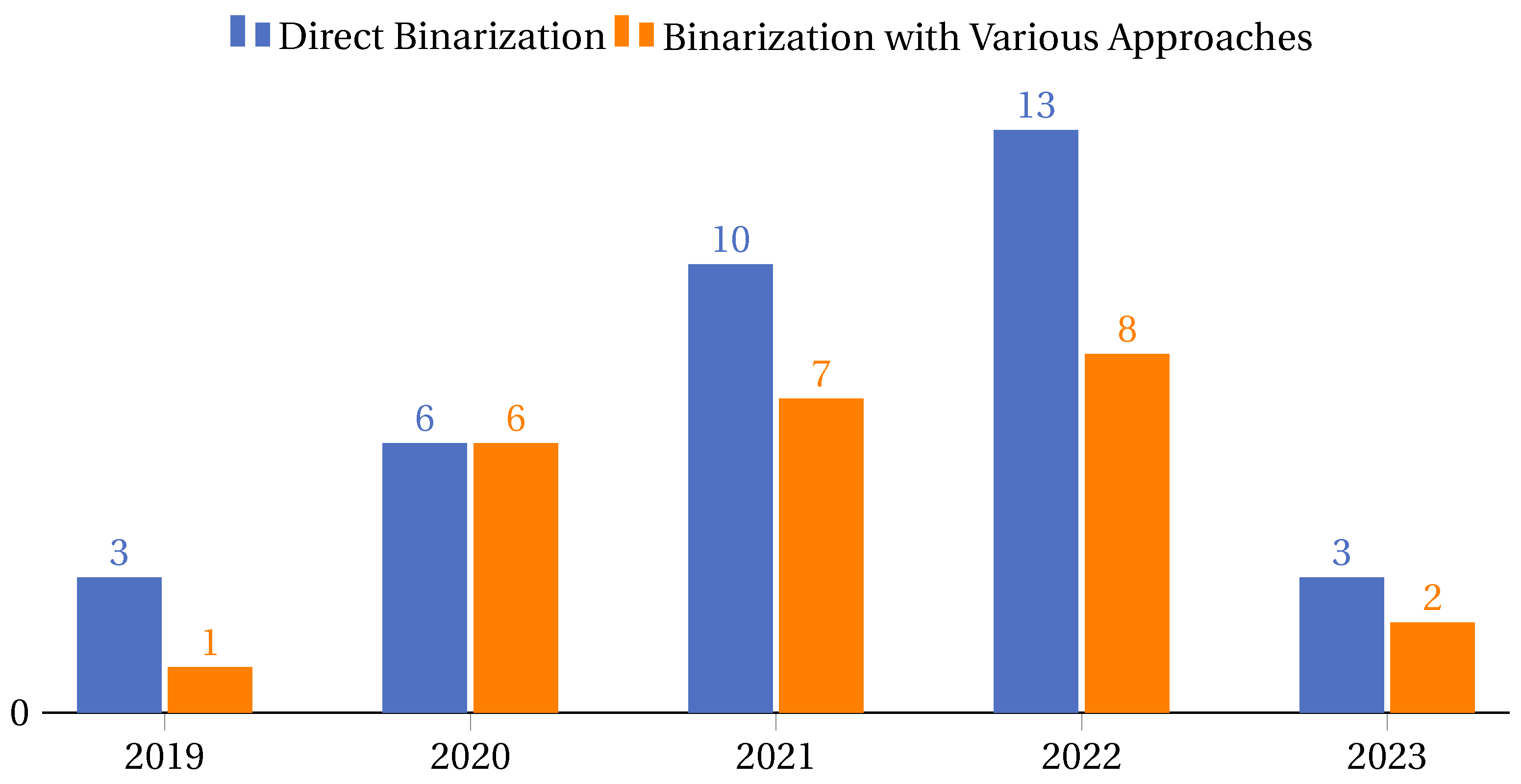
4.4.2. Binarization Approaches in Metaheuristics
- Direct binarization: This approach involves straightforward methods where the binarization process is direct and does not involve extensive testing or evaluation of different techniques. It is often used for its simplicity and efficiency. Cases of this approach are the papers [21,35,36,40,48,63,67,73,80,82,87,94,95,98,101,104,105,108,112,115,118,119,123,133,134,136,149,150,152,153,158,159,161,169,177].
- Binarization with various approaches: This approach involves a comprehensive study and evaluation of multiple binarization techniques to determine the most effective one for a given problem. It is more exhaustive and aims to find the optimal binarization method for specific scenarios. Cases of this approach are the articles [47,71,78,89,91,92,96,106,107,110,114,124,129,131,132,138,142,144,146,154,165,172,176,179].
4.4.3. Hybridization in Metaheuristics
- Most of the metaheuristics, including but not limited to the spotted hyena optimization algorithm (SHO) [85], seagull optimization algorithm (SOA) [86], sine cosine algorithm (SCA) [92], and dwarf mongoose optimization (DMO) [112], have been used once as foundational algorithms. This showcases the diversity of metaheuristics explored by researchers in the hybridization process.
- The wide range of foundational metaheuristics, even those used just once, underscores the richness of the field. It indicates that researchers continuously experiment with different base algorithms to find the most suitable combinations for specific problems.
- A wide array of metaheuristics, including the firefly algorithm (FAA) [144], thermal exchange optimization (TEO) [86], the cuckoo search algorithm (CSA) [88], and harmony search (HS) [154], among others, have been used once. This diversity reflects the rich experimentation in the field, with researchers exploring various combinations to achieve optimal results.
4.4.4. Techniques to Enhance Metaheuristics
- Chaotic maps search function: With a total of 25 instances across the years [34,40,42,53,81,90,93,103,106,114,117,120,121,126,129,134,143,147,156,159,161,162,163,164,183] this technique has seen consistent use, with a noticeable peak in 2022. Its application suggests that researchers find value in its chaotic dynamics to enhance the exploration capabilities of metaheuristics.
- Local search: This technique has been the most frequently employed, with a total of 28 instances [36,40,46,50,69,77,93,99,103,107,111,112,113,120,126,127,128,129,130,134,143,145,147,151,153,161,174,183]. Particularly in 2022, there was a significant surge in its application, indicating its effectiveness in refining solutions and improving convergence rates.
4.4.5. Multi-Objective Approaches in Metaheuristics
- The numbers in 2022 and 2023 (up to April) show a decline, which could be attributed to various factors, including shifts in research focus or the maturation of multi-objective techniques developed in previous years.
- In the context of feature selection, multi-objective metaheuristics are invaluable. Feature selection often involves balancing reducing dimensionality (and thus computational cost) and retaining the most informative features for accurate prediction or classification. Multi-objective approaches provide a framework to navigate these conflicting objectives, ensuring robust and efficient models.
4.4.6. Relationship between Objective Function Formulation and Metaheuristics
4.5. Which Datasets Are Commonly Used as Benchmarks, and Which Are Derived from Real-World Applications?
4.5.1. Overall Trend in Dataset Usage
4.5.2. Real-World Application Datasets and Their Characteristics
- The authors in [178] utilized a dataset constructed from the Twitter API focusing on cancer and drugs, enabling sentiment analysis and text classification.
- For industrial maintenance, The authors in [44] employed a dataset designed for motor fault detection.
- The authors in [164] involved a dataset of 553 drugs bio-transformed in the liver, annotated with toxic effects such as irritant, mutagenic, reproductive, and tumorigenic, each represented by chemical descriptors.
- The authors in [23] used hyperspectral image datasets and spectral data of typical surface features, indicating the application of machine learning in environmental monitoring.
- The authors in [35], a dataset of 500 Arabic email messages from computer science students was analyzed, showing machine learning’s application in language processing and cybersecurity.
- The authors in [21] examined data from Iraqi cancer patients, offering a comprehensive dataset for healthcare research across multiple cancer types.
- The authors in [22] focused on constructing a dataset from Zeek network-based intrusion detection logs, underscoring machine learning’s role in network security.
- The authors in [187] presented a dataset related to medical treatment for cardiogenic shock, highlighting the intersection of machine learning and medical research.
4.5.3. Prevalent Datasets and Their Defining Characteristics
- Source name: The standardized name or label of the dataset.
- Subject area: The domain or field from which the dataset originates, which reveals a significant leaning towards the medical and biological areas but also showcases diversity, with datasets from physical science, politics, games, and synthetic sources.
- Instances/samples: The number of individual data points or samples in each dataset.
- Features/characteristics: The number of attributes or characteristics each sample in the dataset has.
- Classes: The number of unique labels or outcomes into which the samples can be categorized.
- Reference: Based on DOI, a digital object identifier that provides a persistent link to a dataset.
- Repository: The platform or database from which the dataset can be accessed.
4.5.4. A Glimpse into Data Sources
- UCI Repository: Standing as a stalwart in the academic community, the UCI Repository was referenced by a substantial 127 articles [21,23,27,28,29,31,34,36,37,38,40,41,42,44,45,46,48,49,51,53,54,58,62,63,64,66,67,68,69,70,71,72,73,74,75,77,78,79,80,81,82,83,84,85,86,87,88,89,90,92,93,95,96,97,98,99,100,101,102,103,104,105,108,109,111,112,113,114,116,118,119,120,121,124,125,126,127,128,129,130,131,132,133,134,135,136,137,139,140,141,142,143,145,146,148,149,150,152,153,154,155,156,157,158,159,160,161,163,164,165,166,168,169,170,171,173,179,183,184,185]. A testament to its vast collection and diverse range of datasets, UCI has proven to be an indispensable resource.
- Miscellaneous repositories: Several repositories mentioned in two articles were Ke Chen—Ph.D. Candidate Datasets Repository [36,38], which caters to specific academic projects; the UNB CIC [34,56] and UNSW Repositories [22,56], known for cybersecurity and network datasets; and the Mulan Library [41,63], emphasizing multi-label learning datasets.
4.6. Closing of Discussions
- Objective function formulation (RQ1): Our review revealed a diversity of objective functions used in feature selection, generally classified as single-objective or multi-objective functions. We observed that while single-objective functions focus on optimizing a single criterion, multi-objective functions, including pure and weighted types, cater to multiple criteria simultaneously. Weighted multi-objective functions were more prevalent in our dataset, suggesting their broader applicability in complex scenarios.
- Performance metrics (RQ2): We classified the performance metrics used in feature selection research into four main categories: classifier metrics, metaheuristic metrics, feature metrics, and statistical tests. Classifier metrics are the most frequently used, emphasizing the importance of the machine learning technique’s performance. The significant use of metaheuristic metrics and feature metrics underscores the complexity of evaluating feature selection methods.
- Used machine learning techniques (RQ3): We investigated machine learning techniques that are improved by feature selection. We found that a variety of classifiers are used, with k-nearest neighbor (k-NN) being the most common. The prevalence of techniques such as SVM, naive Bayes, and decision tree classifiers, including DT C4.5 and random forest, illustrates the wide applicability of feature selection across different learning paradigms.
- Metaheuristics (RQ4): Our study highlights the significant role of metaheuristics in feature selection, particularly particle swarm optimization (PSO), grey wolf optimizer (GWO), and genetic algorithm (GA). Their frequent use points to a preference for adaptive, population-based algorithms adept at handling the complex aspects of feature selection. This observation not only confirms the effectiveness of these methods but also suggests promising directions for future research in enhancing feature selection procedures.
- Practical applications and trends (RQ5): Our analysis of dataset usage trends in feature selection research reveals a slight increase in the number of datasets used per article over time. This shift, along with the dominant use of benchmark datasets and a focus on real-world applications, reflects the escalating complexity and practical significance of feature selection studies. The variety of dataset sources, especially the frequent citation of the UCI Repository, demonstrates the extensive applicability of feature selection in diverse domains.
5. Conclusions
- Selection of Objective Function: It is interesting to note that the same optimization problem can be represented through three different types of objective functions, each increasing the complexity of the problem. For researchers who are just starting in the field of feature selection, we recommend starting by solving the problem from a single-objective perspective, then moving on to weighted multi-objective, and finally to pure multi-objective.
- Selection of Evaluation Metrics: Regarding metrics, we can observe that there are 4 major groups which are classifier metrics, metaheuristic metrics, feature metrics, and statistical tests. For robustness in future research, we recommend incorporating at least one metric from each of the reported categories.
- −
- For classifier metrics, we recommend using Accuracy, Error Rate, Precision, Recall, and F-score.
- −
- For the case of metaheuristic metrics, we recommend using the computational time, the fitness in the case of using a mono-objective function or weighted multi-objective function, and the hyper-volume metric in the case of using a pure multi-objective function.
- −
- In the case of feature metrics, we recommend reporting the number of features selected and which features were selected.
- −
- For the case of statistical test, we recommend advocating for a balanced application of both non-parametric tests, such as the Wilcoxon and Friedman tests, and parametric tests like the T-test, supplemented by rigorous post hoc analyses for in-depth insights
A metric that, in our opinion, should be included in all research is indicating the solution vector, that is, indicating which features were selected by the metaheuristics. - Selection of classifier: The choice of classifier will depend closely on the dataset used where the important issues to be considered are the unbalance of the target classes, whether it is multi-class or binary-class, and the number of samples. In this sense, we recommend experimenting with more than one classifier to express robust results and can use the KNN, Random Forest, or Xgboost.
- Selection of Benchmark Dataset: Guided by a curated list of the top 20 datasets, ensuring that experimentation and comparison are grounded in both established and innovative contexts.
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Witten, I.H.; Frank, E. Data mining: Practical machine learning tools and techniques with Java implementations. ACM Sigmod Rec. 2002, 31, 76–77. [Google Scholar] [CrossRef]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Agrawal, P.; Abutarboush, H.F.; Ganesh, T.; Mohamed, A.W. Metaheuristic Algorithms on Feature Selection: A Survey of One Decade of Research (2009–2019). IEEE Access 2021, 9, 26766–26791. [Google Scholar] [CrossRef]
- Nssibi, M.; Manita, G.; Korbaa, O. Advances in nature-inspired metaheuristic optimization for feature selection problem: A comprehensive survey. Comput. Sci. Rev. 2023, 49, 100559. [Google Scholar] [CrossRef]
- Kurman, S.; Kisan, S. An in-depth and contrasting survey of meta-heuristic approaches with classical feature selection techniques specific to cervical cancer. Knowl. Inf. Syst. 2023, 65, 1881–1934. [Google Scholar] [CrossRef]
- Pham, T.H.; Raahemi, B. Bio-Inspired Feature Selection Algorithms with Their Applications: A Systematic Literature Review. IEEE Access 2023, 11, 43733–43758. [Google Scholar] [CrossRef]
- Sadeghian, Z.; Akbari, E.; Nematzadeh, H.; Motameni, H. A review of feature selection methods based on meta-heuristic algorithms. J. Exp. Theor. Artif. Intell. 2023, 1–51. [Google Scholar] [CrossRef]
- Arun Kumar, R.; Vijay Franklin, J.; Koppula, N. A Comprehensive Survey on Metaheuristic Algorithm for Feature Selection Techniques. Mater. Today Proc. 2022, 64, 435–441. [Google Scholar] [CrossRef]
- Akinola, O.O.; Ezugwu, A.E.; Agushaka, J.O.; Zitar, R.A.; Abualigah, L. Multiclass feature selection with metaheuristic optimization algorithms: A review. Neural Comput. Appl. 2022, 34, 19751–19790. [Google Scholar] [CrossRef]
- Dokeroglu, T.; Deniz, A.; Kiziloz, H.E. A comprehensive survey on recent metaheuristics for feature selection. Neurocomputing 2022, 494, 269–296. [Google Scholar] [CrossRef]
- Abu Khurma, R.; Aljarah, I.; Sharieh, A.; Abd Elaziz, M.; Damaševičius, R.; Krilavičius, T. A Review of the Modification Strategies of the Nature Inspired Algorithms for Feature Selection Problem. Mathematics 2022, 10, 464. [Google Scholar] [CrossRef]
- Yab, L.Y.; Wahid, N.; Hamid, R.A. A Meta-Analysis Survey on the Usage of Meta-Heuristic Algorithms for Feature Selection on High-Dimensional Datasets. IEEE Access 2022, 10, 122832–122856. [Google Scholar] [CrossRef]
- Abiodun, E.O.; Alabdulatif, A.; Abiodun, O.I.; Alawida, M.; Alabdulatif, A.; Alkhawaldeh, R.S. A systematic review of emerging feature selection optimization methods for optimal text classification: The present state and prospective opportunities. Neural Comput. Appl. 2021, 33, 15091–15118. [Google Scholar] [CrossRef] [PubMed]
- Al-Tashi, Q.; Abdulkadir, S.J.; Rais, H.M.; Mirjalili, S.; Alhussian, H. Approaches to Multi-Objective Feature Selection: A Systematic Literature Review. IEEE Access 2020, 8, 125076–125096. [Google Scholar] [CrossRef]
- Talbi, E.G. Metaheuristics: From Design to Implementation; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Rajwar, K.; Deep, K.; Das, S. An exhaustive review of the metaheuristic algorithms for search and optimization: Taxonomy, applications, and open challenges. Artif. Intell. Rev. 2023, 56, 13187–13257. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.C.; Pepyne, D.L. Simple explanation of the no-free-lunch theorem and its implications. J. Optim. Theory Appl. 2002, 115, 549–570. [Google Scholar] [CrossRef]
- Igel, C. No free lunch theorems: Limitations and perspectives of metaheuristics. In Theory and Principled Methods for the Design of Metaheuristics; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–23. [Google Scholar] [CrossRef]
- Wolpert, D.; Macready, W. No free lunch theorems for optimization. IEEE Trans. Evol. Comput. 1997, 1, 67–82. [Google Scholar] [CrossRef]
- Becerra-Rozas, M.; Lemus-Romani, J.; Cisternas-Caneo, F.; Crawford, B.; Soto, R.; Astorga, G.; Castro, C.; García, J. Continuous metaheuristics for binary optimization problems: An updated systematic literature review. Mathematics 2022, 11, 129. [Google Scholar] [CrossRef]
- Ibrahim, H.T.; Mazher, W.J.; Jassim, E.M. Feature Selection: Binary Harris Hawk Optimizer Based Biomedical Datasets. Intel. Artif. 2022, 25, 33–49. [Google Scholar] [CrossRef]
- Chohra, A.; Shirani, P.; Karbab, E.B.; Debbabi, M. Chameleon: Optimized feature selection using particle swarm optimization and ensemble methods for network anomaly detection. Comput. Secur. 2022, 117, 102684. [Google Scholar] [CrossRef]
- Wan, Y.; Ma, A.; Zhong, Y.; Hu, X.; Zhang, L. Multiobjective Hyperspectral Feature Selection Based on Discrete Sine Cosine Algorithm. IEEE Trans. Geosci. Remote Sens. 2020, 58, 3601–3618. [Google Scholar] [CrossRef]
- Kitchenham, B. Procedures for performing systematic reviews. Keele UK Keele Univ. 2004, 33, 1–26. [Google Scholar]
- Aria, M.; Cuccurullo, C. bibliometrix: An R-tool for comprehensive science mapping analysis. J. Inf. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- Boyd, S.P.; Vandenberghe, L. Convex Optimization; Cambridge University Press: Cambridge, UK, 2004. [Google Scholar]
- Engelbrecht, A.P.; Grobler, J.; Langeveld, J. Set based particle swarm optimization for the feature selection problem. Eng. Appl. Artif. Intell. 2019, 85, 324–336. [Google Scholar] [CrossRef]
- Xue, Y.; Xue, B.; Zhang, M. Self-Adaptive Particle Swarm Optimization for Large-Scale Feature Selection in Classification. ACM Trans. Knowl. Discov. Data 2019, 13, 1–27. [Google Scholar] [CrossRef]
- Huang, Z.; Yang, C.; Zhou, X.; Huang, T. A Hybrid Feature Selection Method Based on Binary State Transition Algorithm and ReliefF. IEEE Biomed. Health Inform. 2019, 23, 1888–1898. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, H.G.; Wang, Q.; Peng, C. A filter-based bare-bone particle swarm optimization algorithm for unsupervised feature selection. Appl. Intell. 2019, 49, 2889–2898. [Google Scholar] [CrossRef]
- Mohammed, T.A.; Bayat, O.; Uçan, O.N.; Alhayali, S. Hybrid efficient genetic algorithm for big data feature selection problems. Found. Sci. 2020, 25, 1009–1025. [Google Scholar] [CrossRef]
- Tan, P.; Wang, X.; Wang, Y. Dimensionality reduction in evolutionary algorithms-based feature selection for motor imagery brain-computer interface. Swarm Evol. Comput. 2020, 52, 100597. [Google Scholar] [CrossRef]
- Alsaleh, A.; Binsaeedan, W. The Influence of Salp Swarm Algorithm-Based Feature Selection on Network Anomaly Intrusion Detection. IEEE Access 2021, 9, 112466–112477. [Google Scholar] [CrossRef]
- Wang, L.; Gao, Y.; Li, J.; Wang, X. A feature selection method by using chaotic cuckoo search optimization algorithm with elitist preservation and uniform mutation for data classification. Discret. Dyn. Nat. Soc. 2021, 2021, 1–19. [Google Scholar] [CrossRef]
- BinSaeedan, W.; Alramlawi, S. CS-BPSO: Hybrid feature selection based on chi-square and binary PSO algorithm for Arabic email authorship analysis. Knowl.-Based Syst. 2021, 227, 107224. [Google Scholar] [CrossRef]
- Yang, J.Q.; Chen, C.H.; Li, J.Y.; Liu, D.; Li, T.; Zhan, Z.H. Compressed-Encoding Particle Swarm Optimization with Fuzzy Learning for Large-Scale Feature Selection. Symmetry 2022, 14, 1142. [Google Scholar] [CrossRef]
- Long, W.; Jiao, J.; Xu, M.; Tang, M.; Wu, T.; Cai, S. Lens-imaging learning Harris hawks optimizer for global optimization and its application to feature selection. Expert Syst. Appl. 2022, 202, 117255. [Google Scholar] [CrossRef]
- Yang, J.Q.; Yang, Q.T.; Du, K.J.; Chen, C.H.; Wang, H.; Jeon, S.W.; Zhang, J.; Zhan, Z.H. Bi-Directional Feature Fixation-Based Particle Swarm Optimization for Large-Scale Feature Selection. IEEE Trans. Big Data 2023, 9, 1004–1017. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Gao, X.; Gong, D.; Song, X.; Guo, Y.; Wang, J. A federated feature selection algorithm based on particle swarm optimization under privacy protection. Knowl.-Based Syst. 2023, 260, 110122. [Google Scholar] [CrossRef]
- Feizi-Derakhsh, M.R.; Kadhim, E.A. An Improved Binary Cuckoo Search Algorithm For Feature Selection Using Filter Method And Chaotic Map. J. Appl. Sci. Eng. 2022, 26, 895–901. [Google Scholar] [CrossRef]
- Park, J.; Park, M.W.; Kim, D.W.; Lee, J. Multi-Population Genetic Algorithm for Multilabel Feature Selection Based on Label Complementary Communication. Entropy 2020, 22, 876. [Google Scholar] [CrossRef]
- Du, Z.; Han, D.; Li, K.C. Improving the performance of feature selection and data clustering with novel global search and elite-guided artificial bee colony algorithm. J. Supercomput. 2019, 75, 5189–5226. [Google Scholar] [CrossRef]
- Zakeri, A.; Hokmabadi, A. Efficient feature selection method using real-valued grasshopper optimization algorithm. Expert Syst. Appl. 2019, 119, 61–72. [Google Scholar] [CrossRef]
- Pourpanah, F.; Lim, C.P.; Wang, X.; Tan, C.J.; Seera, M.; Shi, Y. A hybrid model of fuzzy min–max and brain storm optimization for feature selection and data classification. Neurocomputing 2019, 333, 440–451. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R.; Mohd Saad, N.; Tee, W. EMG feature selection and classification using a Pbest-guide binary particle swarm optimization. Computation 2019, 7, 12. [Google Scholar] [CrossRef]
- Tubishat, M.; Idris, N.; Shuib, L.; Abushariah, M.A.; Mirjalili, S. Improved Salp Swarm Algorithm based on opposition based learning and novel local search algorithm for feature selection. Expert Syst. Appl. 2020, 145, 113122. [Google Scholar] [CrossRef]
- Slezkin, A.; Hodashinsky, I.A.; Shelupanov, A.A. Binarization of the Swallow swarm optimization for feature selection. Program. Comput. Softw. 2021, 47, 374–388. [Google Scholar] [CrossRef]
- Akinola, O.A.; Agushaka, J.O.; Ezugwu, A.E. Binary dwarf mongoose optimizer for solving high-dimensional feature selection problems. PLoS ONE 2022, 17, e0274850. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, Y.; Hong, M.; He, H.; Huang, S. A self-adaptive level-based learning artificial bee colony algorithm for feature selection on high-dimensional classification. Soft Comput. 2022, 26, 9665–9687. [Google Scholar] [CrossRef]
- Tubishat, M.; Rawshdeh, Z.; Jarrah, H.; Elgamal, Z.M.; Elnagar, A.; Alrashdan, M.T. Dynamic generalized normal distribution optimization for feature selection. Neural Comput. Appl. 2022, 34, 17355–17370. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R.; Mohd Saad, N.; Mohd Ali, N.; Tee, W. A New Competitive Binary Grey Wolf Optimizer to Solve the Feature Selection Problem in EMG Signals Classification. Computers 2018, 7, 58. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Ehrlich, R.; Full, W. FCM: The fuzzy c-means clustering algorithm. Comput. Geosci. 1984, 10, 191–203. [Google Scholar] [CrossRef]
- Anter, A.M.; Ali, M. Feature selection strategy based on hybrid crow search optimization algorithm integrated with chaos theory and fuzzy c-means algorithm for medical diagnosis problems. Soft Comput. 2020, 24, 1565–1584. [Google Scholar] [CrossRef]
- Qiu, C.; Xiang, F. Feature selection using a set based discrete particle swarm optimization and a novel feature subset evaluation criterion. Intell. Data Anal. 2019, 23, 5–21. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Wang, J. A neurodynamic optimization approach to supervised feature selection via fractional programming. Neural Netw. 2021, 136, 194–206. [Google Scholar] [CrossRef]
- Halim, Z.; Yousaf, M.N.; Waqas, M.; Sulaiman, M.; Abbas, G.; Hussain, M.; Ahmad, I.; Hanif, M. An effective genetic algorithm-based feature selection method for intrusion detection systems. Comput. Secur. 2021, 110, 102448. [Google Scholar] [CrossRef]
- Hanbay, K. A new standard error based artificial bee colony algorithm and its applications in feature selection. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 4554–4567. [Google Scholar] [CrossRef]
- Liang, J.; Ma, M. FS-MOEA: A Novel Feature Selection Algorithm for IDSs in Vehicular Networks. IEEE Trans. Intell. Transp. Syst. 2022, 23, 368–382. [Google Scholar] [CrossRef]
- Benkessirat, A.; Benblidia, N. A novel feature selection approach based on constrained eigenvalues optimization. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 4836–4846. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Pal, N.R. Supervised Feature Selection via Collaborative Neurodynamic Optimization. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–15. [Google Scholar] [CrossRef]
- Moghaddam, A.A.; Seifi, A.; Niknam, T.; Alizadeh Pahlavani, M.R. Multi-objective operation management of a renewable MG (micro-grid) with back-up micro-turbine/fuel cell/battery hybrid power source. Energy 2011, 36, 6490–6507. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, S.; Shi, Y.; wei Gong, D.; Zhao, X. Cost-sensitive feature selection using two-archive multi-objective artificial bee colony algorithm. Expert Syst. Appl. 2019, 137, 46–58. [Google Scholar] [CrossRef]
- Asilian Bidgoli, A.; Ebrahimpour-komleh, H.; Rahnamayan, S. A novel binary many-objective feature selection algorithm for multi-label data classification. Int. J. Mach. Learn. Cybern. 2021, 12, 2041–2057. [Google Scholar] [CrossRef]
- Xu, H.; Xue, B.; Zhang, M. A Duplication Analysis-Based Evolutionary Algorithm for Biobjective Feature Selection. IEEE Trans. Evol. Comput. 2021, 25, 205–218. [Google Scholar] [CrossRef]
- Han, F.; Chen, W.T.; Ling, Q.H.; Han, H. Multi-objective particle swarm optimization with adaptive strategies for feature selection. Swarm Evol. Comput. 2021, 62, 100847. [Google Scholar] [CrossRef]
- Niu, B.; Yi, W.; Tan, L.; Geng, S.; Wang, H. A multi-objective feature selection method based on bacterial foraging optimization. Nat. Comput. 2021, 20, 63–76. [Google Scholar] [CrossRef]
- Li, T.; Zhan, Z.H.; Xu, J.C.; Yang, Q.; Ma, Y.Y. A binary individual search strategy-based bi-objective evolutionary algorithm for high-dimensional feature selection. Inf. Sci. 2022, 610, 651–673. [Google Scholar] [CrossRef]
- Pan, J.S.; Liu, N.; Chu, S.C. A competitive mechanism based multi-objective differential evolution algorithm and its application in feature selection. Knowl.-Based Syst. 2022, 245, 108582. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, D.; Jiang, L.; Ma, H. A particle swarm optimization based multiobjective memetic algorithm for high-dimensional feature selection. Memetic Comput. 2022, 14, 77–93. [Google Scholar] [CrossRef]
- Hosseini, F.; Gharehchopogh, F.S.; Masdari, M. MOAEOSCA: An enhanced multi-objective hybrid artificial ecosystem-based optimization with sine cosine algorithm for feature selection in botnet detection in IoT. Multimed. Tools Appl. 2023, 82, 13369–13399. [Google Scholar] [CrossRef]
- Al-Tashi, Q.; Abdulkadir, S.J.; Rais, H.M.; Mirjalili, S.; Alhussian, H.; Ragab, M.G.; Alqushaibi, A. Binary Multi-Objective Grey Wolf Optimizer for Feature Selection in Classification. IEEE Access 2020, 8, 106247–106263. [Google Scholar] [CrossRef]
- Usman, A.M.; Yusof, U.K.; Naim, S. Filter-Based Multi-Objective Feature Selection Using NSGA III and Cuckoo Optimization Algorithm. IEEE Access 2020, 8, 76333–76356. [Google Scholar] [CrossRef]
- Xue, Y.; Zhu, H.; Liang, J.; Słowik, A. Adaptive crossover operator based multi-objective binary genetic algorithm for feature selection in classification. Knowl.-Based Syst. 2021, 227, 107218. [Google Scholar] [CrossRef]
- Li, W.; Chai, Z.; Tang, Z. A decomposition-based multi-objective immune algorithm for feature selection in learning to rank. Knowl.-Based Syst. 2021, 234, 107577. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Z.; Liu, X.; Chen, W.T.; Horng, D.J.; Lai, K.K. Feature selection based on a large-scale many-objective evolutionary algorithm. Comput. Intell. Neurosci. 2021, 2021, 9961727. [Google Scholar] [CrossRef] [PubMed]
- Ishibuchi, H.; Murata, T. A multi-objective genetic local search algorithm and its application to flowshop scheduling. IEEE Trans. Syst. Man Cybern. Part (Appl. Rev.) 1998, 28, 392–403. [Google Scholar] [CrossRef]
- Qiu, C. A novel multi-swarm particle swarm optimization for feature selection. Genet. Program. Evolvable Mach. 2019, 20, 503–529. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R.; Mohd Saad, N. A New Quadratic Binary Harris Hawk Optimization for Feature Selection. Electronics 2019, 8, 1130. [Google Scholar] [CrossRef]
- Hodashinsky, I.A.; Sarin, K.S. Feature selection: Comparative analysis of binary metaheuristics and population based algorithm with adaptive memory. Program. Comput. Softw. 2019, 45, 221–227. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R.; Mohd Saad, N. A New Co-Evolution Binary Particle Swarm Optimization with Multiple Inertia Weight Strategy for Feature Selection. Informatics 2019, 6, 21. [Google Scholar] [CrossRef]
- Hegazy, A.E.; Makhlouf, M.; El-Tawel, G.S. Feature selection using chaotic salp swarm algorithm for data classification. Arab. J. Sci. Eng. 2019, 44, 3801–3816. [Google Scholar] [CrossRef]
- Arora, S.; Anand, P. Binary butterfly optimization approaches for feature selection. Expert Syst. Appl. 2019, 116, 147–160. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R.; Mohd Saad, N. Hybrid binary particle swarm optimization differential evolution-based feature selection for EMG signals classification. Axioms 2019, 8, 79. [Google Scholar] [CrossRef]
- Tu, Q.; Chen, X.; Liu, X. Hierarchy Strengthened Grey Wolf Optimizer for Numerical Optimization and Feature Selection. IEEE Access 2019, 7, 78012–78028. [Google Scholar] [CrossRef]
- Jia, H.; Li, J.; Song, W.; Peng, X.; Lang, C.; Li, Y. Spotted Hyena Optimization Algorithm With Simulated Annealing for Feature Selection. IEEE Access 2019, 7, 71943–71962. [Google Scholar] [CrossRef]
- Jia, H.; Xing, Z.; Song, W. A New Hybrid Seagull Optimization Algorithm for Feature Selection. IEEE Access 2019, 7, 49614–49631. [Google Scholar] [CrossRef]
- Al-Tashi, Q.; Abdul Kadir, S.J.; Rais, H.M.; Mirjalili, S.; Alhussian, H. Binary Optimization Using Hybrid Grey Wolf Optimization for Feature Selection. IEEE Access 2019, 7, 39496–39508. [Google Scholar] [CrossRef]
- Arora, S.; Singh, H.; Sharma, M.; Sharma, S.; Anand, P. A New Hybrid Algorithm Based on Grey Wolf Optimization and Crow Search Algorithm for Unconstrained Function Optimization and Feature Selection. IEEE Access 2019, 7, 26343–26361. [Google Scholar] [CrossRef]
- Awadallah, M.A.; Al-Betar, M.A.; Hammouri, A.I.; Alomari, O.A. Binary JAYA algorithm with adaptive mutation for feature selection. Arab. J. Sci. Eng. 2020, 45, 10875–10890. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R. Chaotic atom search optimization for feature selection. Arab. J. Sci. Eng. 2020, 45, 6063–6079. [Google Scholar] [CrossRef]
- Baş, E.; Ülker, E. An efficient binary social spider algorithm for feature selection problem. Expert Syst. Appl. 2020, 146, 113185. [Google Scholar] [CrossRef]
- Hans, R.; Kaur, H. Hybrid binary Sine Cosine Algorithm and Ant Lion Optimization (SCALO) approaches for feature selection problem. Int. J. Comput. Mater. Sci. Eng. 2020, 9, 1950021. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, Y.; Yu, C.; Heidari, A.A.; Li, S.; Chen, H.; Li, C. Gaussian mutational chaotic fruit fly-built optimization and feature selection. Expert Syst. Appl. 2020, 141, 112976. [Google Scholar] [CrossRef]
- Tawhid, M.A.; Ibrahim, A.M. Feature selection based on rough set approach, wrapper approach, and binary whale optimization algorithm. Int. J. Mach. Learn. Cybern. 2020, 11, 573–602. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; El-Shahat, D.; El-henawy, I.; de Albuquerque, V.; Mirjalili, S. A new fusion of grey wolf optimizer algorithm with a two-phase mutation for feature selection. Expert Syst. Appl. 2020, 139, 112824. [Google Scholar] [CrossRef]
- Nadimi-Shahraki, M.H.; Banaie-Dezfouli, M.; Zamani, H.; Taghian, S.; Mirjalili, S. B-MFO: A binary moth-flame optimization for feature selection from medical datasets. Computers 2021, 10, 136. [Google Scholar] [CrossRef]
- Chantar, H.; Tubishat, M.; Essgaer, M.; Mirjalili, S. Hybrid binary dragonfly algorithm with simulated annealing for feature selection. SN Comput. Sci. 2021, 2, 295. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gao, Y.; Gao, S.; Yong, X. A new feature selection method based on a self-variant genetic algorithm applied to android malware detection. Symmetry 2021, 13, 1290. [Google Scholar] [CrossRef]
- Elgamal, Z.M.; Yasin, N.M.; Sabri, A.Q.M.; Sihwail, R.; Tubishat, M.; Jarrah, H. Improved equilibrium optimization algorithm using elite opposition-based learning and new local search strategy for feature selection in medical datasets. Computation 2021, 9, 68. [Google Scholar] [CrossRef]
- Mostert, W.; Malan, K.M.; Engelbrecht, A.P. A feature selection algorithm performance metric for comparative analysis. Algorithms 2021, 14, 100. [Google Scholar] [CrossRef]
- Kitonyi, P.M.; Segera, D.R. Hybrid gradient descent grey wolf optimizer for optimal feature selection. BioMed Res. Int. 2021, 2021, 2555622. [Google Scholar] [CrossRef]
- Elminaam, D.S.A.; Nabil, A.; Ibraheem, S.A.; Houssein, E.H. An Efficient Marine Predators Algorithm for Feature Selection. IEEE Access 2021, 9, 60136–60153. [Google Scholar] [CrossRef]
- Assiri, A.S. On the performance improvement of Butterfly Optimization approaches for global optimization and Feature Selection. PLoS ONE 2021, 16, e0242612. [Google Scholar] [CrossRef]
- Liu, W.; Wang, J. Recursive elimination–election algorithms for wrapper feature selection. Appl. Soft Comput. 2021, 113, 107956. [Google Scholar] [CrossRef]
- Too, J.; Abdullah, A.R. Opposition based competitive grey wolf optimizer for EMG feature selection. Evol. Intell. 2021, 14, 1691–1705. [Google Scholar] [CrossRef]
- Mohmmadzadeh, H.; Gharehchopogh, F.S. An efficient binary chaotic symbiotic organisms search algorithm approaches for feature selection problems. J. Supercomput. 2021, 77, 9102–9144. [Google Scholar] [CrossRef]
- Al-Betar, M.A.; Hammouri, A.I.; Awadallah, M.A.; Abu Doush, I. Binary β-hill climbing optimizer with S-shape transfer function for feature selection. J. Ambient Intell. Humaniz. Comput. 2021, 12, 7637–7665. [Google Scholar] [CrossRef]
- Agrawal, P.; Ganesh, T.; Mohamed, A.W. A novel binary gaining–sharing knowledge-based optimization algorithm for feature selection. Neural Comput. Appl. 2021, 33, 5989–6008. [Google Scholar] [CrossRef]
- Long, W.; Jiao, J.; Liang, X.; Wu, T.; Xu, M.; Cai, S. Pinhole-imaging-based learning butterfly optimization algorithm for global optimization and feature selection. Appl. Soft Comput. 2021, 103, 107146. [Google Scholar] [CrossRef]
- Chaudhuri, A.; Sahu, T.P. Feature selection using Binary Crow Search Algorithm with time varying flight length. Expert Syst. Appl. 2021, 168, 114288. [Google Scholar] [CrossRef]
- Jaddi, N.S.; Abdullah, S.; Nazri, M.Z.A. A Recurrence Population-based Great Deluge Algorithm with Independent Quality Estimation for Feature Selection from Academician Data. Appl. Artif. Intell. 2021, 35, 1081–1105. [Google Scholar] [CrossRef]
- Akinola, O.A.; Ezugwu, A.E.; Oyelade, O.N.; Agushaka, J.O. A hybrid binary dwarf mongoose optimization algorithm with simulated annealing for feature selection on high dimensional multi-class datasets. Sci. Rep. 2022, 12, 14945. [Google Scholar] [CrossRef]
- Alsmadi, M.K.; Alzaqebah, M.; Jawarneh, S.; Brini, S.; Al-Marashdeh, I.; Briki, K.; Alrefai, N.; Alghamdi, F.A.; Al-Rashdan, M.T. Cuckoo algorithm with great deluge local-search for feature selection problems. Int. J. Electr. Comput. Eng. (2088-8708) 2022, 12, 4315–4326. [Google Scholar]
- Feng, J.; Kuang, H.; Zhang, L. EBBA: An enhanced binary bat algorithm integrated with chaos theory and lévy flight for feature selection. Future Internet 2022, 14, 178. [Google Scholar] [CrossRef]
- Hichem, H.; Elkamel, M.; Rafik, M.; Mesaaoud, M.T.; Ouahiba, C. A new binary grasshopper optimization algorithm for feature selection problem. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 316–328. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, T.; Li, W. An Efficient Hybrid Feature Selection Method Using the Artificial Immune Algorithm for High-Dimensional Data. Comput. Intell. Neurosci. 2022, 2022, 1452301. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, S.; Yin, D.; Chen, D.; Dong, X. Two-stage feature selection for classification of gene expression data based on an improved Salp Swarm Algorithm. Math. Biosci. Eng. 2022, 19, 13747–13781. [Google Scholar] [CrossRef]
- Keleş, M.K.; Kiliç, Ü. Binary Black Widow Optimization Approach for Feature Selection. IEEE Access 2022, 10, 95936–95948. [Google Scholar] [CrossRef]
- Singh, H.; Sharma, S.; Khurana, M.; Kaur, M.; Lee, H.N. Binary Drone Squadron Optimization Approaches for Feature Selection. IEEE Access 2022, 10, 87099–87114. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Zhang, C.; Zhou, C. Multiobjective Harris Hawks Optimization With Associative Learning and Chaotic Local Search for Feature Selection. IEEE Access 2022, 10, 72973–72987. [Google Scholar] [CrossRef]
- Elgamal, Z.; Sabri, A.Q.M.; Tubishat, M.; Tbaishat, D.; Makhadmeh, S.N.; Alomari, O.A. Improved Reptile Search Optimization Algorithm Using Chaotic Map and Simulated Annealing for Feature Selection in Medical Field. IEEE Access 2022, 10, 51428–51446. [Google Scholar] [CrossRef]
- Takieldeen, A.E.; El-kenawy, E.S.M.; Hadwan, M.; Zaki, R.M. Dipper Throated Optimization Algorithm for Unconstrained Function and Feature Selection. Comput. Mater. Contin. 2022, 72, 1465–1481. [Google Scholar] [CrossRef]
- Kalra, M.; Kumar, V.; Kaur, M.; Idris, S.A.; Öztürk, Ş.; Alshazly, H. A Novel Binary Emperor Penguin Optimizer for Feature Selection Tasks. Comput. Mater. Contin. 2022, 70, 6239–6255. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Dhanalakshmi, R.; Seetharaman, G. S-shaped and V-shaped binary African vulture optimization algorithm for feature selection. Expert Syst. 2022, 39, e13079. [Google Scholar] [CrossRef]
- Too, J.; Liang, G.; Chen, H. Memory-based Harris hawk optimization with learning agents: A feature selection approach. Eng. Comput. 2022, 38, 4457–4478. [Google Scholar] [CrossRef]
- Agrawal, U.; Rohatgi, V.; Katarya, R. Normalized Mutual Information-based equilibrium optimizer with chaotic maps for wrapper-filter feature selection. Expert Syst. Appl. 2022, 207, 118107. [Google Scholar] [CrossRef]
- Isuwa, J.; Abdullahi, M.; Sahabi Ali, Y.; Abdulrahim, A. Hybrid particle swarm optimization with sequential one point flipping algorithm for feature selection. Concurr. Comput. Pract. Exp. 2022, 34, e7239. [Google Scholar] [CrossRef]
- Preeti; Deep, K. A random walk Grey wolf optimizer based on dispersion factor for feature selection on chronic disease prediction. Expert Syst. Appl. 2022, 206, 117864. [Google Scholar] [CrossRef]
- Khosravi, H.; Amiri, B.; Yazdanjue, N.; Babaiyan, V. An improved group teaching optimization algorithm based on local search and chaotic map for feature selection in high-dimensional data. Expert Syst. Appl. 2022, 204, 117493. [Google Scholar] [CrossRef]
- Samieiyan, B.; MohammadiNasab, P.; Mollaei, M.A.; Hajizadeh, F.; Kangavari, M. Novel optimized crow search algorithm for feature selection. Expert Syst. Appl. 2022, 204, 117486. [Google Scholar] [CrossRef]
- Beheshti, Z. BMPA-TVSinV: A Binary Marine Predators Algorithm using time-varying sine and V-shaped transfer functions for wrapper-based feature selection. Knowl.-Based Syst. 2022, 252, 109446. [Google Scholar] [CrossRef]
- Asghari Varzaneh, Z.; Hossein, S.; Ebrahimi Mood, S.; Javidi, M.M. A new hybrid feature selection based on Improved Equilibrium Optimization. Chemom. Intell. Lab. Syst. 2022, 228, 104618. [Google Scholar] [CrossRef]
- Nadimi-Shahraki, M.H.; Zamani, H.; Mirjalili, S. Enhanced whale optimization algorithm for medical feature selection: A COVID-19 case study. Comput. Biol. Med. 2022, 148, 105858. [Google Scholar] [CrossRef]
- Hu, J.; Heidari, A.A.; Zhang, L.; Xue, X.; Gui, W.; Chen, H.; Pan, Z. Chaotic diffusion-limited aggregation enhanced grey wolf optimizer: Insights, analysis, binarization, and feature selection. Int. J. Intell. Syst. 2022, 37, 4864–4927. [Google Scholar] [CrossRef]
- Bacanin, N.; Bezdan, T.; Al-Turjman, F.; Rashid, T.A. Artificial Flora Optimization Algorithm with Genetically Guided Operators for Feature Selection and Neural Network Training. Int. J. Fuzzy Syst. 2022, 24, 2538–2559. [Google Scholar] [CrossRef]
- Bezdan, T.; Zivkovic, M.; Bacanin, N.; Chhabra, A.; Suresh, M. Feature Selection by Hybrid Brain Storm Optimization Algorithm for COVID-19 Classification. J. Comput. Biol. 2022, 29, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Qaraad, M.; Amjad, S.; Hussein, N.K.; Elhosseini, M.A. Large scale salp-based grey wolf optimization for feature selection and global optimization. Neural Comput. Appl. 2022, 34, 8989–9014. [Google Scholar] [CrossRef]
- Thaher, T.; Chantar, H.; Too, J.; Mafarja, M.; Turabieh, H.; Houssein, E.H. Boolean Particle Swarm Optimization with various Evolutionary Population Dynamics approaches for feature selection problems. Expert Syst. Appl. 2022, 195, 116550. [Google Scholar] [CrossRef]
- Hu, G.; Du, B.; Wang, X.; Wei, G. An enhanced black widow optimization algorithm for feature selection. Knowl.-Based Syst. 2022, 235, 107638. [Google Scholar] [CrossRef]
- Wang, X.; Dong, X.; Zhang, Y.; Chen, H. Crisscross Harris Hawks Optimizer for Global Tasks and Feature Selection. J. Bionic Eng. 2023, 20, 1153–1174. [Google Scholar] [CrossRef]
- Balakrishnan, K.; Dhanalakshmi, R.; Akila, M.; Sinha, B.B. Improved equilibrium optimization based on Levy flight approach for feature selection. Evol. Syst. 2023, 14, 735–746. [Google Scholar] [CrossRef]
- Agrawal, P.; Ganesh, T.; Oliva, D.; Mohamed, A.W. S-shaped and V-shaped gaining-sharing knowledge-based algorithm for feature selection. Appl. Intell. 2022, 52, 81–112. [Google Scholar] [CrossRef]
- Chhabra, A.; Hussien, A.G.; Hashim, F.A. Improved bald eagle search algorithm for global optimization and feature selection. Alex. Eng. J. 2023, 68, 141–180. [Google Scholar] [CrossRef]
- Oyelade, O.N.; Agushaka, J.O.; Ezugwu, A.E. Evolutionary binary feature selection using adaptive ebola optimization search algorithm for high-dimensional datasets. PLoS ONE 2023, 18, e0282812. [Google Scholar] [CrossRef] [PubMed]
- Ewees, A.A.; Ismail, F.H.; Sahlol, A.T. Gradient-based optimizer improved by Slime Mould Algorithm for global optimization and feature selection for diverse computation problems. Expert Syst. Appl. 2023, 213, 118872. [Google Scholar] [CrossRef]
- Devi, R.M.; Premkumar, M.; Kiruthiga, G.; Sowmya, R. IGJO: An Improved Golden Jackel Optimization Algorithm Using Local Escaping Operator for Feature Selection Problems. Neural Process. Lett. 2023, 55, 6443–6531. [Google Scholar] [CrossRef]
- Yu, W.; Kang, H.; Sun, G.; Liang, S.; Li, J. Bio-Inspired Feature Selection in Brain Disease Detection via an Improved Sparrow Search Algorithm. IEEE Trans. Instrum. Meas. 2023, 72, 1–15. [Google Scholar] [CrossRef]
- Yong, X.; Gao, Y.l. Improved firefly algorithm for feature selection with the ReliefF-based initialization and the weighted voting mechanism. Neural Comput. Appl. 2023, 35, 275–301. [Google Scholar] [CrossRef]
- Shaddeli, A.; Gharehchopogh, F.S.; Masdari, M.; Solouk, V. BFRA: A New Binary Hyper-Heuristics Feature Ranks Algorithm for Feature Selection in High-Dimensional Classification Data. Int. J. Inf. Technol. Decis. Mak. 2023, 22, 471–536. [Google Scholar] [CrossRef]
- Tanha, J.; Zarei, Z. The Bombus-terrestris bee optimization algorithm for feature selection. Appl. Intell. 2023, 53, 470–490. [Google Scholar] [CrossRef]
- Tubishat, M.; Alswaitti, M.; Mirjalili, S.; Al-Garadi, M.A.; Alrashdan, M.T.; Rana, T.A. Dynamic Butterfly Optimization Algorithm for Feature Selection. IEEE Access 2020, 8, 194303–194314. [Google Scholar] [CrossRef]
- Zhong, C.; Chen, Y.; Peng, J. Feature Selection Based on a Novel Improved Tree Growth Algorithm. Int. J. Comput. Intell. Syst. 2020, 13, 247–258. [Google Scholar] [CrossRef]
- Ji, B.; Lu, X.; Sun, G.; Zhang, W.; Li, J.; Xiao, Y. Bio-Inspired Feature Selection: An Improved Binary Particle Swarm Optimization Approach. IEEE Access 2020, 8, 85989–86002. [Google Scholar] [CrossRef]
- Bhattacharyya, T.; Chatterjee, B.; Singh, P.K.; Yoon, J.H.; Geem, Z.W.; Sarkar, R. Mayfly in Harmony: A New Hybrid Meta-Heuristic Feature Selection Algorithm. IEEE Access 2020, 8, 195929–195945. [Google Scholar] [CrossRef]
- Tawhid, M.A.; Dsouza, K.B. Hybrid binary dragonfly enhanced particle swarm optimization algorithm for solving feature selection problems. Math. Found. Comput. 2018, 1, 181–200. [Google Scholar] [CrossRef]
- Pichai, S.; Sunat, K.; Chiewchanwattana, S. An Asymmetric Chaotic Competitive Swarm Optimization Algorithm for Feature Selection in High-Dimensional Data. Symmetry 2020, 12, 1782. [Google Scholar] [CrossRef]
- El-Kenawy, E.S.M.; Eid, M.M.; Saber, M.; Ibrahim, A. MbGWO-SFS: Modified Binary Grey Wolf Optimizer Based on Stochastic Fractal Search for Feature Selection. IEEE Access 2020, 8, 107635–107649. [Google Scholar] [CrossRef]
- Li, J.; Kang, H.; Sun, G.; Feng, T.; Li, W.; Zhang, W.; Ji, B. IBDA: Improved Binary Dragonfly Algorithm With Evolutionary Population Dynamics and Adaptive Crossover for Feature Selection. IEEE Access 2020, 8, 108032–108051. [Google Scholar] [CrossRef]
- Elgamal, Z.M.; Yasin, N.B.M.; Tubishat, M.; Alswaitti, M.; Mirjalili, S. An Improved Harris Hawks Optimization Algorithm With Simulated Annealing for Feature Selection in the Medical Field. IEEE Access 2020, 8, 186638–186652. [Google Scholar] [CrossRef]
- Wu, H.; Du, S.; Zhang, Y.; Zhang, Q.; Duan, K.; Lin, Y. Threshold Binary Grey Wolf Optimizer Based on Multi-Elite Interaction for Feature Selection. IEEE Access 2023, 11, 34332–34348. [Google Scholar] [CrossRef]
- Adamu, A.; Abdullahi, M.; Junaidu, S.B.; Hassan, I.H. An hybrid particle swarm optimization with crow search algorithm for feature selection. Mach. Learn. Appl. 2021, 6, 100108. [Google Scholar] [CrossRef]
- Ewees, A.A.; El Aziz, M.A.; Hassanien, A.E. Chaotic multi-verse optimizer-based feature selection. Neural Comput. Appl. 2019, 31, 991–1006. [Google Scholar] [CrossRef]
- Sayed, G.I.; Hassanien, A.E.; Azar, A.T. Feature selection via a novel chaotic crow search algorithm. Neural Comput. Appl. 2019, 31, 171–188. [Google Scholar] [CrossRef]
- Sayed, G.I.; Tharwat, A.; Hassanien, A.E. Chaotic dragonfly algorithm: An improved metaheuristic algorithm for feature selection. Appl. Intell. 2019, 49, 188–205. [Google Scholar] [CrossRef]
- Manita, G.; Korbaa, O. Binary Political Optimizer for Feature Selection Using Gene Expression Data. Comput. Intell. Neurosci. 2020, 2020, 8896570. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; He, S.; Wang, L.; Zeng, W.; Yang, J. Feature Selection Using an Improved Gravitational Search Algorithm. IEEE Access 2019, 7, 114440–114448. [Google Scholar] [CrossRef]
- Ibrahim, R.A.; Abd Elaziz, M.; Oliva, D.; Lu, S. An improved runner-root algorithm for solving feature selection problems based on rough sets and neighborhood rough sets. Appl. Soft Comput. 2020, 97, 105517. [Google Scholar] [CrossRef]
- Ding, Y.; Zhou, K.; Bi, W. Feature selection based on hybridization of genetic algorithm and competitive swarm optimizer. Soft Comput. 2020, 24, 11663–11672. [Google Scholar] [CrossRef]
- Mandal, A.K.; Sen, R.; Chakraborty, B. Feature selection in classification using self-adaptive owl search optimization algorithm with elitism and mutation strategies. J. Intell. Fuzzy Syst. 2021, 40, 535–550. [Google Scholar] [CrossRef]
- Shen, C.; Zhang, K. Two-stage improved Grey Wolf optimization algorithm for feature selection on high-dimensional classification. Complex Intell. Syst. 2022, 8, 2769–2789. [Google Scholar] [CrossRef]
- Kundu, R.; Chattopadhyay, S.; Cuevas, E.; Sarkar, R. AltWOA: Altruistic Whale Optimization Algorithm for feature selection on microarray datasets. Comput. Biol. Med. 2022, 144, 105349. [Google Scholar] [CrossRef]
- Pashaei, E.; Pashaei, E. An efficient binary chimp optimization algorithm for feature selection in biomedical data classification. Neural Comput. Appl. 2022, 34, 6427–6451. [Google Scholar] [CrossRef]
- Segera, D.; Mbuthia, M.; Nyete, A. An Innovative Excited-ACS-IDGWO Algorithm for Optimal Biomedical Data Feature Selection. BioMed Res. Int. 2020, 2020, 8506365. [Google Scholar] [CrossRef]
- Xue, Y.; Tang, T.; Pang, W.; Liu, A.X. Self-adaptive parameter and strategy based particle swarm optimization for large-scale feature selection problems with multiple classifiers. Appl. Soft Comput. 2020, 88, 106031. [Google Scholar] [CrossRef]
- Ahn, G.; Hur, S. Efficient genetic algorithm for feature selection for early time series classification. Comput. Ind. Eng. 2020, 142, 106345. [Google Scholar] [CrossRef]
- Sadeghian, Z.; Akbari, E.; Nematzadeh, H. A hybrid feature selection method based on information theory and binary butterfly optimization algorithm. Eng. Appl. Artif. Intell. 2021, 97, 104079. [Google Scholar] [CrossRef]
- Adel, A.; Omar, N.; Abdullah, S.; Al-Shabi, A. Co-Operative Binary Bat Optimizer with Rough Set Reducts for Text Feature Selection. Appl. Sci. 2022, 12, 11296. [Google Scholar] [CrossRef]
- Anuprathibha, T.; Kanimozhiselvi, C. Penguin search optimization based feature selection for automated opinion mining. Int. J. Recent Technol. Eng. 2019, 8, 648–653. [Google Scholar] [CrossRef]
- Sharafi, Y.; Teshnehlab, M. Opposition-based binary competitive optimization algorithm using time-varying V-shape transfer function for feature selection. Neural Comput. Appl. 2021, 33, 17497–17533. [Google Scholar] [CrossRef]
- Long, W.; Jiao, J.; Wu, T.; Xu, M.; Cai, S. A balanced butterfly optimization algorithm for numerical optimization and feature selection. Soft Comput. 2022, 26, 11505–11523. [Google Scholar] [CrossRef]
- Bhadra, T.; Maulik, U. Unsupervised Feature Selection Using Iterative Shrinking and Expansion Algorithm. IEEE Trans. Emerg. Top. Comput. Intell. 2022, 6, 1453–1462. [Google Scholar] [CrossRef]
- Hashemi, A.; Joodaki, M.; Joodaki, N.Z.; Dowlatshahi, M.B. Ant colony optimization equipped with an ensemble of heuristics through multi-criteria decision making: A case study in ensemble feature selection. Appl. Soft Comput. 2022, 124, 109046. [Google Scholar] [CrossRef]
- Hussien, A.G.; Amin, M. A self-adaptive Harris Hawks optimization algorithm with opposition-based learning and chaotic local search strategy for global optimization and feature selection. Int. J. Mach. Learn. Cybern. 2022, 13, 309–336. [Google Scholar] [CrossRef]
- Karimi, F.; Dowlatshahi, M.B.; Hashemi, A. SemiACO: A semi-supervised feature selection based on ant colony optimization. Expert Syst. Appl. 2023, 214, 119130. [Google Scholar] [CrossRef]
- Durgam, R.; Devarakonda, N. A Quasi-Oppositional Based Flamingo Search Algorithm Integrated with Generalized Ring Crossover for Effective Feature Selection. IETE J. Res. 2023, 0, 1–17. [Google Scholar] [CrossRef]
- Azar, A.T.; Khan, Z.I.; Amin, S.U.; Fouad, K.M. Hybrid Global Optimization Algorithm for Feature Selection. Comput. Mater. Contin. 2023, 74, 2021–2037. [Google Scholar] [CrossRef]
- Li, Z.; Du, J.; Nie, B.; Xiong, W.; Xu, G.; Luo, J. A new two-stage hybrid feature selection algorithm and its application in Chinese medicine. Int. J. Mach. Learn. Cybern. 2022, 13, 1243–1264. [Google Scholar] [CrossRef]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Witten, D.; James, G. An Introduction to Statistical Learning with Applications in R; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Alpaydin, E. Introduction to Machine Learning; MIT Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Song, H.; Triguero, I.; Özcan, E. A review on the self and dual interactions between machine learning and optimisation. Prog. Artif. Intell. 2019, 8, 143–165. [Google Scholar] [CrossRef]
- Kennedy, J.; Eberhart, R. Particle swarm optimization. In Proceedings of the ICNN’95—International Conference on Neural Networks, Perth, WA, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey Wolf Optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Holland, J.H. Genetic algorithms. Sci. Am. 1992, 267, 66–73. [Google Scholar] [CrossRef]
- Crawford, B.; Soto, R.; Astorga, G.; García, J.; Castro, C.; Paredes, F. Putting continuous metaheuristics to work in binary search spaces. Complexity 2017, 2017, 8404231. [Google Scholar] [CrossRef]
- Coello Coello, C.A. Evolutionary algorithms for solving multi-objective problems. Genet. Program. Evolvable Mach. 2007, 8, 221–252. [Google Scholar]
- Li, J.; Cheng, K.; Wang, S.; Morstatter, F.; Trevino, R.P.; Tang, J.; Liu, H. Feature selection: A data perspective. ACM Comput. Surv. (CSUR) 2018, 50, 94. [Google Scholar] [CrossRef]
- Cios, K.; Kurgan, L.; Goodenday, L. SPECT Heart. UCI Machine Learning Repository. 2001. [Google Scholar] [CrossRef]
- Ilter, N.; Guvenir, H. Dermatology. UCI Machine Learning Repository. 1998. [Google Scholar] [CrossRef]
- Chapman, D.; Jain, A. Musk (Version 1). UCI Machine Learning Repository. 1994. [Google Scholar] [CrossRef]
- Wolberg, W.; Street, W.N.; Mangasarian, O. Breast Cancer Diagnosis and Prognosis via Linear Programming; Technical Report; University of Wisconsin–Madison: Madison, WI, USA, 1994. [Google Scholar]
- Wolberg, W. Breast Cancer Wisconsin (Original). UCI Machine Learning Repository. 1992. [Google Scholar] [CrossRef]
- Hong, Z.; Yang, J. Lung Cancer. UCI Machine Learning Repository. 1992. [Google Scholar] [CrossRef]
- Aeberhard, S.; Forina, M. Wine. UCI Machine Learning Repository. 1991. [Google Scholar] [CrossRef]
- Aha, D. Tic-Tac-Toe Endgame. UCI Machine Learning Repository. 1991. [Google Scholar] [CrossRef]
- Forsyth, R. Zoo. UCI Machine Learning Repository. 1990. [Google Scholar] [CrossRef]
- Sigillito, V.G.; Wing, S.P.; Hutton, L.V.; Baker, K.B. Classification of radar returns from the ionosphere using neural networks. Johns Hopkins APL Tech. Dig. 1989, 10, 262–266. [Google Scholar]
- Shapiro, A. Chess (King-Rook vs. King-Pawn). UCI Machine Learning Repository. 1989. [Google Scholar] [CrossRef]
- Breiman, L.; Stone, C. Waveform Database Generator (Version 2). UCI Machine Learning Repository. 1988. [Google Scholar] [CrossRef]
- Zwitter, M.; Soklic, M. Lymphography. UCI Machine Learning Repository. 1988. [Google Scholar] [CrossRef]
- Congressional Voting Records. UCI Machine Learning Repository. 1987. [CrossRef]
- Sejnowski, T.; Gorman, R. Connectionist Bench (Sonar, Mines vs. Rocks). UCI Machine Learning Repository. [CrossRef]
- Statlog (Heart). UCI Machine Learning Repository. [CrossRef]
- Lemus-Romani, J.; Becerra-Rozas, M.; Crawford, B.; Soto, R.; Cisternas-Caneo, F.; Vega, E.; Castillo, M.; Tapia, D.; Astorga, G.; Palma, W.; et al. A Novel Learning-Based Binarization Scheme Selector for Swarm Algorithms Solving Combinatorial Problems. Mathematics 2021, 9, 2887. [Google Scholar] [CrossRef]
- Crawford, B.; Soto, R.; Lemus-Romani, J.; Becerra-Rozas, M.; Lanza-Gutiérrez, J.M.; Caballé, N.; Castillo, M.; Tapia, D.; Cisternas-Caneo, F.; García, J.; et al. Q-Learnheuristics: Towards Data-Driven Balanced Metaheuristics. Mathematics 2021, 9, 1839. [Google Scholar] [CrossRef]
- Becerra-Rozas, M.; Lemus-Romani, J.; Cisternas-Caneo, F.; Crawford, B.; Soto, R.; García, J. Swarm-Inspired Computing to Solve Binary Optimization Problems: A Backward Q-Learning Binarization Scheme Selector. Mathematics 2022, 10, 4776. [Google Scholar] [CrossRef]
- Becerra-Rozas, M.; Cisternas-Caneo, F.; Crawford, B.; Soto, R.; García, J.; Astorga, G.; Palma, W. Embedded Learning Approaches in the Whale Optimizer to Solve Coverage Combinatorial Problems. Mathematics 2022, 10, 4529. [Google Scholar] [CrossRef]
- Lemus-Romani, J.; Crawford, B.; Cisternas-Caneo, F.; Soto, R.; Becerra-Rozas, M. Binarization of Metaheuristics: Is the Transfer Function Really Important? Biomimetics 2023, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- García, J.; Moraga, P.; Crawford, B.; Soto, R.; Pinto, H. Binarization Technique Comparisons of Swarm Intelligence Algorithm: An Application to the Multi-Demand Multidimensional Knapsack Problem. Mathematics 2022, 10, 3183. [Google Scholar] [CrossRef]
- García, J.; Leiva-Araos, A.; Crawford, B.; Soto, R.; Pinto, H. Exploring Initialization Strategies for Metaheuristic Optimization: Case Study of the Set-Union Knapsack Problem. Mathematics 2023, 11, 2695. [Google Scholar] [CrossRef]
- Figueroa-Torrez, P.; Durán, O.; Crawford, B.; Cisternas-Caneo, F. A Binary Black Widow Optimization Algorithm for Addressing the Cell Formation Problem Involving Alternative Routes and Machine Reliability. Mathematics 2023, 11, 3475. [Google Scholar] [CrossRef]



























| Paper | Year | Objective Function | Evaluation Metrics | Optimization Techniques | Classifier | Benchmark Application | Real-Word Application |
|---|---|---|---|---|---|---|---|
| [4] | 2023 | ✓ | ✓ | ✓ | |||
| [5] | 2023 | ✓ | |||||
| [6] | 2023 | ✓ | ✓ | ✓ | ✓ | ||
| [7] | 2023 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| [8] | 2022 | ✓ | |||||
| [9] | 2022 | ✓ | ✓ | ✓ | ✓ | ||
| [10] | 2022 | ✓ | ✓ | ✓ | ✓ | ✓ | |
| [11] | 2022 | ✓ | ✓ | ✓ | |||
| [12] | 2022 | ✓ | ✓ | ||||
| [13] | 2021 | ✓ | ✓ | ✓ | |||
| [3] | 2021 | ✓ | ✓ | ✓ | ✓ | ||
| [14] | 2020 | ✓ | ✓ | ✓ | ✓ | ||
| Our Work | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
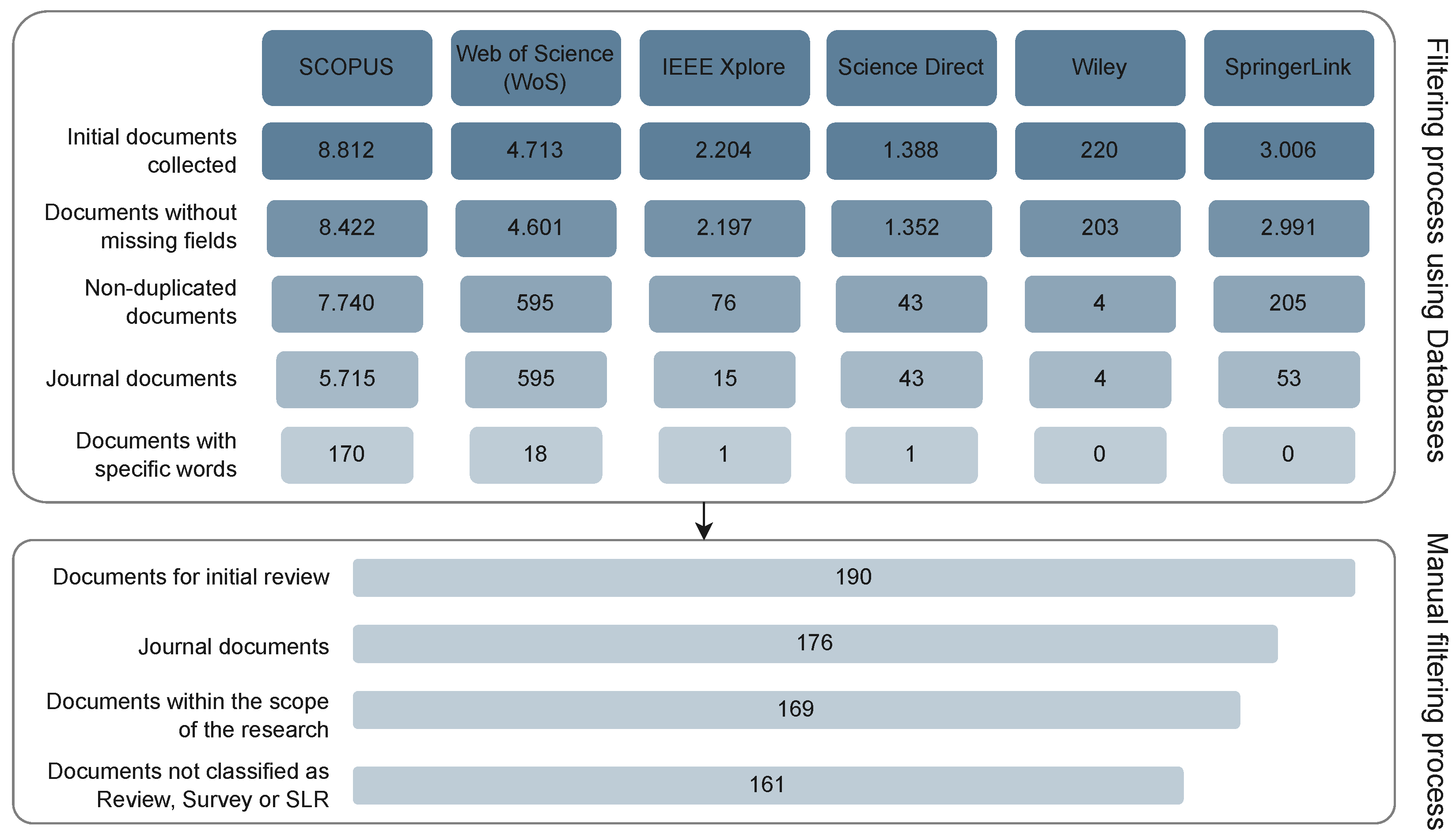
| Database | Query | #Result |
|---|---|---|
| • IEEE Xplore | (“Document Title”:“Feature Selection”) and Filters Applied: 2019–2023 | 2204 |
| • ScienceDirect by Elsevier | Title field: “Feature Selection” and Year field: “2019–2023” | 1388 |
| • Scopus | TITLE (“feature selection”) AND PUBYEAR > 2018 AND PUBYEAR < 2024 AND (LIMIT-TO (LANGUAGE, “English”) | 8812 |
| • SpringerLink | Title field: “Feature Selection” | 3006 |
| • Web of Sciences | (TI=(“feature selection”)) AND (DT==(“ARTICLE” OR “REVIEW”) AND LA==(“ENGLISH”) AND PY==(“2023” OR “2022” OR “2021” OR “2020” OR “2019”)) | 4713 |
| • Wiley | [Publication Title: “feature selection”] AND [Earliest: (01/01/2019 TO 04/20/2023)] | 220 |
| Predicted Negative | Predicted Positive | |
|---|---|---|
| Actual negative | TN | FP |
| Actual positive | FN | TP |
| Classifier | Description |
|---|---|
| Adaptive Boosting (ADABOOST) | Ensemble technique adjusting weights on misclassified instances for improved accuracy. |
| Artificial Neural Network (ANN) | Model inspired by the brain, with interconnected neurons for data processing. |
| Decision Tree (DT) | Divides data into branches by evaluating feature values, arriving at decisions at each internal node, and assigning class labels to leaf nodes. |
| Decision Tree C4.5 (DT C4.5) | Refined algorithm dividing data based on features, selecting attributes via info gain, handling varied types, missing values, and pruning. |
| Decision Tree J48 (DT J48) | Improved C4.5 in Weka, selects attributes with info gain, handles varied attributes, missing data, and pruning. |
| Discriminant Analysis (DA) | Technique finding linear combinations of features for class separation and dimensionality reduction. |
| Extreme Gradient Boosting (XGBOOST) | Boosting algorithm that builds strong learners by focusing on instances with poor previous learner performance. |
| Extreme Learning Machine (ELM) | Single-hidden-layer neural network that randomly assigns weights and determines output weights analytically. |
| Fuzzy Classifier (FC) | Classifier using fuzzy logic to handle uncertainty in data. |
| Fuzzy Min–Max Neuronal Network (FMM) | Fuzzy system for classification, handling uncertainty using membership functions. |
| Gaussian Naive Bayes (GNB) | Naive Bayes variation assuming Gaussian distribution of feature values. |
| Growing Hierarchical Self-Organizing Map (GHSOM) | Neural-network-based algorithm for clustering and visualization of high-dimensional data. |
| K-Nearest Neighbor (k-NN) | Assigns labels based on the majority class of k nearest neighbors. |
| Kernel Extreme Learning Machine (KELM) | ELM variant using kernel methods for nonlinear classification in high-dimensional space. |
| Kstar Classifier (KSTAR) | Lazy learning algorithm classifying new instances based on closest neighbors. |
| Latent Dirichlet Allocation (LDA) | Generative model used for topic modeling in text data, revealing hidden topic structures. |
| Light Gradient Boosting (LightGBM) | Gradient boosting with histogram-based training for efficiency and accuracy. |
| Logistic Model Tree (LMT) | Decision tree with leaf nodes containing logistic regression models. |
| Logistic Regression (LR) | Linear model estimating the probability of binary classification. |
| Multi-Label KNN (ML-KNN) | Extends k-NN for multi-label classification, allowing instances to have multiple labels. |
| Multi-Label Naive Bayes (MLNB) | Naive Bayes extension for multi-label classification problems. |
| Multilayer Perceptron (MLP) | Neural network with multiple layers for complex nonlinear mappings. |
| Naive Bayes (NB) | Probabilistic classifier based on Bayes’ theorem, assuming feature independence. |
| Oblique Random Forest Heterogeneous (OblRF(H)) | Variant of random forest using oblique splits for decision trees. |
| Optimum-Path Forest (OPF) | Pattern recognition algorithm constructing decision boundaries through graph-based approach. |
| Random Forest (RF) | Ensemble classifier that combines multiple decision trees to improve accuracy. |
| Standard Voting Classifier (SVC) | Ensemble technique combining classifier predictions through majority voting. |
| Support Vector Machine (SVM) | Finds a hyperplane to separate classes, maximizing the margin between them. |
| Year | Ref. | Algorithm | Focus | Innovation | Validation |
|---|---|---|---|---|---|
| 2023 | [70] | MOAEOSCA | Botnet detection in IoT opposition-based learning | Hybridization of AEO and SCA; bitwise operations | Achieved acceptable accuracy in Botnet detection in IoT |
| 2022 | [68] | CMODE | Multi-objective optimization and crowding distance | Rank based on non-dominated sorting optimization algorithms | Outperformed six state-of-the-art multi-objective algorithms |
| 2022 | [69] | PSOMMFS | High-dimensional feature selection adaptive local search | Information entropy-based initialization | Improved quality of Pareto front |
| 2022 | [120] | MOHHOAC | Feature selection using HHO chaotic local search | Associative learning; grey wolf optimization | Effective feature selection on sixteen UCI datasets |
| 2022 | [172] | BChOA | Biomedical data classification operator for enhanced exploration | Two binary variants of ChOA; crossover | Effective feature selection on biomedical datasets |
| 2021 | [63] | NSGA-III | Multi-label data feature selection maximizing feature-label correlation | Incorporation of additional objectives | Outperformed other algorithms on eight multi-label datasets |
| 2021 | [64] | DAEA | Bi-objective feature selection in classification diversity-based selection method | Duplication analysis method | Superior performance on 20 classification datasets |
| 2021 | [65] | MOPSO-ASFS | High-dimensional feature selection particle selection mechanism | Adaptive penalty mechanism; adaptive leading | Enhanced performance on high-dimensional datasets |
| 2021 | [66] | MOBIFS | Multi-objective feature selection roulette wheel mechanism | Bacterial foraging optimization algorithm | Effective removal of redundant features |
| 2021 | [73] | MOBGA-AOS | Feature selection as a pre-processing technique five crossover operators | Adaptive operator selection mechanism | Outperformed other evolutionary multi-objective algorithms |
| 2021 | [74] | MOIA/D-FSRank | Feature selection in L2R clonal selection and mutation operators | Tchebycheff decomposition; elite selection strategy | Significant improvements on public LETOR datasets |
| 2021 | [179] | OBCOOA | Wrapper-based feature selection; opposition-based learning mechanism | Time-varying V-shape transfer function | Applied to 27 benchmark datasets |
| 2020 | [23] | MOSCA_FS | Hyperspectral imagery feature selection Jeffries–Matusita distance and mutual information | Novel discrete SCA framework; ratio between | Tested on diverse datasets |
| 2020 | [71] | BMOGW | Feature selection | Multi-objective grey wolf optimizer | Effective feature selection with reduced classification error rates |
| 2020 | [72] | BCNSG3 & BCNSG2 | Multi-objective feature selection | Cuckoo optimization algorithm | Achieved non-dominated solutions with reduced error rates |
| 2020 | [175] | EGA | Early time-series classification mathematical model targeting classification performance | Emphasis on the starting time of classification | Outperformed a general genetic algorithm |
| 2019 | [62] | TMABC-FS | Cost-sensitive feature selection diversity-guiding searches; dual-archive system | Introduction of convergence and | Demonstrated robustness on UCI datasets |
| Year | Ref. | Description | Field | Instances | Features | Classes | Repository |
|---|---|---|---|---|---|---|---|
| 2018 | [199] | Colon | Medical | 62 | 2000 | 2 | ASU |
| 2001 | [200] | SPECT Heart | Medical | 267 | 22 | 2 | UCI |
| 1998 | [201] | Dermatology | Medical | 366 | 34 | 6 | UCI |
| 1994 | [202] | Musk (Version 1) | Chemistry | 476 | 166 | 2 | UCI |
| 1994 | [203] | Breast Cancer Wisconsin (Diagnostic) | Medical | 569 | 30 | 2 | UCI |
| 1992 | [204] | Breast Cancer Wisconsin (Original) | Medical | 699 | 9 | 2 | UCI |
| 1992 | [205] | Lung Cancer | Medical | 32 | 56 | 3 | UCI |
| 1991 | [206] | Wine | Biology/Chemistry | 178 | 13 | 3 | UCI |
| 1991 | [207] | Tic-Tac-Toe Endgame | Game | 958 | 9 | 2 | UCI |
| 1990 | [208] | Zoo | Biology | 101 | 16 | 7 | UCI |
| 1989 | [209] | Ionosphere | Physical Science | 351 | 34 | 2 | UCI |
| 1989 | [210] | Chess (King-Rook vs. King-Pawn) | Game | 3196 | 36 | 2 | UCI |
| 1988 | [211] | Waveform Database Generator (Version 2) | Synthetic | 5000 | 40 | 3 | UCI |
| 1988 | [212] | Lymphography | Medical | 148 | 18 | 4 | UCI |
| 1987 | [213] | Congressional Voting Records | Politics | 435 | 16 | 2 | UCI |
| - | [214] | Sonar | Physical Science | 208 | 60 | 2 | UCI |
| - | [215] | Statlog heart | Medical | 270 | 13 | 2 | UCI |
| - | n.a. | Exactly | n.d. | 1000 | 13 | 2 | UCI |
| - | n.a. | Exactly2 | n.d. | 1000 | 13 | 2 | UCI |
| - | n.a. | m-of-n | Biological | 1000 | 13 | 2 | UCI |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barrera-García, J.; Cisternas-Caneo, F.; Crawford, B.; Gómez Sánchez, M.; Soto, R. Feature Selection Problem and Metaheuristics: A Systematic Literature Review about Its Formulation, Evaluation and Applications. Biomimetics 2024, 9, 9. https://doi.org/10.3390/biomimetics9010009
Barrera-García J, Cisternas-Caneo F, Crawford B, Gómez Sánchez M, Soto R. Feature Selection Problem and Metaheuristics: A Systematic Literature Review about Its Formulation, Evaluation and Applications. Biomimetics. 2024; 9(1):9. https://doi.org/10.3390/biomimetics9010009
Chicago/Turabian StyleBarrera-García, José, Felipe Cisternas-Caneo, Broderick Crawford, Mariam Gómez Sánchez, and Ricardo Soto. 2024. "Feature Selection Problem and Metaheuristics: A Systematic Literature Review about Its Formulation, Evaluation and Applications" Biomimetics 9, no. 1: 9. https://doi.org/10.3390/biomimetics9010009
APA StyleBarrera-García, J., Cisternas-Caneo, F., Crawford, B., Gómez Sánchez, M., & Soto, R. (2024). Feature Selection Problem and Metaheuristics: A Systematic Literature Review about Its Formulation, Evaluation and Applications. Biomimetics, 9(1), 9. https://doi.org/10.3390/biomimetics9010009









