Biomineralization through a Symmetry-Controlled Oligomeric Peptide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis and Purification
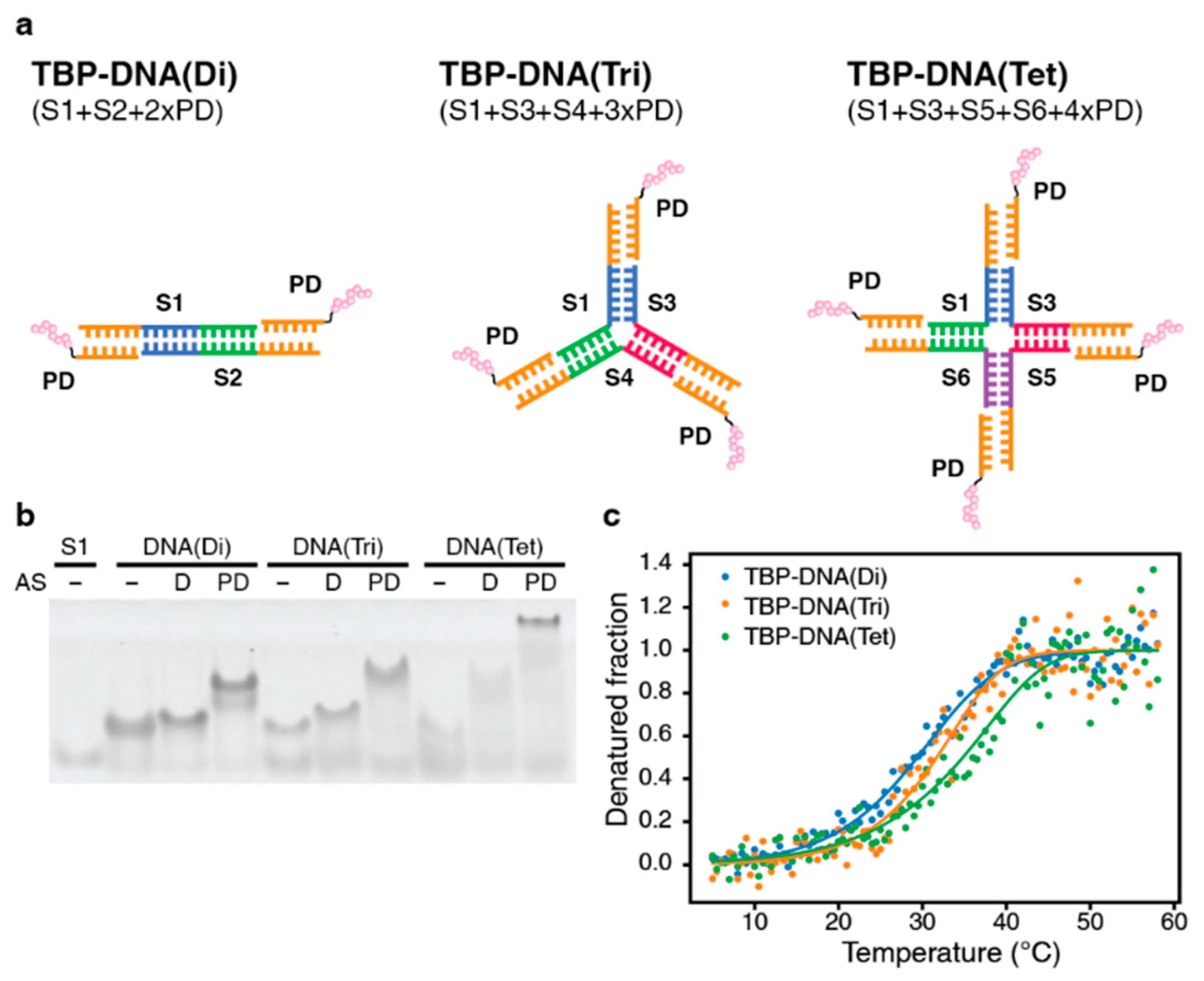
2.2. Synthesis and Purification of TBP–DNA Fragment and Oligomer Formation
2.3. Confirmation of TBP–DNA Structure Formation by Native-PAGE
2.4. Thermal Stability Analysis by Circular Dichroism (CD) Spectroscopy
2.5. Biomineralization Reaction
2.6. Electron Microscopy
3. Results
3.1. Design and Synthesis of the Oligomeric minTBP-1
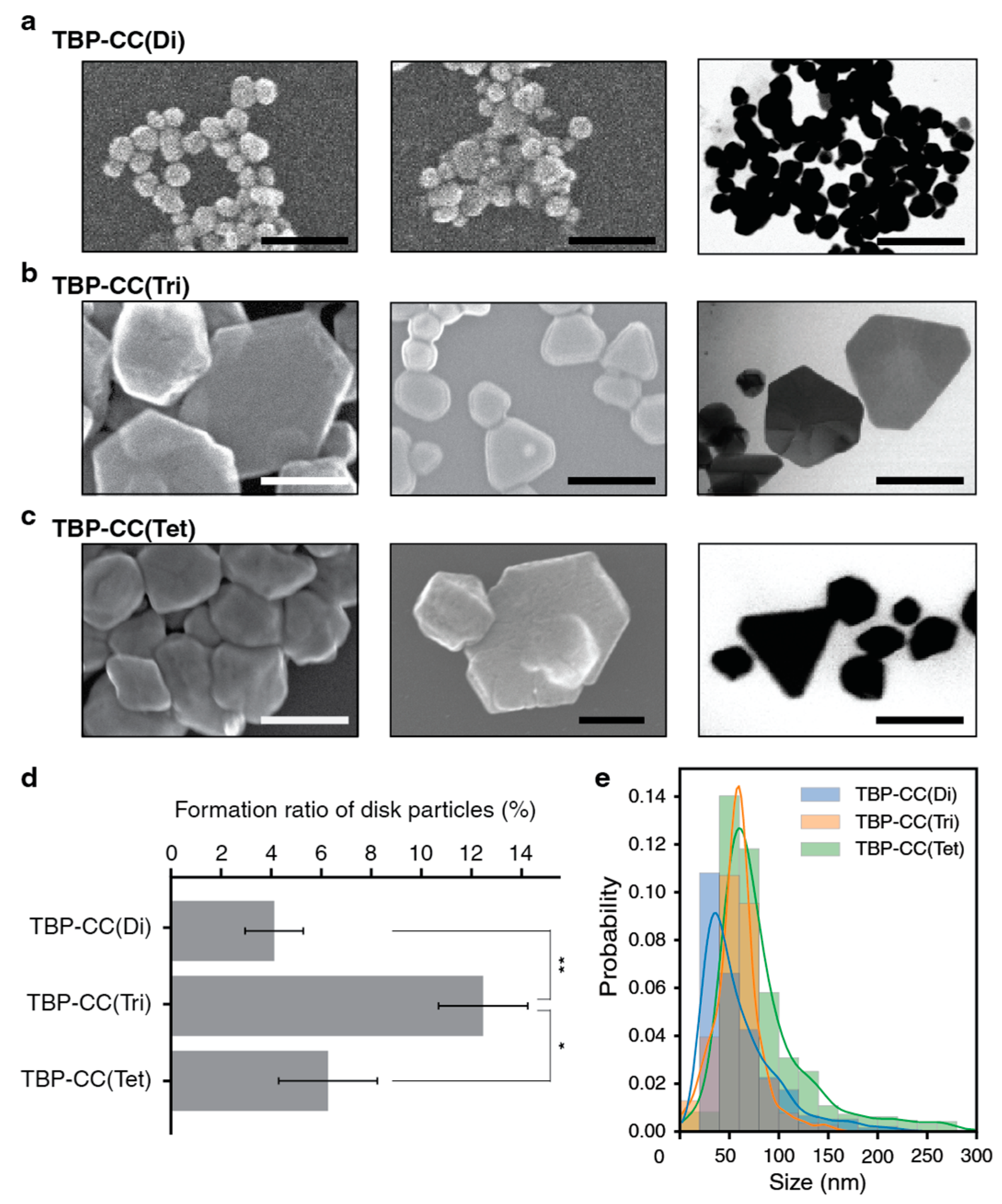
3.1.1. Coiled-Coil-Based Oligomer
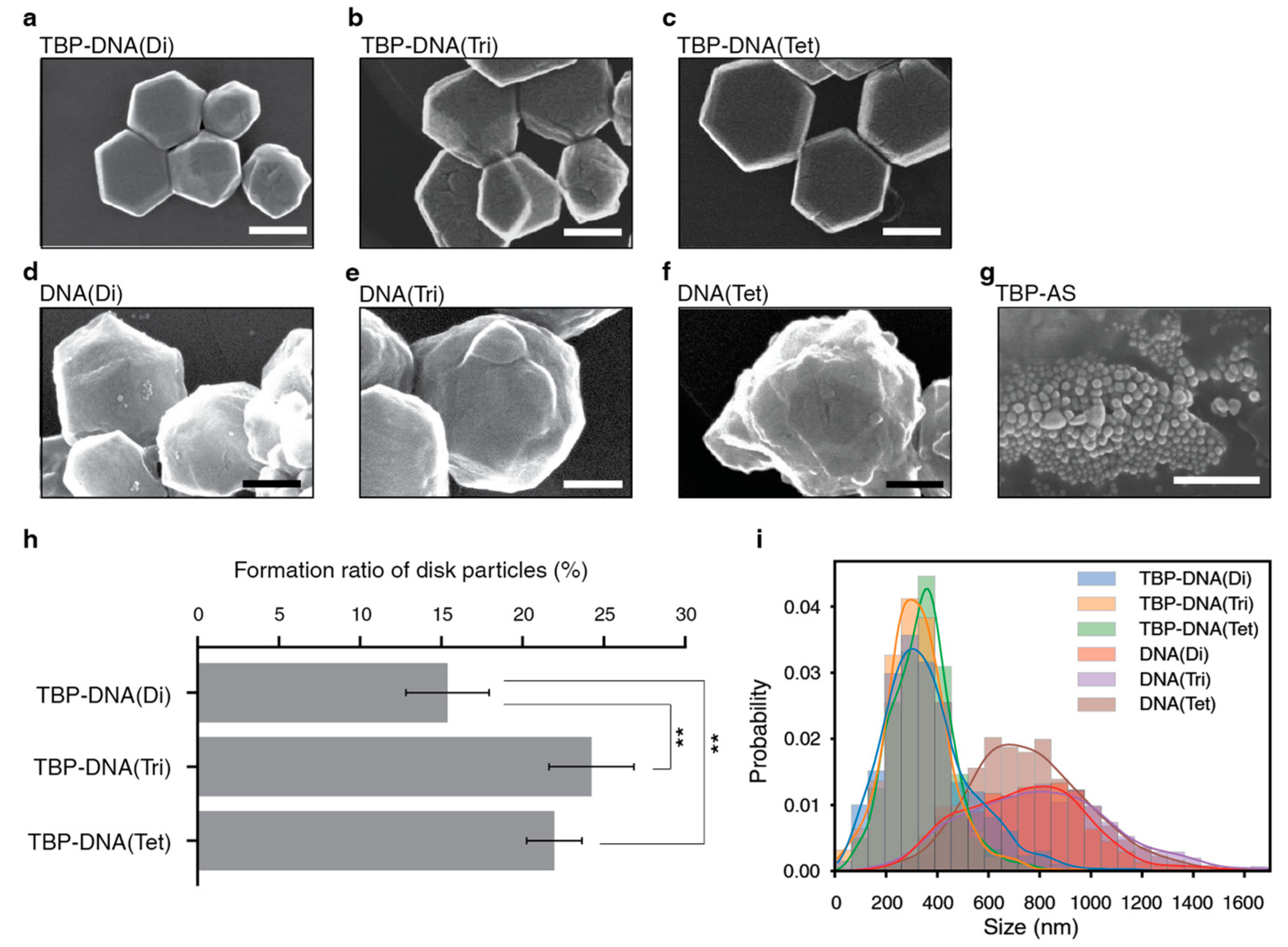
3.1.2. DNA-Based Oligomer
3.2. Silver Nanostructure Formation
3.2.1. Coiled-Coil-Based Oligomer
3.2.2. DNA-Based Oligomer
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Burda, C.; Chen, X.; Narayanan, R.; El-Sayed, M.A. Chemistry and Properties of Nanocrystals of Different Shapes. Chem. Rev. 2005, 105, 1025–1102. [Google Scholar] [CrossRef]
- Xia, Y.; Xiong, Y.; Lim, B.; Skrabalak, S.E. Shape-Controlled Synthesis of Metal Nanocrystals: Simple Chemistry Meets Complex Physics? Angew. Chem. Int. Ed. Engl. 2009, 48, 60–103. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.C. Size and Shape Dependent Second Order Nonlinear Optical Properties of Nanomaterials and Their Application in Biological and Chemical Sensing. Chem. Rev. 2010, 110, 5332–5365. [Google Scholar] [CrossRef] [PubMed]
- Albanese, A.; Tang, P.S.; Chan, W.C.W. The Effect of Nanoparticle Size, Shape, and Surface Chemistry on Biological Systems. Annu. Rev. Biomed. Eng. 2012, 14, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kelly, K.L.; Coronado, E.; Zhao, L.L.; Schatz, G.C. The Optical Properties of Metal Nanoparticles: The Influence of Size, Shape, and Dielectric Environment. J. Phys. Chem. B 2003, 107, 668–677. [Google Scholar] [CrossRef]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum Dot Bioconjugates for Imaging, Labelling and Sensing. Nat. Mater. 2005, 4, 435–446. [Google Scholar] [CrossRef]
- Schenning, A.P.H.J.; Meijer, E.W. Supramolecular Electronics; Nanowires from Self-Assembled Pi-Conjugated Systems. Chem. Commun. 2005, 14, 3245–3258. [Google Scholar] [CrossRef]
- Subhramannia, M.; Pillai, V.K. Shape-Dependent Electrocatalytic Activity of Platinum Nanostructures. J. Mater. Chem. 2008, 18, 5858–5870. [Google Scholar] [CrossRef]
- Schlögl, R.; Abd Hamid, S.B. Nanocatalysis: Mature Science Revisited or Something Really New? Angew. Chem. Int. Ed. Engl. 2004, 43, 1628–1637. [Google Scholar] [CrossRef]
- Piao, M.J.; Kang, K.A.; Lee, I.K.; Kim, H.S.; Kim, S.; Choi, J.Y.; Choi, J.; Hyun, J.W. Silver Nanoparticles Induce Oxidative Cell Damage in Human Liver Cells through Inhibition of Reduced Glutathione and Induction of Mitochondria-Involved Apoptosis. Toxicol. Lett. 2011, 201, 92–100. [Google Scholar] [CrossRef]
- Sakaguchi, T.; Mine, K.; Kudoh, F.; Kamada, R.; Sakaguchi, K. Antiproliferative Activity of Silver Nanoplates on Human Promyelocytic Leukemia Cell Lines. Chem. Lett. 2015, 44, 327–329. [Google Scholar] [CrossRef]
- Walsh, T.R.; Knecht, M.R. Biointerface Structural Effects on the Properties and Applications of Bioinspired Peptide-Based Nanomaterials. Chem. Rev. 2017, 117, 12641–12704. [Google Scholar] [CrossRef] [PubMed]
- Janairo, J.I.B.; Sakaguchi, T.; Mine, K.; Kamada, R.; Sakaguchi, K. Synergic Strategies for the Enhanced Self-Assembly of Biomineralization Peptides for the Synthesis of Functional Nanomaterials. Protein Pept. Lett. 2018, 25, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-L.; Zhang, P.; Rosi, N.L. A New Peptide-Based Method for the Design and Synthesis of Nanoparticle Superstructures: Construction of Highly Ordered Gold Nanoparticle Double Helices. J. Am. Chem. Soc. 2008, 130, 13555–13557. [Google Scholar] [CrossRef] [PubMed]
- Hwang, L.; Zhao, G.; Zhang, P.; Rosi, N.L. Size-Controlled Peptide-Directed Synthesis of Hollow Spherical Gold Nanoparticle Superstructures. Small 2011, 7, 1938–1942. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.D.; Labean, T.H. Coupling Strategies for the Synthesis of Peptide-Oligonucleotide Conjugates for Patterned Synthetic Biomineralization. J. Nucleic Acids 2011, 2011, 926595. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Faivre, D. Diversity of Microbial Metal Sulfide Biomineralization. Chempluschem 2022, 87, e202100457. [Google Scholar] [CrossRef]
- Wong Po Foo, C.; Patwardhan, S.V.; Belton, D.J.; Kitchel, B.; Anastasiades, D.; Huang, J.; Naik, R.R.; Perry, C.C.; Kaplan, D.L. Novel Nanocomposites from Spider Silk-Silica Fusion (Chimeric) Proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 9428–9433. [Google Scholar] [CrossRef]
- Stearns, L.A.; Chhabra, R.; Sharma, J.; Liu, Y.; Petuskey, W.T.; Yan, H.; Chaput, J.C. Template-Directed Nucleation and Growth of Inorganic Nanoparticles on DNA Scaffolds. Angew. Chem. Int. Ed. Engl. 2009, 48, 8494–8496. [Google Scholar] [CrossRef]
- Zhang, T.; Dong, Y.; Sun, Y.; Chen, P.; Yang, Y.; Zhou, C.; Xu, L.; Yang, Z.; Liu, D. DNA Bimodified Gold Nanoparticles. Langmuir 2012, 28, 1966–1970. [Google Scholar] [CrossRef]
- Carter, J.D.; LaBean, T.H. Organization of Inorganic Nanomaterials via Programmable DNA Self-Assembly and Peptide Molecular Recognition. ACS Nano 2011, 5, 2200–2205. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, M.; Imai, T.; Tsuruoka, T.; Sakashita, S.; Tomizaki, K.-Y.; Usui, K. Elemental Composition Control of Gold-Titania Nanocomposites by Site-Specific Mineralization Using Artificial Peptides and DNA. Commun Chem 2021, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, Z.; Li, D.; Zhang, W.; Yu, X.; Liu, W.; Gong, C.; Wei, G.; Su, Z. Biomimetic Ultralight, Highly Porous, Shape-Adjustable, and Biocompatible 3D Graphene Minerals via Incorporation of Self-Assembled Peptide Nanosheets. Adv. Funct. Mater. 2018, 28, 1801056. [Google Scholar] [CrossRef]
- Coppage, R.; Slocik, J.M.; Briggs, B.D.; Frenkel, A.I.; Naik, R.R.; Knecht, M.R. Determining Peptide Sequence Effects That Control the Size, Structure, and Function of Nanoparticles. ACS Nano 2012, 6, 1625–1636. [Google Scholar] [CrossRef]
- Coppage, R.; Slocik, J.M.; Sethi, M.; Pacardo, D.B.; Naik, R.R.; Knecht, M.R. Elucidation of Peptide Effects That Control the Activity of Nanoparticles. Angew. Chem. Int. Ed. Engl. 2010, 49, 3767–3770. [Google Scholar] [CrossRef]
- Addison, W.N.; Miller, S.J.; Ramaswamy, J.; Mansouri, A.; Kohn, D.H.; McKee, M.D. Phosphorylation-Dependent Mineral-Type Specificity for Apatite-Binding Peptide Sequences. Biomaterials 2010, 31, 9422–9430. [Google Scholar] [CrossRef]
- Sakaguchi, T.; Janairo, J.I.B.; Lussier-Price, M.; Wada, J.; Omichinski, J.G.; Sakaguchi, K. Oligomerization Enhances the Binding Affinity of a Silver Biomineralization Peptide and Catalyzes Nanostructure Formation. Sci. Rep. 2017, 7, 1400. [Google Scholar] [CrossRef]
- Janairo, J.I.B.; Sakaguchi, K. Effects of Buffer on the Structure and Catalytic Activity of Palladium Nanomaterials Formed by Biomineralization. Chem. Lett. 2014, 43, 1315–1317. [Google Scholar] [CrossRef]
- Yu, X.; Wang, Z.; Su, Z.; Wei, G. Design, Fabrication, and Biomedical Applications of Bioinspired Peptide–Inorganic Nanomaterial Hybrids. J. Mater. Chem. B Mater. Biol. Med. 2017, 5, 1130–1142. [Google Scholar] [CrossRef]
- Sano, K.I.; Sasaki, H.; Shiba, K. Specificity and Biomineralization Activities of Ti-Binding Peptide-1 (TBP-1). Langmuir 2005, 21, 3090–3095. [Google Scholar] [CrossRef]
- Mateu, M.G.; Fersht, A.R. Nine Hydrophobic Side Chains Are Key Determinants of the Thermodynamic Stability and Oligomerization Status of Tumour Suppressor P53 Tetramerization Domain. EMBO J. 1998, 17, 2748–2758. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.R.; Freire, E.; Morin, P.E.; Arrowsmith, C.H. Thermodynamic Analysis of the Structural Stability of the Tetrameric Oligomerization Domain of P53 Tumor Suppressor. Biochemistry 1995, 34, 5309–5316. [Google Scholar] [CrossRef] [PubMed]
- Mason, J.M.; Arndt, K.M. Coiled Coil Domains: Stability, Specificity, and Biological Implications. Chembiochem 2004, 5, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Woolfson, D.N. The Design of Coiled-Coil Structures and Assemblies. Adv. Protein Chem. 2005, 70, 79–112. [Google Scholar] [PubMed]
- O’Shea, E.K.; Klemm, J.D.; Kim, P.S.; Alber, T. X-Ray Structure of the GCN4 Leucine Zipper, a Two-Stranded, Parallel Coiled Coil. Science 1991, 254, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Bewley, C.A.; Louis, J.M.; Ghirlando, R.; Marius Clore, G. Design of a Novel Peptide Inhibitor of HIV Fusion That Disrupts the Internal Trimeric Coiled-Coil of Gp41. J. Biol. Chem. 2002, 277, 14238–14245. [Google Scholar] [CrossRef] [PubMed]
- Zaccai, N.R.; Chi, B.; Thomson, A.R.; Boyle, A.L.; Bartlett, G.J.; Bruning, M.; Linden, N.; Sessions, R.B.; Booth, P.J.; Brady, R.L.; et al. A de Novo Peptide Hexamer with a Mutable Channel. Nat. Chem. Biol. 2011, 7, 935–941. [Google Scholar] [CrossRef]
- Swasey, S.M.; Leal, L.E.; Lopez-Acevedo, O.; Pavlovich, J.; Gwinn, E.G. Silver (I) as DNA Glue: Ag(+)-Mediated Guanine Pairing Revealed by Removing Watson-Crick Constraints. Sci. Rep. 2015, 5, 10163. [Google Scholar] [CrossRef]
- Volkov, I.L.; Smirnova, A.; Makarova, A.A.; Reveguk, Z.V.; Ramazanov, R.R.; Usachov, D.Y.; Adamchuk, V.K.; Kononov, A.I. DNA with Ionic, Atomic, and Clustered Silver: An XPS Study. J. Phys. Chem. B 2017, 121, 2400–2406. [Google Scholar] [CrossRef]
- Xiong, Y.; Xia, Y. Shape-Controlled Synthesis of Metal Nanostructures: The Case of Palladium. Adv. Mater. 2007, 19, 3385–3391. [Google Scholar] [CrossRef]
- Kim, M.H.; Kwak, S.K.; Im, S.H.; Lee, J.-B.; Choi, K.-Y.; Byun, D.-J. Maneuvering the Growth of Silver Nanoplates: Use of Halide Ions to Promote Vertical Growth. J. Mater. Chem. 2014, 2, 6165–6170. [Google Scholar] [CrossRef]
- Downs, R.T.; Hall-Wallace, M. The American Mineralogist Crystal Structure Database. Am. Mineral. 2003, 88, 247–250. [Google Scholar]
- Poudel, L.; Tamerler, C.; Misra, A.; Ching, W.-Y. Atomic-Scale Quantification of Interfacial Binding between Peptides and Inorganic Crystals: The Case of Calcium Carbonate Binding Peptide on Aragonite. J. Phys. Chem. C 2017, 121, 28354–28363. [Google Scholar] [CrossRef]
- Lou, T.; Bai, X.; He, X.; Yang, Y.; Yuan, C. Molecular Dynamics Simulation of Peptide Attachment on Al-Based Surfaces. Prog. Org. Coat. 2021, 157, 106310. [Google Scholar] [CrossRef]
- Turner, M.; Mutter, S.T.; Deeth, R.J.; Platts, J.A. Ligand Field Molecular Dynamics Simulation of Pt(II)-Phenanthroline Binding to N-Terminal Fragment of Amyloid-β Peptide. PLoS ONE 2018, 13, e0193668. [Google Scholar] [CrossRef]
- Ozaki, M.; Nagai, K.; Nishiyama, H.; Tsuruoka, T.; Fujii, S.; Endoh, T.; Imai, T.; Tomizaki, K.-Y.; Usui, K. Site-Specific Control of Silica Mineralization on DNA Using a Designed Peptide. Chem. Commun. 2016, 52, 4010–4013. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Liu, Z.-G.; Shen, W.; Gurunathan, S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef]
- Akter, M.; Sikder, M.T.; Rahman, M.M.; Ullah, A.K.M.A.; Hossain, K.F.B.; Banik, S.; Hosokawa, T.; Saito, T.; Kurasaki, M. A Systematic Review on Silver Nanoparticles-Induced Cytotoxicity: Physicochemical Properties and Perspectives. J. Advert. Res. 2018, 9, 1–16. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sakaguchi, T.; Nakagawa, N.; Mine, K.; Janairo, J.I.B.; Kamada, R.; Omichinski, J.G.; Sakaguchi, K. Biomineralization through a Symmetry-Controlled Oligomeric Peptide. Biomimetics 2023, 8, 606. https://doi.org/10.3390/biomimetics8080606
Sakaguchi T, Nakagawa N, Mine K, Janairo JIB, Kamada R, Omichinski JG, Sakaguchi K. Biomineralization through a Symmetry-Controlled Oligomeric Peptide. Biomimetics. 2023; 8(8):606. https://doi.org/10.3390/biomimetics8080606
Chicago/Turabian StyleSakaguchi, Tatsuya, Natsumi Nakagawa, Kenta Mine, Jose Isagani B. Janairo, Rui Kamada, James G. Omichinski, and Kazuyasu Sakaguchi. 2023. "Biomineralization through a Symmetry-Controlled Oligomeric Peptide" Biomimetics 8, no. 8: 606. https://doi.org/10.3390/biomimetics8080606
APA StyleSakaguchi, T., Nakagawa, N., Mine, K., Janairo, J. I. B., Kamada, R., Omichinski, J. G., & Sakaguchi, K. (2023). Biomineralization through a Symmetry-Controlled Oligomeric Peptide. Biomimetics, 8(8), 606. https://doi.org/10.3390/biomimetics8080606





