A Visualization Method of Knowledge Graphs for the Computation and Comprehension of Ultrasound Reports
Abstract
:1. Introduction
- Firstly, we propose a novel key term network extraction method for ultrasound reports that achieves the highly accurate decomposition and annotation of the narrative text within reports.
- Secondly, we introduce a knowledge representation framework based on ultrasound reports that provides a structured and intuitive visualization of ultrasound report knowledge.
- Finally, we propose a knowledge graph completion model to address the lack of entities in physicians’ writing habits and improve the accuracy of using ultrasound knowledge.
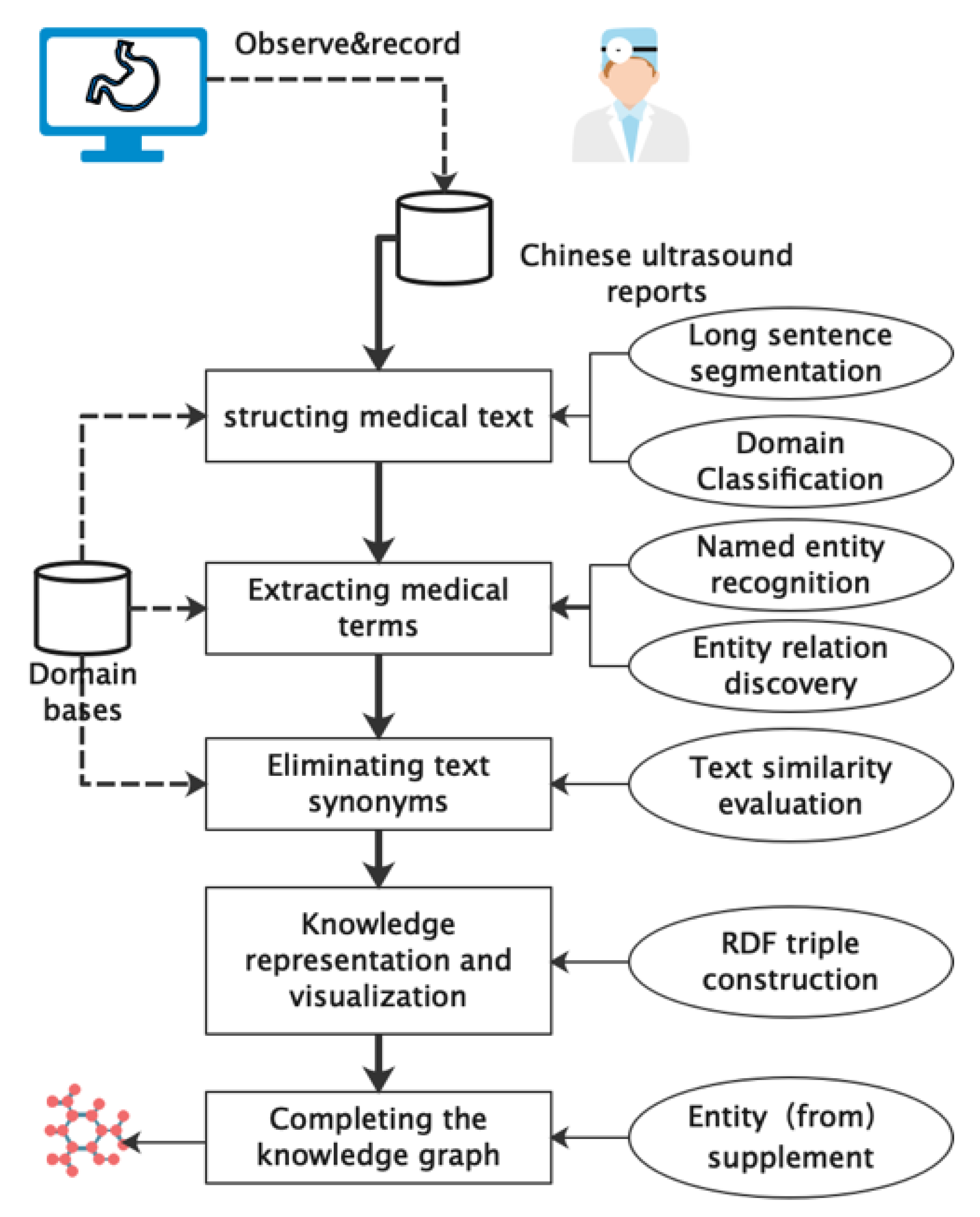
2. Materials and Methods
2.1. Structuring Medical Text
| Algorithm 1. Structuring narrative text from ultrasound reports |
| Input: Narrative text of a report (text) Output: Term network of the report (TN) 1: Let t ← a list of terms through the WSM (text) 2: Let s ← an empty set of concept relationships between two concepts {r: (c1, c2)}, where a concept is defined as a tuple c = (w, nw, pos). 3: for each w ∈ t perform the following: 4: Let ST be the result from the SNLP (w) 5: Let pos ← ST.pos 6: Let nw ← normalize (w) from ST 7: Let c(t) ← a tuple: (w, nw, pos) 8: end for 9: for ∀c1, c2 ∈ t perform the following: 10: If dist(c1(t), c2(t)) = 1 then Append (c1, c2) into s 11: end for 12: return TN (s) |
2.2. Extracting Medical Terms
| Algorithm 2. Constructing simple knowledge graphs based on the term network |
| Input: a term network of a report (TN = {t1, t2, t3, …}) Output: a simple knowledge graph (SKG = {r1, r2, r3, …}) 1: Let R ← a set of {rule: (tag1, tag2, …, tagc} where c is the number of tags in a rule and tag ∈ {pos} ∪ {custom tag} 2: Let M ← a subset of TN where ∀term ∈ M are restricted to rules R. 3: for ∀t1, t2, t3, … tn ∈ M perform the following: 4: if rule of (t1, t2, t3, … tn) ∈ R in Table 1 begins 5: Let r ← a tuple that preserves an entity relation in Table 1 6: if n = 2 then r ← (t1, a predefined relationship, t2) 7: if n = 3 then r ← (t1, t2, t3) 8: if r is not null then Append r into SKG 9: end if 10: end for 11: return SKG |
2.3. Extracting Medical Terms
2.4. Knowledge Representation and Visualization
| <?xml version=“1.0”?> <rdf:RDF xmlns=“#”> <owl:Ontology rdf:about=“”/> <owl:ObjectProperty rdf:about=“#hasProperty”> <rdf:type rdf:resource=“&owl;TransitiveProperty”/> </owl:ObjectProperty> <owl:ObjectProperty rdf:about=“#internal_diameter”/> <owl:Class rdf:about=“#liver”/> <owl:Class rdf:about=“#portal_vein”> <rdfs:subClassOf rdf:resource=“#liver”/> <owl:disjointWith rdf:resource=“#surface”/> </owl:Class> <owl:Class rdf:about=“#surface”> <rdfs:subClassOf rdf:resource=“#liver”/> </owl:Class> <owl:Class rdf:about=“#trunk”> <rdfs:subClassOf rdf:resource=“#portal_vein”/> <rdfs:subClassOf> <owl:Restriction> <owl:onProperty rdf:resource=“#hasProperty”/> <owl:onClass rdf:resource=“#trunk”/> <owl:minQualifiedCardinality rdf:datatype=“&xsd;nonNegativeInteger”>1</owl:minQualifiedCardinality> </owl:Restriction> </rdfs:subClassOf> </owl:Class> <owl:NamedIndividual rdf:about=“#no_smooth”> <rdf:type rdf:resource=“#surface”/> </owl:NamedIndividual> </rdf:RDF> |
2.5. Completing Knowledge Graph
3. Results
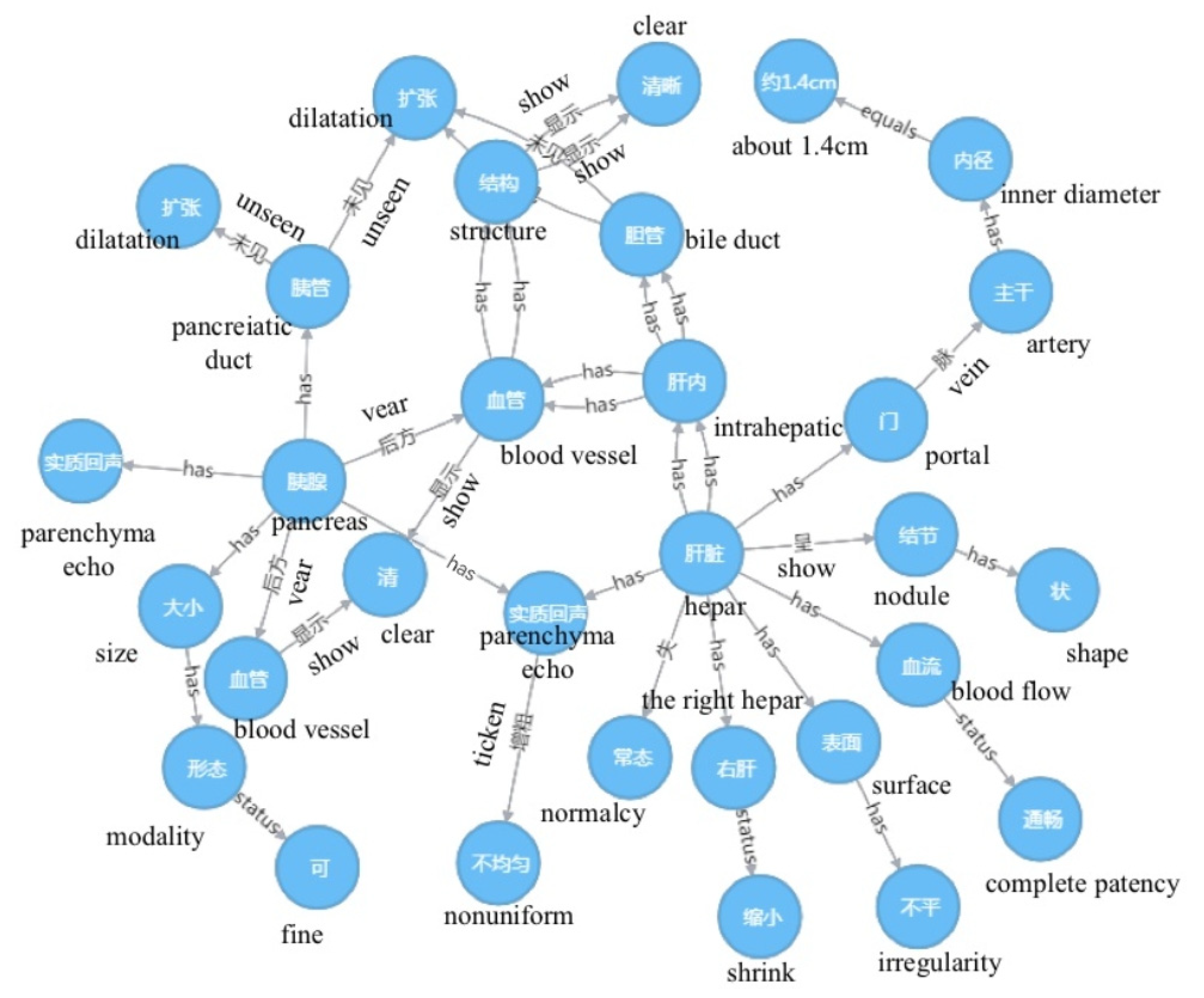
3.1. Visualization of Ultrasound Reports
3.2. Evaluation in Knowledge Extraction
3.3. Evaluation in Knowledge Expression and Visualization
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yadav, S.P.; Yadav, S. Image fusion using hybrid methods in multimodality medical images. Med. Biol. Eng. Comput. 2020, 58, 669–687. [Google Scholar] [CrossRef]
- Bai, J.; Song, S.; Fan, T.; Jiao, L. Medical image denoising based on sparse dictionary learning and cluster ensemble. Soft Comput. 2018, 22, 1467–1473. [Google Scholar] [CrossRef]
- Yang, S.; Niu, J.; Wang, Y.; Liu, X.; Li, Q. Automatic ultrasound image report generation with adaptive multimodal attention mechanism. Neurocomputing 2021, 427, 40–49. [Google Scholar] [CrossRef]
- Ribeiro, R.T.; Marinho, R.T.; Sanches, J.M. An ultrasound-based computer-aided diagnosis tool for steatosis detection. IEEE J. Biomed. Health Inform. 2014, 18, 1397–1403. [Google Scholar] [CrossRef]
- Penning, M.L.; Blach, C.; Walden, A.; Wang, P.; Donovan, K.M.; Garza, M.Y.; Wang, Z.; Frund, J.; Syed, S.; Syed, M.; et al. Near real time EHR data utilization in a clinical study. Stud. Health Technol. Inform. 2020, 270, 337–341. [Google Scholar]
- Inamullah; Hassan, S.; Alrajeh, N.A.; Mohammed, E.A.; Khan, S. Data diversity in convolutional neural network based ensemble model for diabetic retinopathy. Biomimetics 2023, 8, 187. [Google Scholar] [CrossRef]
- Bozkurt, S.; Gimenez, F.; Burnside, E.S.; Gulkesen, K.H.; Rubin, D.L. Using automatically extracted information from mammography reports for decision-support. J. Biomed. Inform. 2016, 62, 224–231. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, J.; Song, J.; Liu, J. A weakly-supervised method for named entity recognition of agricultural knowledge graph. Intell. Autom. Soft Comput. 2023, 37, 833–848. [Google Scholar] [CrossRef]
- Hogan, A.; Blomqvist, E.; Cochez, M.; d’Amato, C.; Melo, G.D.; Gutierrez, C.; Kirrane, S.; Labra Gayo, J.E.; Navigli, R.; Neumaier, S.; et al. Knowledge graphs. ACM Comput. Surv. 2021, 54, 1–37. [Google Scholar] [CrossRef]
- Joneidy, S.; Burke, M. Towards a deeper understanding of meaningful use in electronic health records. Health Inf. Libr. J. 2019, 36, 134–152. [Google Scholar] [CrossRef]
- Sheikhtaheri, A.; Tabatabaee Jabali, S.M.; Bitaraf, E.; TehraniYazdi, A.; Kabir, A. A near real-time electronic health record-based COVID-19 surveillance system: An experience from a developing country. Health Inf. Manag. J. 2022; ahead of print. [Google Scholar] [CrossRef]
- Šubert, M.; Novotný, M.; Tykalová, T.; Srpová, B.; Friedová, L.; Uher, T.; Horáková, D.; Rusz, J. Lexical and syntactic deficits analyzed via automated natural language processing: The new monitoring tool in multiple sclerosis. Ther. Adv. Neurol. Disord. 2023, 6, 17562864231180719. [Google Scholar] [CrossRef]
- Hu, C.; Zhang, S.; Gu, T.; Yan, Z.; Jiang, J. Multi-Task joint learning model for Chinese word segmentation and syndrome differentiation in traditional Chinese medicine. Int. J. Environ. Res. Public Health. 2022, 19, 5601. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, Q.; Qian, S.; Xu, C. Multi-modal multi-relational feature aggregation network for medical knowledge representation learning. In Proceedings of the 28th ACM International Conference on Multimedia, Seattle, WA, USA, 12–16 October 2020. [Google Scholar]
- Ji, S.; Pan, S.; Cambria, E.; Marttinen, P.; Philip, S.Y. A survey on knowledge graphs: Representation, acquisition, and applications. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 494–514. [Google Scholar] [CrossRef]
- Rout, J.K.; Choo, K.K.R.; Dash, A.K.; Bakshi, S.; Jena, S.; Williams, K.L. A model for sentiment and emotion analysis of unstructured social media text. Elec. Commer. Res. 2018, 18, 181–199. [Google Scholar] [CrossRef]
- Maurice, P.; Dhombres, F.; Blondiaux, E.; Friszer, E.; Guilbaud, S.; Lelong, N.; Khoshnood, B.; Charlet, J.; Perrot, N.; Jauniaux, E.; et al. Towards ontology-based decision support systems for complex ultrasound diagnosis in obstetrics and gynecology. J. Gynecol. Obstet. Hum. Reprod. 2017, 46, 423–429. [Google Scholar] [CrossRef]
- Li, X.; Sun, S.; Tang, T.; Lu, J.; Zhang, L.; Yin, J.; Geng, Q.; Wu, Y. Construction of a knowledge graph for breast cancer diagnosis based on Chinese electronic medical records: Development and usability study. BMC Med. Inform. Decis. Mak. 2023, 23, 210. [Google Scholar] [CrossRef]
- An, B. Construction and application of Chinese breast cancer knowledge graph based on multi-source heterogeneous data. Math. Biosci. Eng. 2023, 20, 6776–6799. [Google Scholar] [CrossRef]
- Frei, J.; Kramer, F. German medical named entity recognition model and data set creation using machine translation and word alignment: Algorithm development and validation. JMIR Form. Res. 2023, 7, e39077. [Google Scholar] [CrossRef]
- Kaplar, A.; Stošović, M.; Kaplar, A.; Brković, V.; Naumović, R.; Kovačević, A. Evaluation of clinical named entity recognition methods for Serbian electronic health records. Int. J. Med. Inform. 2022, 64, 104805. [Google Scholar] [CrossRef]
- Colicchio, T.K.; Dissanayake, P.I.; Cimino, J.J. Formal representation of patients’ care context data: The path to improving the electronic health record. J. Am. Med. Inform. Assoc. 2020, 27, 1648–1657. [Google Scholar] [CrossRef]
- Meystre, S.M.; Savova, G.K.; Kipper-Schuler, K.C.; Hurdle, J.F. Extracting information from textual documents in the electronic health record: A review of recent research. Yearb. Med. Inf. 2008, 17, 128–144. [Google Scholar]
- Shi, J.; Sun, M.; Sun, Z.; Li, M.; Gu, Y.; Wensheng, Z. Multi-level semantic fusion network for Chinese medical named entity recognition. J. Biomed. Inform. 2022, 133, 104144. [Google Scholar] [CrossRef]
- Meystre, S.M.; Kim, Y.; Gobbel, G.T.; Matheny, M.E.; Redd, A.; Bray, B.E.; Garvin, J.H. Congestive heart failure information extraction framework for automated treatment performance measures assessment. J. Am. Med. Inform. Assoc. 2017, 24, e40–e46. [Google Scholar] [CrossRef]
- Ji, B.; Li, S.; Yu, J.; Tao, J.; Wu, Q.; Tan, Y.; Liu, H.; Ji, Y. Research on Chinese medical named entity recognition based on collaborative cooperation of multiple neural network models. J. Biomed. Inform. 2020, 104, 103395. [Google Scholar] [CrossRef]
- Rotmensch, M.; Halpern, Y.; Tlimat, A.; Horng, S.; Sontag, D. Learning a health knowledge graph from electronic medical records. Sci. Rep. 2017, 7, 5994. [Google Scholar] [CrossRef]
- Horal, A.; Baisa, V.; Rambousek, A.; Suchomel, V. A new approach for semi-automatic building and extending a multilingual terminology thesaurus. Int. J. Artif. Intell. Tools 2019, 28, 1950008. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Wang, Z.; Zhang, J.W.; Feng, J.L.; Chen, Z. Knowledge graph embedding by translating on hyperplanes. In Proceedings of the AAAI-14: Twenty-Eighth Conference on Artificial Intelligence, Québec, QC, Canada, 27–31 July 2014. [Google Scholar]
- Bordes, A.; Usunier, N.; Garcia-Duran, A.; Westo, J.; Yakhnenko, O. Translating embeddings for modeling multi-relational data. In Proceedings of the 26th International Conference on Neural Information Processing Systems, Lake Tahoe, NV, USA, 5–10 December 2013. [Google Scholar]
- Lin, Y.; Liu, Z.; Sun, M.; Liu, Y.; Zhu, X. Learning entity and relation embeddings for knowledge graph completion. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence, Austin, TX, USA, 25–30 January 2015. [Google Scholar]
- Sun, Z.; Deng, Z.H.; Nie, J.Y.; Tang, J. Rotate: Knowledge graph embedding by relational rotation in complex space. In Proceedings of the Seventh International Conference on Learning Representations, New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Wang, J.; Qu, Z.; Hu, Y.; Ling, Q.; Yu, J.; Jiang, Y. Diagnosis and treatment knowledge graph modeling application based on Chinese Medical Records. Electronics 2023, 12, 3412. [Google Scholar] [CrossRef]
- Doğan, R.I.; Leaman, R.; Lu, Z. NCBI disease corpus: A resource for disease name recognition and concept normalization. J. Biomed. Inform. 2014, 47, 1–10. [Google Scholar] [CrossRef]
- Huang, M.S.; Lai, P.T.; Tsai, R.T.H.; Hsu, W.L. Revised JNLPBA corpus: A revised version of biomedical NER corpus for relation extraction task. Brief. Bioinform. 2020, 21, 2219–2238. [Google Scholar] [CrossRef]




| Id | Rules | Relationships |
|---|---|---|
| 1 | /organ→/verb→/adj | (/organ, /verb, /adj) |
| 2 | /organ→/adj | (/organ, show, /adj) |
| 3 | /organ→/verb | (/organ, show, /verb) |
| 4 | /organ→/n→/adj | (/organ, has, /n) (/n, show, /adj) |
| Item of Report | Definition | Example Content |
|---|---|---|
| patientid | Patient ID | TG923088WY |
| admiss_times | Examination frequency | 1 |
| StudyClass | Types of ultrasound examinations | Abdominal routine |
| Observations of imaging | The condition of the organ tissues is observed and recorded by the doctor through operating the ultrasound device | The liver is abnormal, and the right liver is reduced with an uneven surface, the substantial echo is thickened, uneven, and nodular, and the hepatic vascular structure is clear. The internal diameter of the main portal vein is 1.4 cm, and the blood flow is unobstructed. The intrahepatic bile duct does not expand. The size and shape of the pancreas are observed, the echo is homogeneous, the pancreatic duct is not dilated, and the posterior pancreatic vessels are clear. The splenic thickness and length are approximately 4.6 and 10.7 cm, the echo is homogeneous, and the color blood flow distribution is normal. The size and shape of the double kidneys are homogeneous, the echo is homogeneous, and the renal pelvis, calyx, and ureter do not expand. |
| Result of examination | The ultrasound examination conclusion provided by the doctor | Liver cirrhosis, and portal hypertension are observed; the accessory umbilical vein is open; the spleen is slightly large. |
| Original Text | TN | SKG | SKG with KGCM |
|---|---|---|---|
| The liver is abnormal, and the right liver is reduced with an uneven surface, the substantial echo is thickened, uneven, and nodular, and the hepatic vascular structure is clear. | liver; has no normalcy | {liver, has no normalcy} | {liver, has no normalcy} |
| right liver; shrink; | {right liver; shows shrink;} | {right liver; shows shrink; } | |
| surface; uneven | {@, surface, uneven} | {liver, surface, uneven} | |
| substantial echo; thickens; nonuniform; | {substantial echo, thickens, nonuniform} | {substantial echo, thickens, nonuniform} | |
| show; nodule shape | {@, show, nodule shape} | {substantial echo, show, nodule shape} | |
| Intrahepatic; blood vessel; structure; show; clear | {liver, has, blood vessel}; {blood vessel, has, structure}; {structure, show; clear} | {liver has, blood vessel); {blood vessel, has, structure); {structure, show; clear} |
| Entity 1 | Relationship | Entity 2 | % |
|---|---|---|---|
| size | has | form | 3.14 |
| form | status | suitable | 2.99 |
| liver | has | inside liver | 2.88 |
| inside liver | has | blood vessel | 2.37 |
| parenchymal | echo | average | 2.31 |
| colour | has | blood stream | 1.96 |
| inside liver | has | bile duct | 1.96 |
| bile duct | unseen | expand | 1.88 |
| gallbladder | has | intracavity | 1.68 |
| blood vessel | has | structure | 1.38 |
| renal pelvis | has | calyx | 1.15 |
| outside liver | has | bile duct | 1.12 |
| gallbladder | has | wall | 1.11 |
| liver | has | parenchymal | 1.11 |
| spleen | has | parenchymal | 1.07 |
| parenchymal | status | echo | 1.02 |
| gallbladder | has | outside liver | 1.00 |
| liver | has | envelope | 1.00 |
| size | has | form | 3.14 |
| Semantically Similar Words | Cosine Similarity |
|---|---|
| Unsmooth | 0.937705 |
| Rough | 0.851965 |
| Less smooth | 0.816471 |
| Uneven | 0.74689 |
| Generally flat | 0.588166 |
| Methods | NCBI | JLNPBA | Ultrasound Reports | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | |
| TransE | 0.75 | 0.77 | 0.76 | 0.73 | 0.72 | 0.73 | 0.80 | 0.82 | 0.81 |
| TransH | 0.76 | 0.74 | 0.75 | 0.77 | 0.79 | 0.78 | 0.84 | 0.86 | 0.85 |
| TransR | 0.78 | 0.79 | 0.79 | 0.74 | 0.72 | 0.73 | 0.83 | 0.84 | 0.84 |
| RotatE | 0.82 | 0.81 | 0.81 | 0.78 | 0.75 | 0.77 | 0.81 | 0.84 | 0.83 |
| EEM-CMR | 0.76 | 0.75 | 0.76 | 0.71 | 0.70 | 0.71 | 0.82 | 0.81 | 0.82 |
| KGCM | 0.83 | 0.84 | 0.84 | 0.78 | 0.80 | 0.79 | 0.85 | 0.89 | 0.87 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, J.; Zhang, R.; Chen, D.; Shi, L. A Visualization Method of Knowledge Graphs for the Computation and Comprehension of Ultrasound Reports. Biomimetics 2023, 8, 560. https://doi.org/10.3390/biomimetics8080560
Feng J, Zhang R, Chen D, Shi L. A Visualization Method of Knowledge Graphs for the Computation and Comprehension of Ultrasound Reports. Biomimetics. 2023; 8(8):560. https://doi.org/10.3390/biomimetics8080560
Chicago/Turabian StyleFeng, Jiayi, Runtong Zhang, Donghua Chen, and Lei Shi. 2023. "A Visualization Method of Knowledge Graphs for the Computation and Comprehension of Ultrasound Reports" Biomimetics 8, no. 8: 560. https://doi.org/10.3390/biomimetics8080560
APA StyleFeng, J., Zhang, R., Chen, D., & Shi, L. (2023). A Visualization Method of Knowledge Graphs for the Computation and Comprehension of Ultrasound Reports. Biomimetics, 8(8), 560. https://doi.org/10.3390/biomimetics8080560






