Heterogeneous Nucleation in Protein Crystallization
Abstract
1. Introduction
2. Protein Crystallization Nucleation
2.1. Homogeneous Nucleation
2.2. Heterogeneous Nucleation
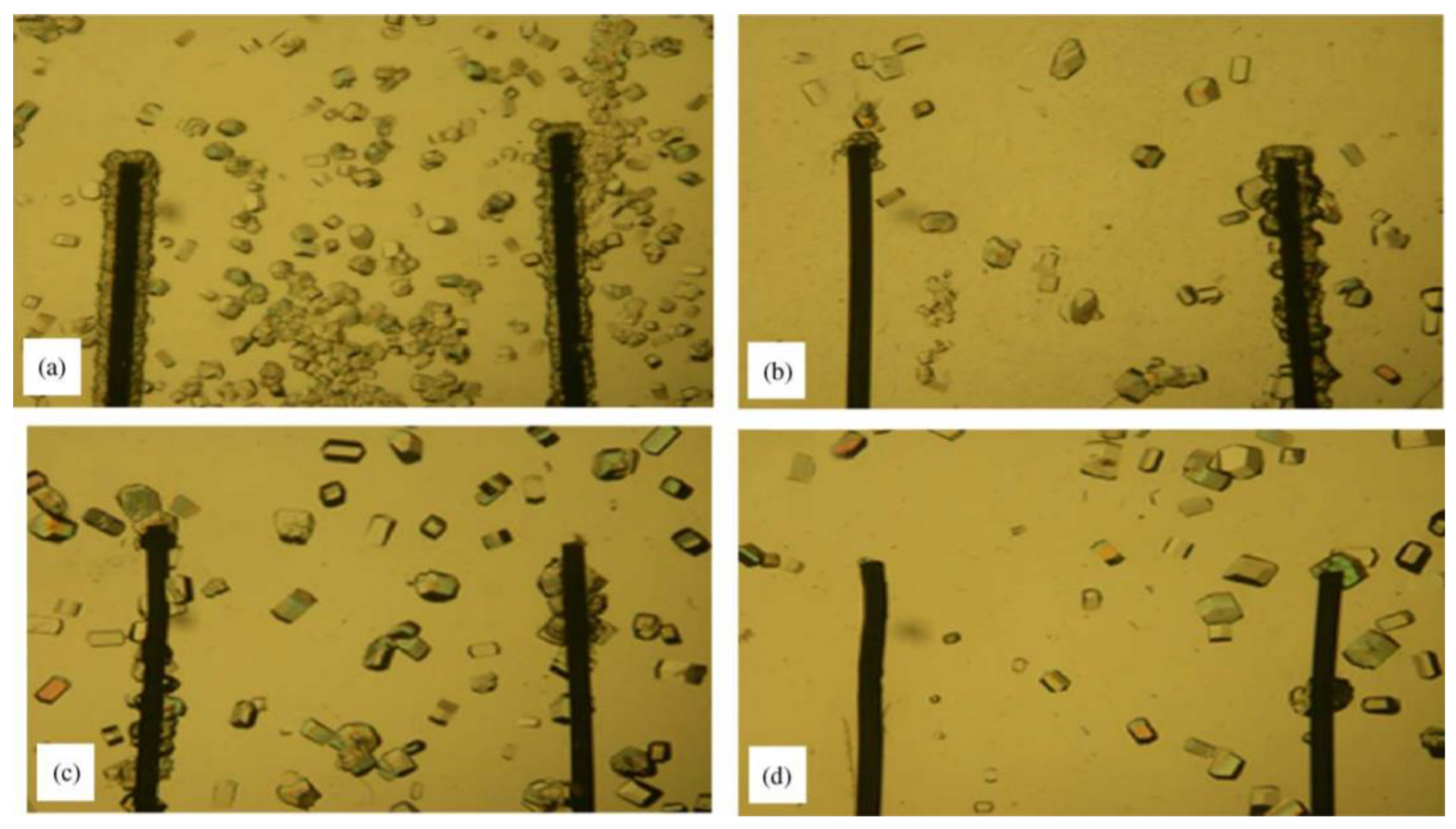
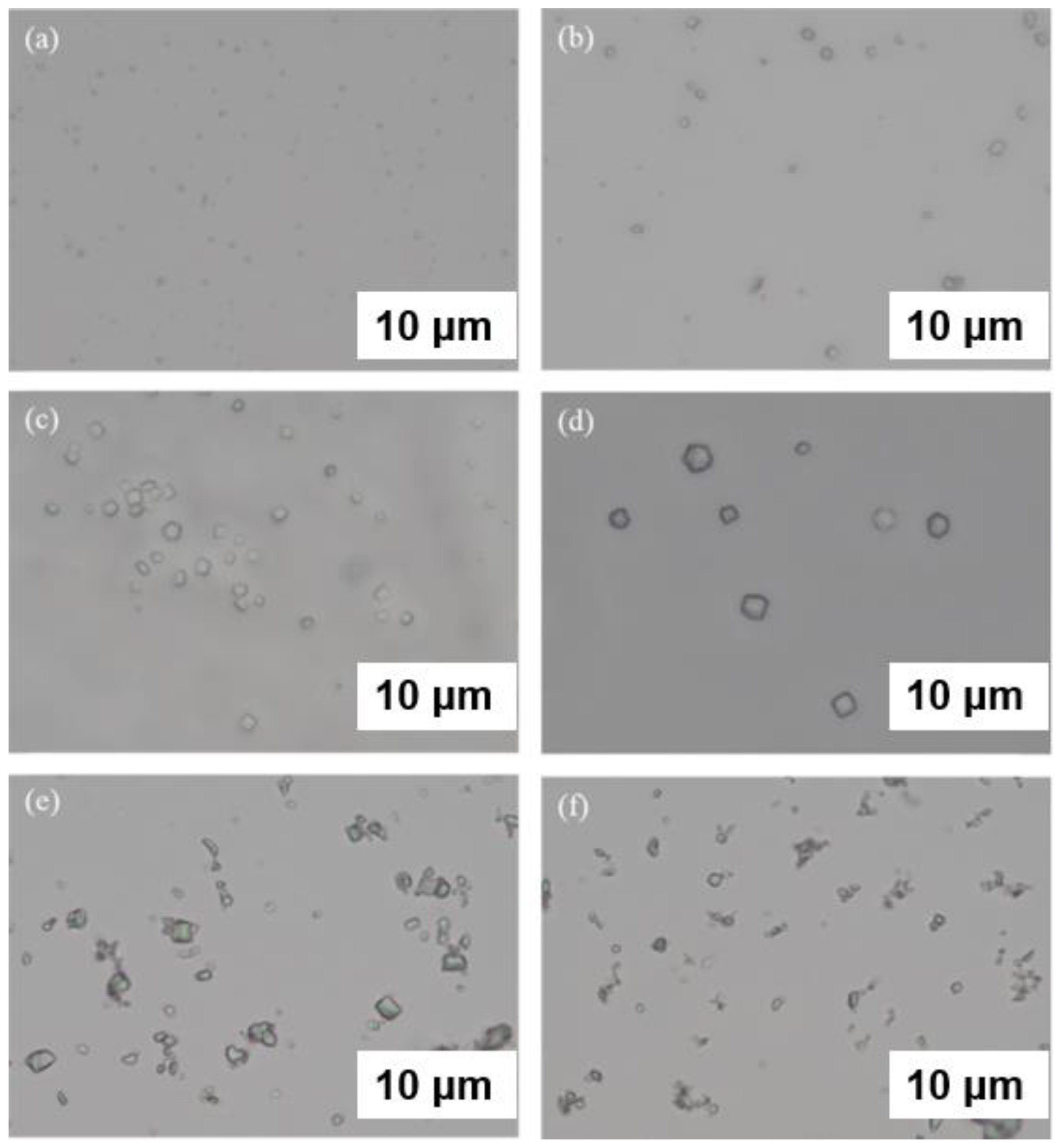
3. Heterogeneous Nucleating Agents
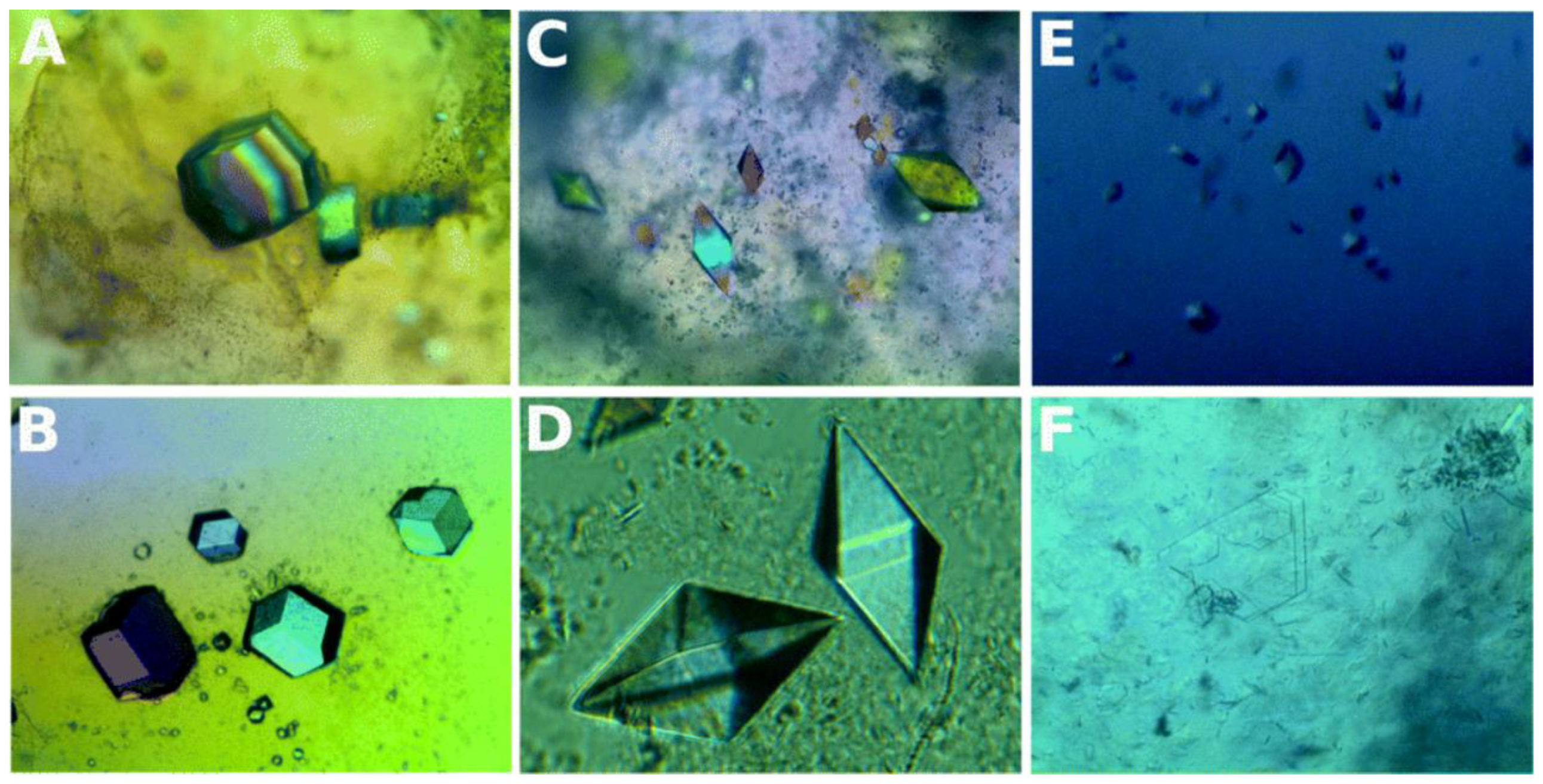
3.1. Nucleating Agents from Natural Sources
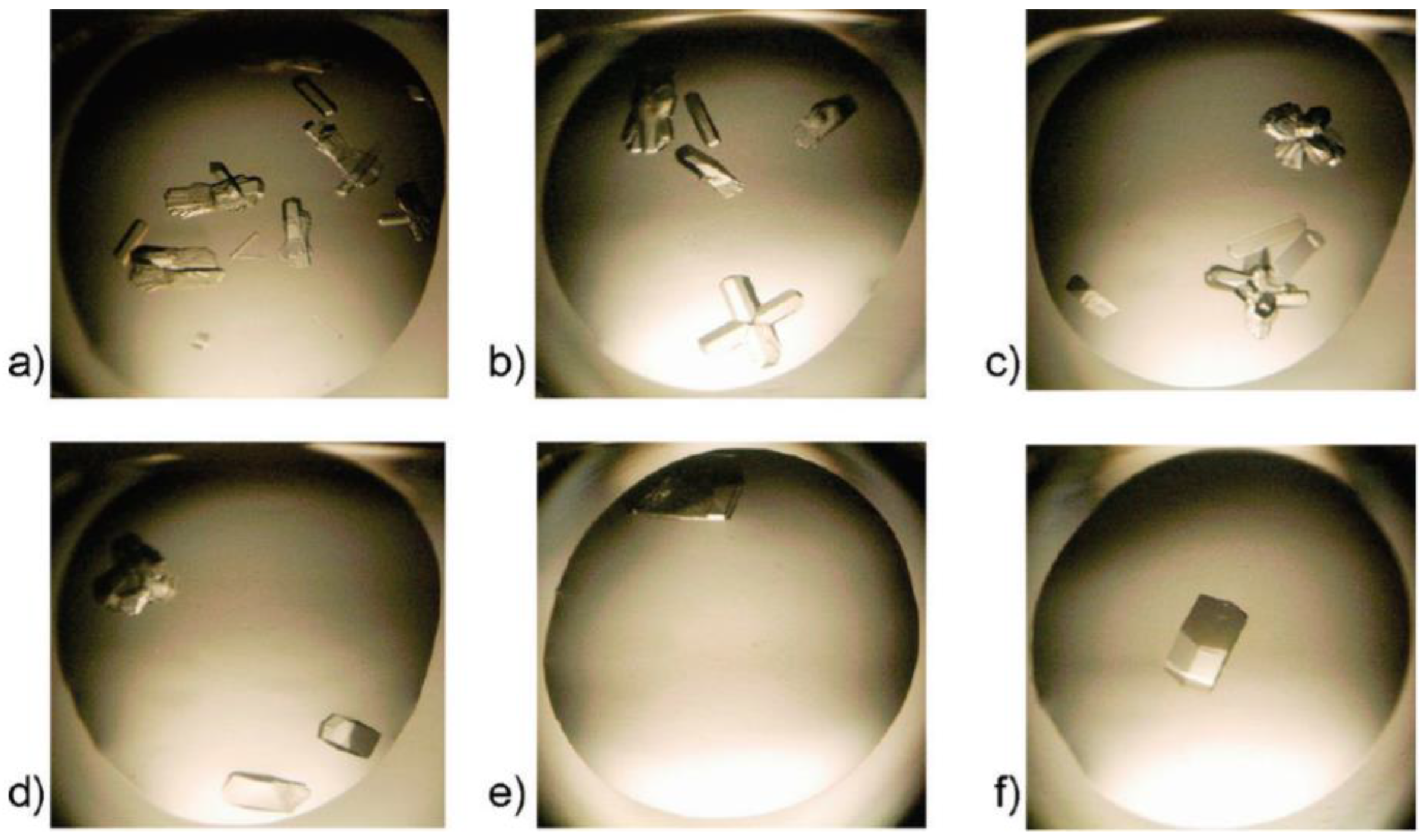
3.2. Short Peptide Supramolecular Hydrogels
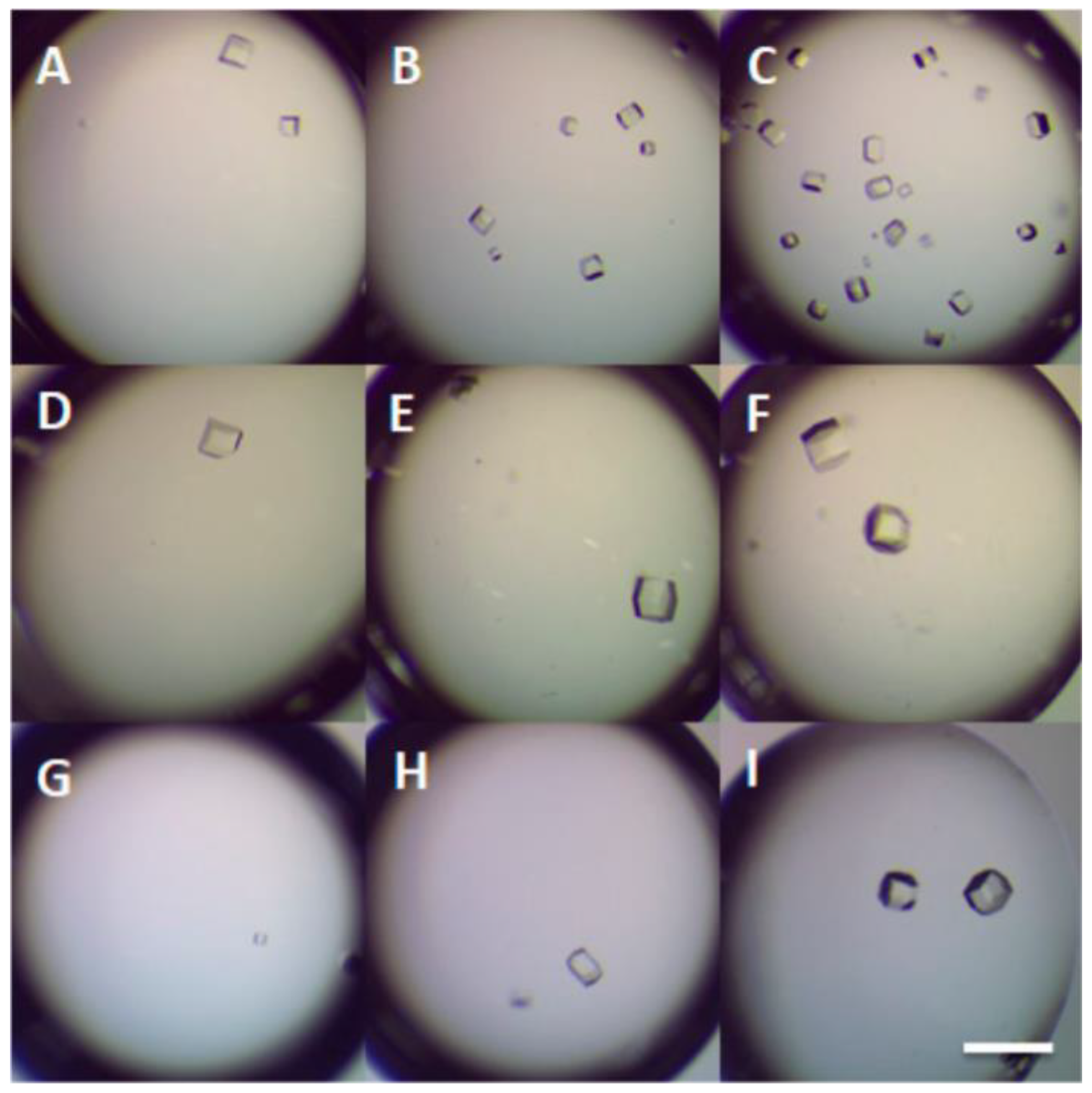
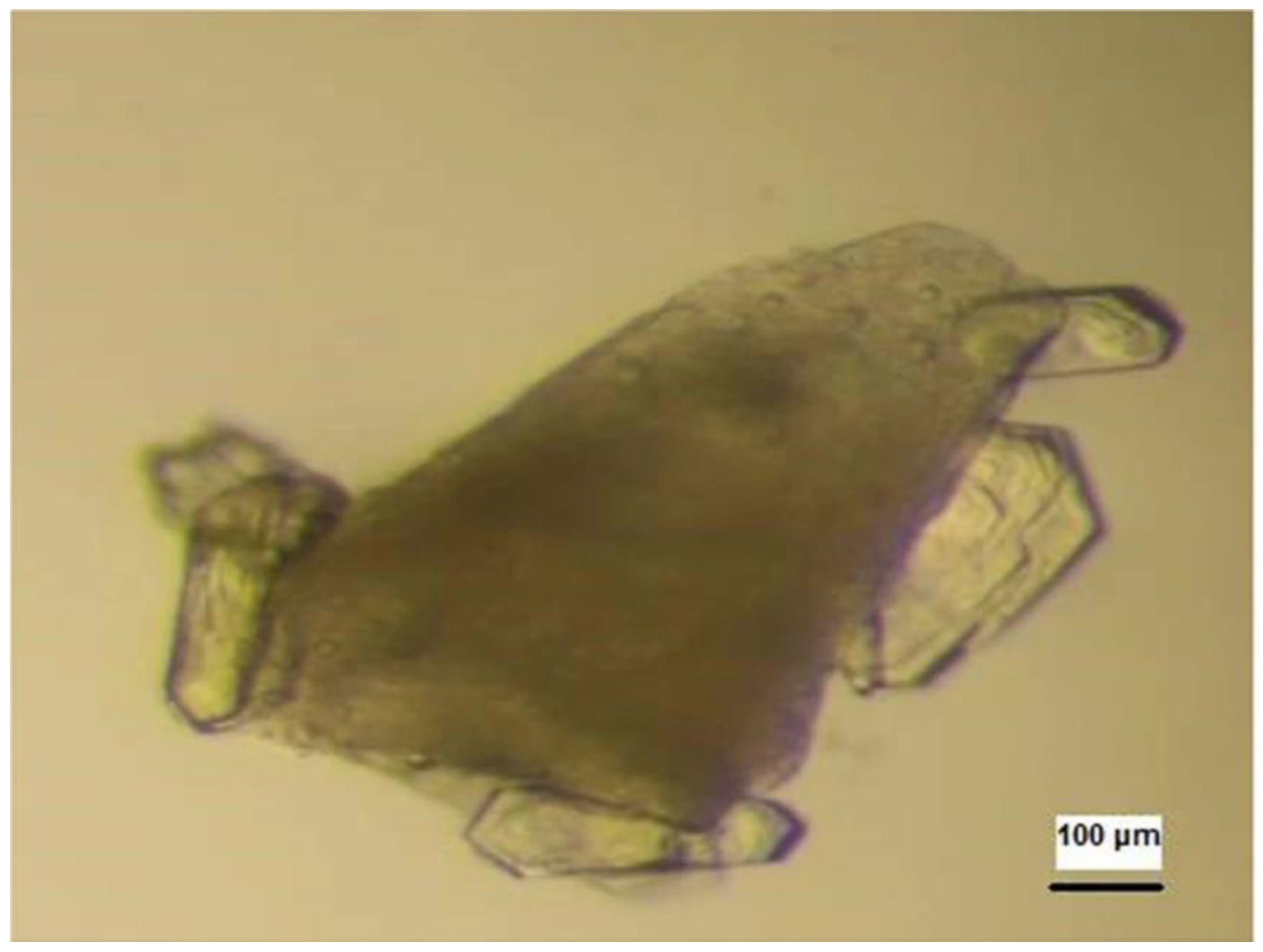
3.3. DNA
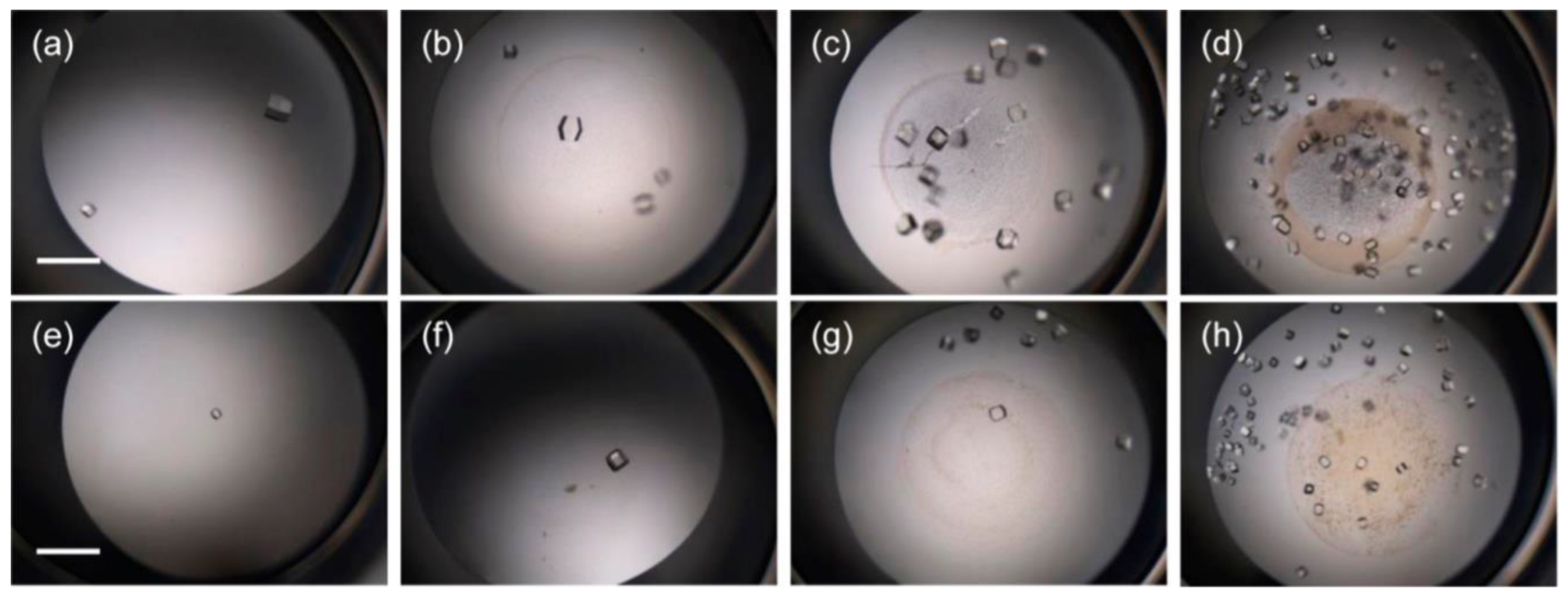
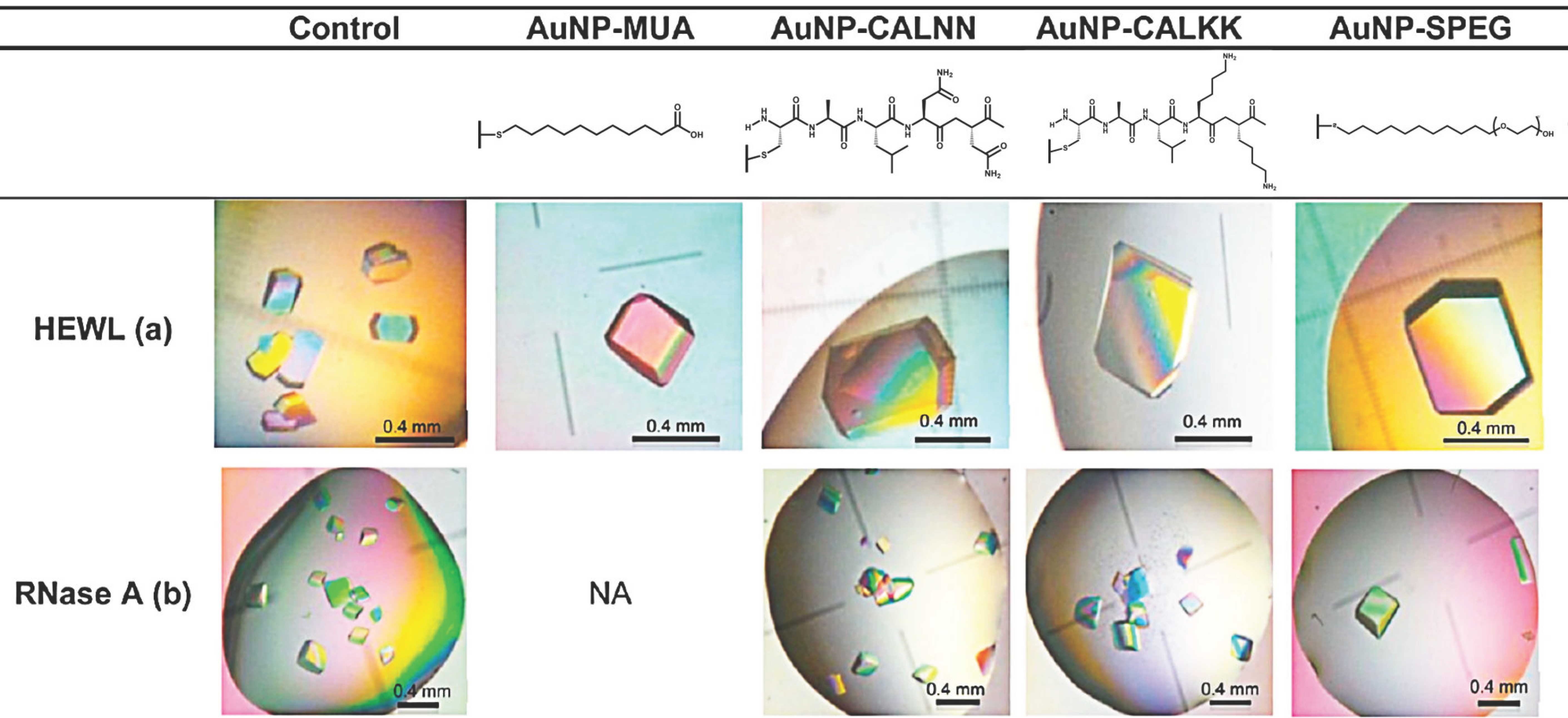
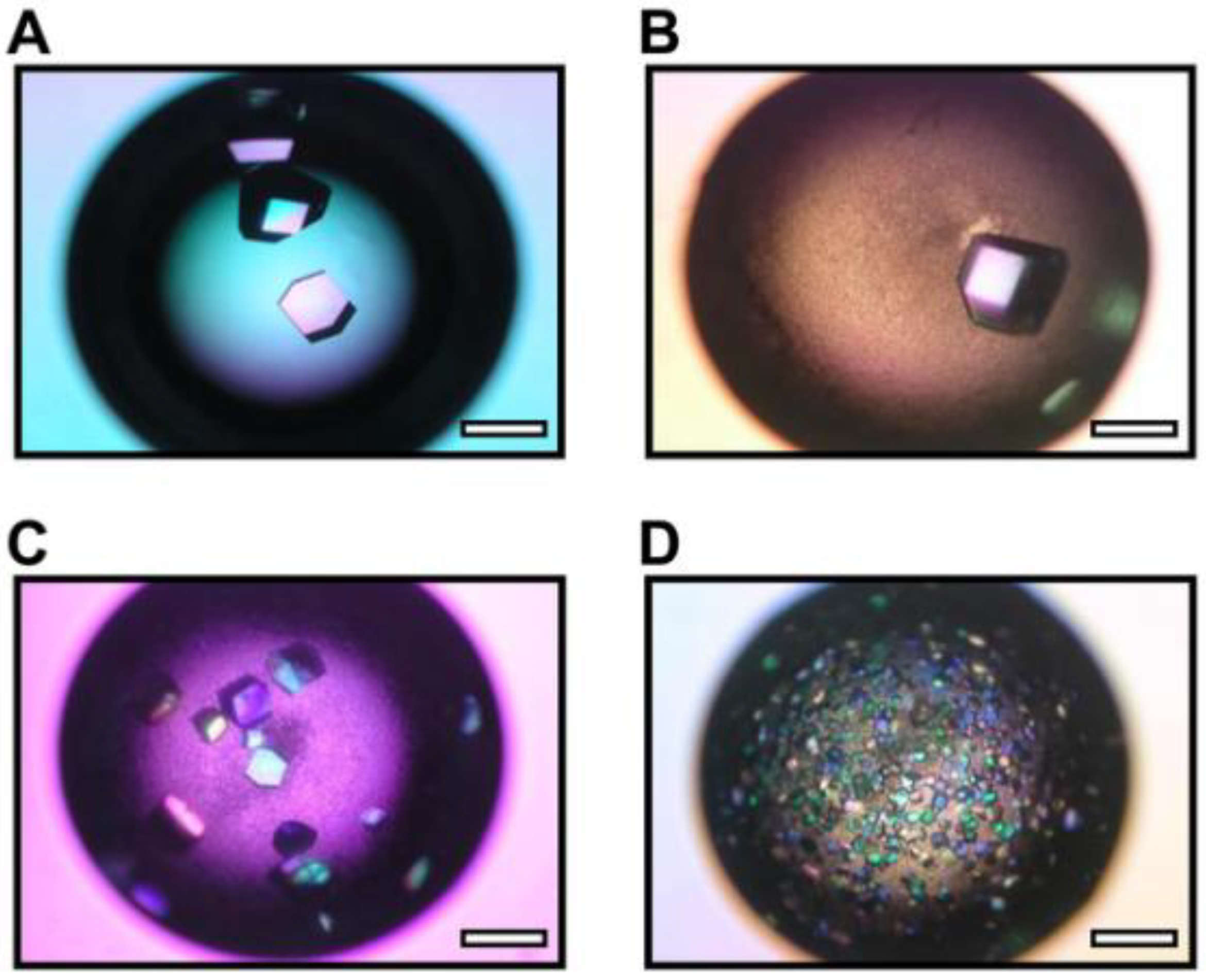
3.4. Nanoparticles
3.5. Ionic Liquids
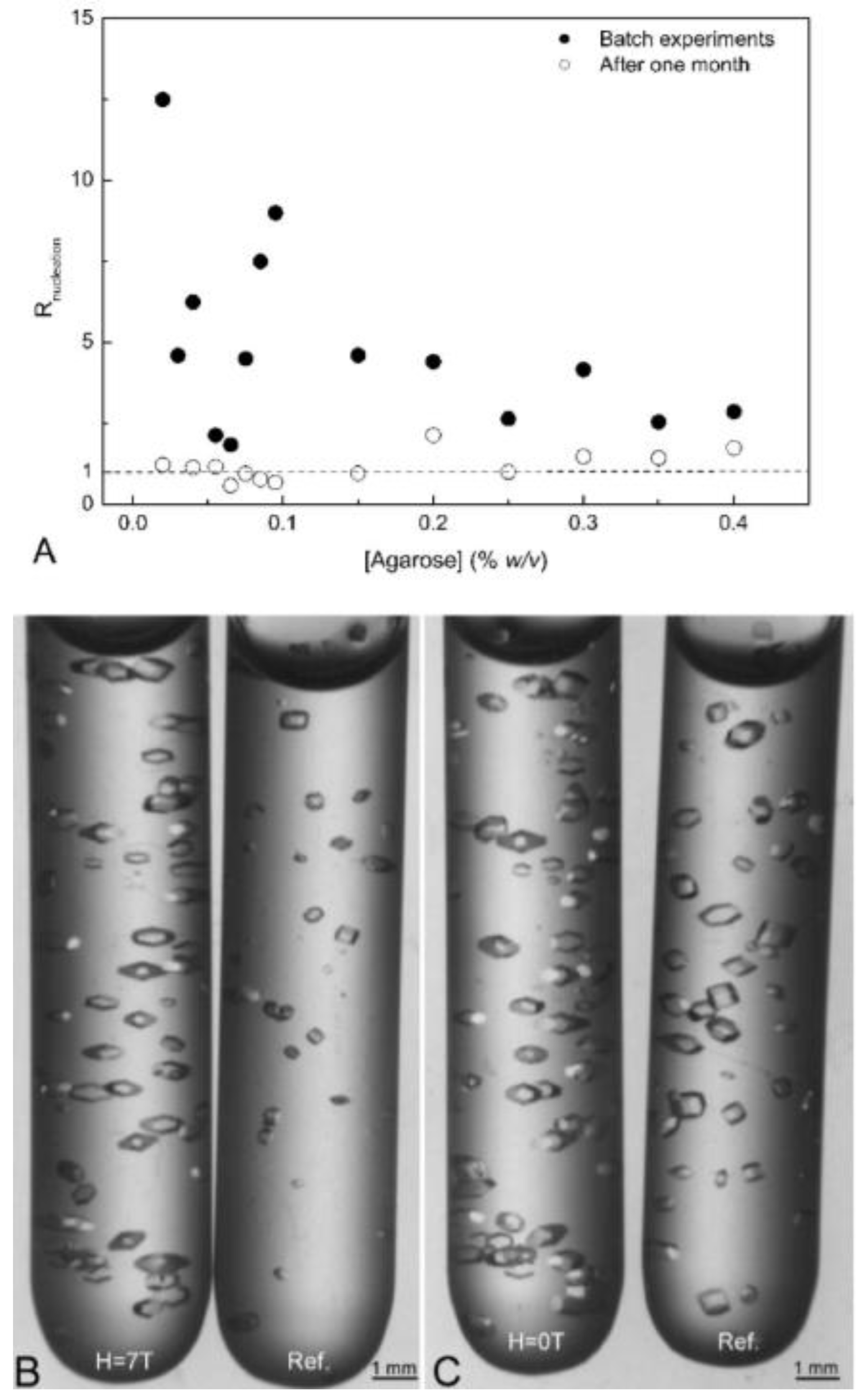
3.6. Porous Materials
4. Crystallization Strategies
4.1. Functional Interfaces
4.2. Electricity and Magnetic Fields
4.3. Ultrasonic Field
5. Applications of Protein Crystallization
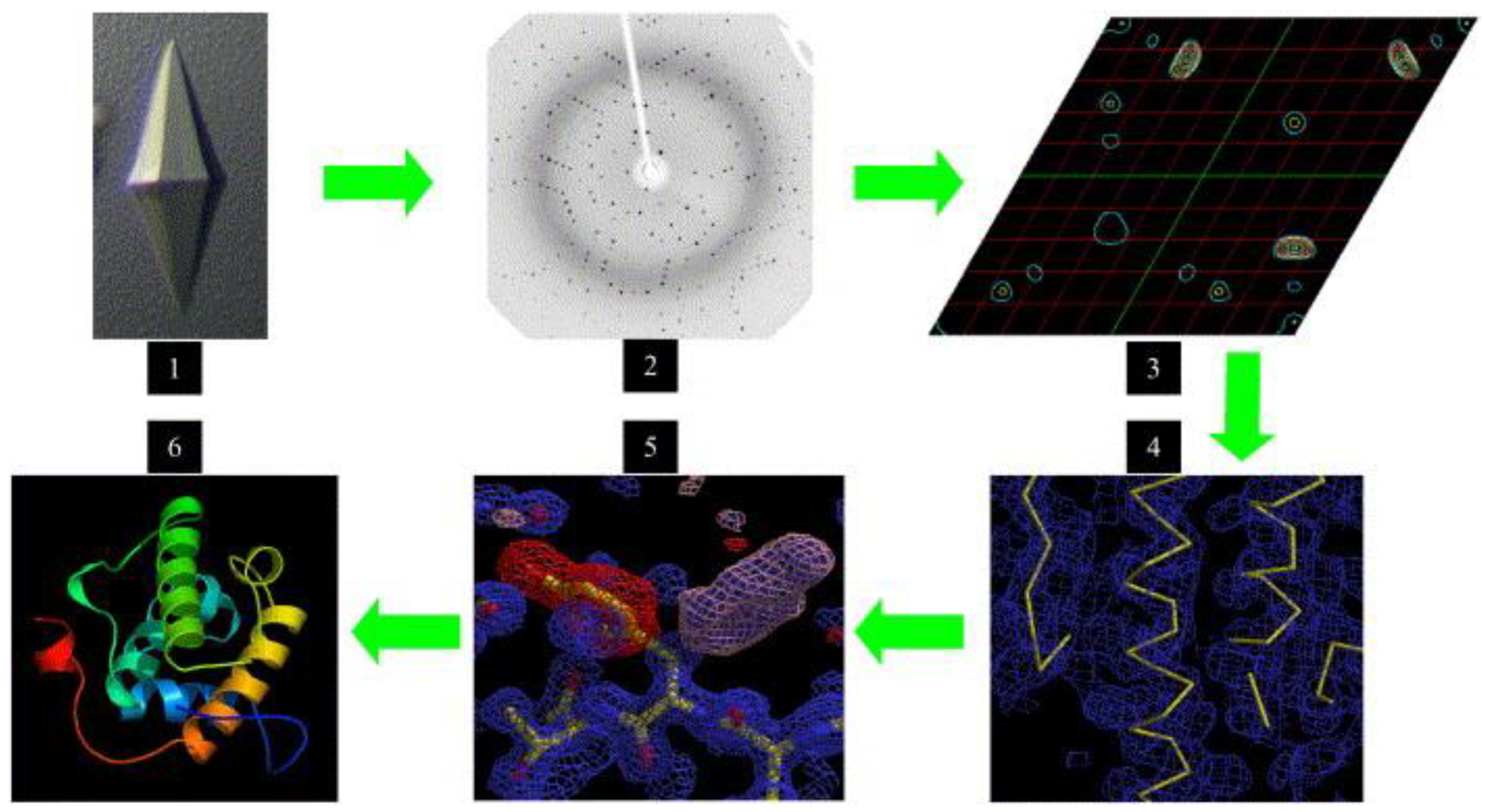
5.1. X-ray Crystallography
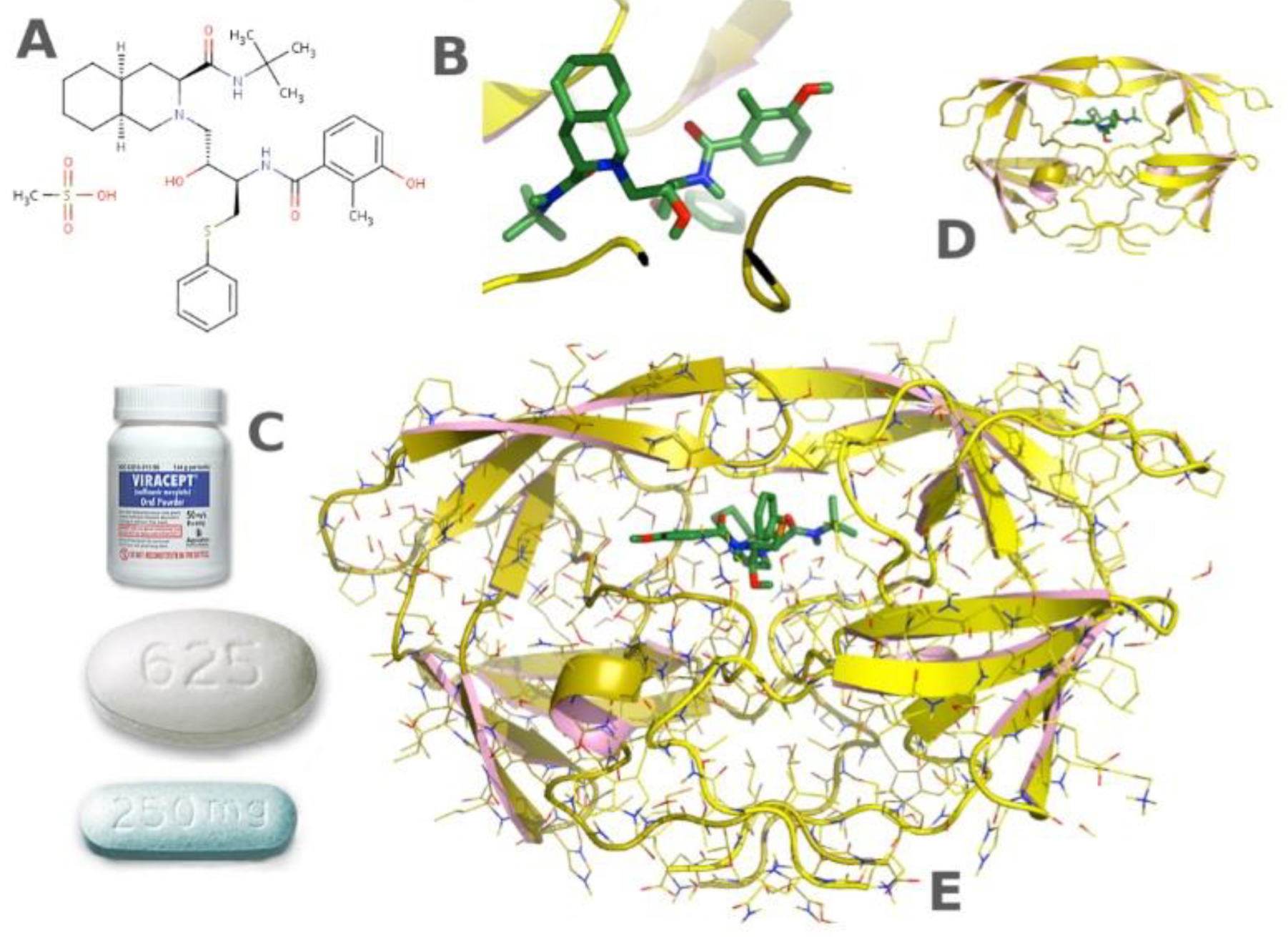
5.2. Pharmaceuticals
6. Discussion and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanvictores, T.; Farci, F. Biochemistry, Primary Protein Structure. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- McPherson, A. A brief history of protein crystal growth. J. Cryst. Growth 1991, 110, 1–10. [Google Scholar] [CrossRef]
- McPherson, A. Two approaches to the rapid screening of crystallization conditions. J. Cryst. Growth 1992, 122, 161–167. [Google Scholar] [CrossRef]
- Dale, G.E.; Oefner, C.; D’Arcy, A. The protein as a variable in protein crystallization. J. Struct. Biol. 2003, 142, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Chayen, N.E.; Saridakis, E. Protein crystallization: From purified protein to diffraction-quality crystal. Nat. Methods 2008, 5, 147–153. [Google Scholar] [CrossRef]
- Wlodawer, A.; Dauter, Z.; Jaskolski, M. Collection of X-Ray Diffraction Data from Macromolecular Crystals. In Protein Crystallography. Methods in Molecular Biology; Wlodawer, A., Dauter, Z., Jaskolski, M., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1607, pp. 165–184. [Google Scholar]
- Saridakis, E.; Chayen, N.E. Towards a “universal” nucleant for protein crystallization. Trends Biotechnol. 2009, 27, 99–106. [Google Scholar] [CrossRef]
- Löffelmann, M.; Mersmann, A. How to measure supersaturation? Chem. Eng. Sci. 2002, 57, 4301–4310. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Wu, Z.Q.; Yin, D.C.; Zhou, B.R.; Guo, Y.Z.; Lu, H.M.; Zhou, R.B.; Shang, P. A strategy for selecting the pH of protein solutions to enhance crystallization. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 821–826. [Google Scholar] [CrossRef]
- Zhang, C.-Y.; Yin, D.-C.; Lu, Q.-Q.; Guo, Y.; Guo, W.; Wang, X.; Li, H.; Lu, H.; Ye, Y. Cycling Temperature Strategy: A Method to Improve the Efficiency of Crystallization Condition Screening of Proteins. Cryst. Growth. Des. 2008, 8, 4227–4232. [Google Scholar] [CrossRef]
- Liu, J.; Yin, D.C.; Guo, Y.Z.; Wang, X.K.; Xie, S.X.; Lu, Q.Q.; Liu, Y.M. Selecting temperature for protein crystallization screens using the temperature dependence of the second virial coefficient. PLoS ONE 2011, 6, e17950. [Google Scholar] [CrossRef]
- Saridakis, E.; Dierks, K.; Moreno, A.; Dieckmann, M.W.M.; Chayen, N.E. Separating nucleation and growth in protein crystallization using dynamic light scattering. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 1597–1600. [Google Scholar] [CrossRef]
- Majeed, S.; Ofek, G.; Belachew, A.; Huang, C.-C.; Zhou, T.; Kwong, P.D. Enhancing Protein Crystallization through Precipitant Synergy. Structure 2003, 11, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A. Crystallization of Macromolecules: General Principles. In Crystallization and Treatment of Crystals; Academic Press: Cambridge, MA, USA, 1985. [Google Scholar]
- Muschol, M.; Rosenberger, F. Liquid–liquid phase separation in supersaturated lysozyme solutions and associated precipitate formation/crystallization. J. Chem. Phys. 1997, 107, 1953–1962. [Google Scholar] [CrossRef]
- Kundrot, C.E.; Judge, R.A.; Pusey, M.L.; Snell, E.H. Microgravity and Macromolecular Crystallography. Cryst. Growth. Des. 2001, 1, 87–99. [Google Scholar] [CrossRef]
- Cao, H.L.; Sun, L.H.; Li, J.; Tang, L.; Lu, H.M.; Guo, Y.Z.; He, J.; Liu, Y.M.; Xie, X.Z.; Shen, H.F.; et al. A quality comparison of protein crystals grown under containerless conditions generated by diamagnetic levitation, silicone oil and agarose gel. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, H.; Uda, S.; Fujiwara, K.; Nozawa, J. Control of effect on the nucleation rate for hen egg white lysozyme crystals under application of an external ac electric field. Langmuir 2011, 27, 8333–8338. [Google Scholar] [CrossRef]
- Koopmann, R.; Cupelli, K.; Redecke, L.; Nass, K.; Deponte, D.P.; White, T.A.; Stellato, F.; Rehders, D.; Liang, M.; Andreasson, J.; et al. In vivo protein crystallization opens new routes in structural biology. Nat. Methods 2012, 9, 259–262. [Google Scholar] [CrossRef]
- Otalora, F.; Gavira, J.A.; Ng, J.D.; Garcia-Ruiz, J.M. Counterdiffusion methods applied to protein crystallization. Prog. Biophys. Mol. Biol. 2009, 101, 26–37. [Google Scholar] [CrossRef]
- Jarmer, D.J.; Lengsfeld, C.S.; Anseth, K.S.; Randolph, T.W. Supercritical fluid crystallization of griseofulvin: Crystal habit modification with a selective growth inhibitor. J. Pharm. Sci. 2005, 94, 2688–2702. [Google Scholar] [CrossRef]
- Carruthers, C.W., Jr.; Gerdts, C.; Johnson, M.D.; Webb, P. A microfluidic, high throughput protein crystal growth method for microgravity. PLoS ONE 2013, 8, e82298. [Google Scholar] [CrossRef]
- Moreno, A. Advanced Methods of Protein Crystallization. Methods Mol. Biol. 2017, 1607, 51–76. [Google Scholar] [CrossRef]
- Biertümpfel, C.; Basquin, J.; Suck, D.; Sauter, C. Crystallization of biological macromolecules using agarose gel. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Mueller-Dieckmann, J. Automation in biological crystallization. Acta Crystallogr. F Struct. Biol. Commun. 2014, 70, 686–696. [Google Scholar] [CrossRef]
- Stewart, L.; Clark, R.; Behnke, C. High-throughput crystallization and structure determination in drug discovery. Drug Discov. Today 2002, 7, 187–196. [Google Scholar] [CrossRef]
- Zhou, R.-B.; Cao, H.-L.; Zhang, C.-Y.; Yin, D.-C. A review on recent advances for nucleants and nucleation in protein crystallization. CrystEngComm 2017, 19, 1143–1155. [Google Scholar] [CrossRef]
- Vekilov, P.G. Nucleation. Cryst. Growth Des. 2010, 10, 5007–5019. [Google Scholar] [CrossRef] [PubMed]
- Tosi, G.; Fermani, S.; Falini, G.; Gavira, J.A.; Garcia Ruiz, J.M. Hetero- vs Homogeneous Nucleation of Protein Crystals Discriminated by Supersaturation. Cryst. Growth Des. 2011, 11, 1542–1548. [Google Scholar] [CrossRef]
- Nanev, C. Recent Insights into Protein Crystal Nucleation. Crystals 2018, 8, 219. [Google Scholar] [CrossRef]
- Wolde, P.R.T.; Frenkel, D. Enhancement of protein crystal nucleation by critical density fluctuations. Science 1997, 277, 1975–1978. [Google Scholar] [CrossRef]
- Erdemir, D.; Lef, A.Y.; Myerson, A.S. Nucleation of Crystals from Solution: Classical and Two-Step Models. Acc. Chem. Res. 2009, 42, 621–629. [Google Scholar] [CrossRef]
- Shi, Z.; Wei, Y.; Zhu, C.; Sun, J.; Li, Z. Crystallization-Driven Two-Dimensional Nanosheet from Hierarchical Self-Assembly of Polypeptoid-Based Diblock Copolymers. Macromolecules 2018, 51, 6344–6351. [Google Scholar] [CrossRef]
- Sun, J.; Wang, Z.; Zhu, C.; Wang, M.; Shi, Z.; Wei, Y.; Fu, X.; Chen, X.; Zuckermann, R.N. Hierarchical supramolecular assembly of a single peptoid polymer into a planar nanobrush with two distinct molecular packing motifs. Proc. Natl. Acad. Sci. USA 2020, 117, 31639–31647. [Google Scholar] [CrossRef] [PubMed]
- Manuel García-Ruiz, J. Nucleation of protein crystals. J. Struct. Biol. 2003, 142, 22–31. [Google Scholar] [CrossRef]
- Sear, R.P. Nucleation: Theory and applications to protein solutions and colloidal suspensions. J. Phys. Condens. Matter 2007, 19, 033101. [Google Scholar] [CrossRef]
- Tuovinen, S.; Kontkanen, J.; Jiang, J.; Kulmala, M. Investigating the effectiveness of condensation sink based on heterogeneous nucleation theory. J. Aerosol Sci. 2020, 149, 105613. [Google Scholar] [CrossRef]
- Khurshid, S.; Saridakis, E.; Govada, L.; Chayen, N.E. Porous nucleating agents for protein crystallization. Nat. Protoc. 2014, 9, 1621–1633. [Google Scholar] [CrossRef]
- Thakur, A.S.; Robin, G.; Guncar, G.; Saunders, N.F.; Newman, J.; Martin, J.L.; Kobe, B. Improved success of sparse matrix protein crystallization screening with heterogeneous nucleating agents. PLoS ONE 2007, 2, e1091. [Google Scholar] [CrossRef]
- McPherson, A.; Shlichta, P. Heterogeneous and Epitaxial Nucleation of Protein Crystals on Mineral Surfaces. Science 1988, 239, 385–387. [Google Scholar] [CrossRef]
- Artusio, F.; Castellví, A.; Sacristán, A.; Pisano, R.; Gavira, J.A. Agarose Gel as a Medium for Growing and Tailoring Protein Crystals. Cryst. Growth. Des. 2020, 20, 5564–5571. [Google Scholar] [CrossRef]
- Artusio, F.; Castellví, A.; Pisano, R.; Gavira, J.A. Tuning Transport Phenomena in Agarose Gels for the Control of Protein Nucleation Density and Crystal Form. Crystals 2021, 11, 466. [Google Scholar] [CrossRef]
- Conejero-Muriel, M.; Contreras-Montoya, R.; Díaz-Mochón, J.J.; Álvarez de Cienfuegos, L.; Gavira, J.A. Protein crystallization in short-peptide supramolecular hydrogels: A versatile strategy towards biotechnological composite materials. CrystEngComm 2015, 17, 8072–8078. [Google Scholar] [CrossRef]
- Qing, G.; Shan, X.; Chen, W.; Lv, Z.; Xiong, P.; Sun, T. Solvent-Driven Chiral-Interaction Reversion for Organogel Formation. Angew. Chem. Int. Ed. 2014, 53, 2124–2129. [Google Scholar] [CrossRef]
- Conejero-Muriel, M.; Gavira, J.A.; Pineda-Molina, E.; Belsom, A.; Bradley, M.; Moral, M.; Garcia-Lopez Duran Jde, D.; Luque Gonzalez, A.; Diaz-Mochon, J.J.; Contreras-Montoya, R.; et al. Influence of the chirality of short peptide supramolecular hydrogels in protein crystallogenesis. Chem. Commun. 2015, 51, 3862–3865. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Montoya, R.; Arredondo-Amador, M.; Escolano-Casado, G.; Manas-Torres, M.C.; Gonzalez, M.; Conejero-Muriel, M.; Bhatia, V.; Diaz-Mochon, J.J.; Martinez-Augustin, O.; de Medina, F.S.; et al. Insulin Crystals Grown in Short-Peptide Supramolecular Hydrogels Show Enhanced Thermal Stability and Slower Release Profile. ACS Appl. Mater. Interfaces 2021, 13, 11672–11682. [Google Scholar] [CrossRef]
- Tao, K.; Levin, A.; Adler-Abramovich, L.; Gazit, E. Fmoc-modified amino acids and short peptides: Simple bio-inspired building blocks for the fabrication of functional materials. Chem. Soc. Rev. 2016, 45, 3935–3953. [Google Scholar] [CrossRef]
- Fleming, S.; Ulijn, R.V. Design of nanostructures based on aromatic peptide amphiphiles. Chem. Soc. Rev. 2014, 43, 8150–8177. [Google Scholar] [CrossRef]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 17068. [Google Scholar] [CrossRef]
- Seeman, N.C. Nucleic Acid Junctions and Lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Zhang, B.; Mei, A.R.; Isbell, M.A.; Wang, D.; Wang, Y.; Tan, S.F.; Teo, X.L.; Xu, L.; Yang, Z.; Heng, J.Y.Y. DNA Origami as Seeds for Promoting Protein Crystallization. ACS Appl. Mater. Interfaces 2018, 10, 44240–44246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Y.; Thi, S.; Toong, V.; Luo, P.; Fan, S.; Xu, L.; Yang, Z.Q.; Heng, J. Enhancement of Lysozyme Crystallization Using DNA as a Polymeric Additive. Crystals 2019, 9, 186. [Google Scholar] [CrossRef]
- Tosi, G.; Fermani, S.; Falini, G.; Gavira Gallardo, J.A.; Garcia Ruiz, J.M. Crystallization of proteins on functionalized surfaces. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 1054–1061. [Google Scholar] [CrossRef]
- Mohanraj, V.; Chen, Y. Nanoparticles—A Review. Trop. J. Pharm. Res. 2006, 5, 561–573. [Google Scholar] [CrossRef]
- Hodzhaoglu, F.; Kurniawan, F.; Mirsky, V.; Nanev, C. Gold nanoparticles induce protein crystallization. Cryst. Res. Technol. 2008, 43, 588–593. [Google Scholar] [CrossRef]
- Holcomb, J.; Spellmon, N.; Zhang, Y.; Doughan, M.; Li, C.; Yang, Z. Protein crystallization: Eluding the bottleneck of X-ray crystallography. AIMS Biophys. 2017, 4, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Mafuné, F. Induction of protein crystallization by platinum nanoparticles. Chem. Phys. Lett. 2016, 647, 181–184. [Google Scholar] [CrossRef]
- Chen, W.; Cheng, T.N.H.; Khaw, L.F.; Li, X.; Yang, H.; Ouyang, J.; Heng, J.Y.Y. Protein purification with nanoparticle-enhanced crystallisation. Sep. Purif. Technol. 2021, 255, 117384. [Google Scholar] [CrossRef]
- Chen, Y.W.; Lee, C.H.; Wang, Y.L.; Li, T.L.; Chang, H.C. Nanodiamonds as Nucleating Agents for Protein Crystallization. Langmuir 2017, 33, 6521–6527. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Lee, G.; Jang, K.; Jeong, D.W.; Lee, J.-O.; Kim, H.; Kim, Y.J. Graphene Quantum Dots as Nucleants for Protein Crystallization. Cryst. Growth Des. 2021, 22, 269–276. [Google Scholar] [CrossRef]
- Ribeiro, D.; Kulakova, A.; Quaresma, P.; Pereira, E.; Bonifácio, C.; Romão, M.J.; Franco, R.; Carvalho, A.L. Use of Gold Nanoparticles as Additives in Protein Crystallization. Cryst. Growth Des. 2013, 14, 222–227. [Google Scholar] [CrossRef]
- Belviso, B.D.; Caliandro, R.; Salehi, S.M.; Di Profio, G.; Caliandro, R. Protein Crystallization in Ionic-Liquid Hydrogel Composite Membranes. Crystals 2019, 9, 253. [Google Scholar] [CrossRef]
- Garlitz, J.A.; Summers, C.A.; Flowers, R.A., II; Borgstahl, G.E.O. Ethylammonium nitrate: A protein crystallization reagent. Acta Crystallogr. D Biol. Crystallogr. 1999, D55, 2037–2038. [Google Scholar] [CrossRef]
- Judge, R.A.; Takahashi, S.; Longenecker, K.L.; Fry, E.H.; Abad-Zapatero, C.; Chiu, M.L. The Effect of Ionic Liquids on Protein Crystallization and X-ray Diffraction Resolution. Cryst. Growth Des. 2009, 2009, 3463–3469. [Google Scholar] [CrossRef]
- Schroder, C. Proteins in Ionic Liquids: Current Status of Experiments and Simulations. Top. Curr. Chem. 2017, 375, 25. [Google Scholar] [CrossRef]
- Kowacz, M.; Marchel, M.; Juknaitė, L.; Esperança, J.M.S.S.; Romão, M.J.; Carvalho, A.L.; Rebelo, L.P.N. Ionic-Liquid-Functionalized Mineral Particles for Protein Crystallization. Cryst. Growth Des. 2015, 15, 2994–3003. [Google Scholar] [CrossRef]
- Chayen, E.N.; Saridakis, E.; Sear, R.P. Experiment and theory for heterogeneous nucleation of protein crystals in a porous medium. Proc. Natl. Acad. Sci. USA 2006, 103, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Z.; Sun, L.H.; Oberthuer, D.; Zhang, C.Y.; Shi, J.Y.; Di, J.L.; Zhang, B.L.; Cao, H.L.; Liu, Y.M.; Li, J.; et al. Utilisation of adsorption and desorption for simultaneously improving protein crystallization success rate and crystal quality. Sci. Rep. 2014, 4, 7308. [Google Scholar] [CrossRef] [PubMed]
- Chayen, N.E.; Saridakis, E.; El-Bahar, R.; Nemirovsky, Y. Porous silicon: An effective nucleation-inducing material for protein crystallization. J. Mol. Biol. 2001, 312, 591–595. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, X.; Zhang, J.; Fan, X.; Chen, M.; Yang, H. Nucleation, crystallization and biological activity of Na2O-CaO-P2O5-SiO2 bioactive glass. J. Non-Cryst. Solids 2021, 568, 120929. [Google Scholar] [CrossRef]
- Nanev, C.N.; Saridakis, E.; Chayen, N.E. Protein crystal nucleation in pores. Sci. Rep. 2017, 7, 35821. [Google Scholar] [CrossRef]
- Saridakis, E.; Khurshid, S.; Govada, L.; Phan, Q.; Hawkins, D.; Crichlow, G.V.; Lolis, E.; Reddy, S.M.; Chayen, N.E. Protein crystallization facilitated by molecularly imprinted polymers. Proc. Natl. Acad. Sci. USA 2011, 108, 11081–11086. [Google Scholar] [CrossRef]
- Xing, Y.; Hu, Y.; Jiang, L.; Gao, Z.; Chen, Z.; Chen, Z.; Ren, X. Zwitterion-Immobilized Imprinted Polymers for Promoting the Crystallization of Proteins. Cryst. Growth Des. 2015, 15, 4932–4937. [Google Scholar] [CrossRef]
- Saridakis, E.; Chayen, N.E. Imprinted polymers assisting protein crystallization. Trends Biotechnol. 2013, 31, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-Y.; Shen, H.-F.; Wang, Q.-J.; Guo, Y.-Z.; He, J.; Cao, H.-L.; Liu, Y.-M.; Shang, P.; Yin, D.-C. An Investigation of the Effects of Self-Assembled Monolayers on Protein Crystallisation. Int. J. Mol. Sci. 2013, 14, 12329–12345. [Google Scholar] [CrossRef]
- Ino, K.; Udagawa, I.; Iwabata, K.; Takakusagi, Y.; Kubota, M.; Kurosaka, K.; Arai, K.; Seki, Y.; Nogawa, M.; Tsunoda, T.; et al. Heterogeneous nucleation of protein crystals on fluorinated layered silicate. PLoS ONE 2011, 6, e22582. [Google Scholar] [CrossRef]
- Tang, L.; Huang, Y.B.; Liu, D.Q.; Li, J.L.; Mao, K.; Liu, L.; Cheng, Z.J.; Gong, W.M.; Hu, J.; He, J.H. Effects of the silanized mica surface on protein crystallization. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 53–59. [Google Scholar] [CrossRef]
- Hemming, S.A.; Bochkarev, A.; Kornberg, R.D.; Ala, P.; Edwards, A.M. The Mechanism of Protein Crystal Growth from Lipid Layers. J. Mol. Biol. 1995, 246, 308–316. [Google Scholar] [CrossRef]
- Wallace, E.; Dranow, D.; Laible, P.D.; Christensen, J.; Nollert, P. Monoolein lipid phases as incorporation and enrichment materials for membrane protein crystallization. PLoS ONE 2011, 6, e24488. [Google Scholar] [CrossRef] [PubMed]
- Fermani, S.; Falini, G.; Minnucci, M.; Ripamonti, A. Protein crystallization on polymeric film surfaces. J. Cryst. Growth 2001, 224, 327–334. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Wang, X.-J.; Lu, J.; Ching, C.-B. Influence of the Roughness, Topography, and Physicochemical Properties of Chemically Modified Surfaces on the Heterogeneous Nucleation of Protein Crystals. J. Phys. Chem. B 2007, 111, 13971–19378. [Google Scholar] [CrossRef]
- Guo, Y.-Z.; Yin, D.-C.; Lu, Q.-Q.; Wang, X.-K.; Liu, J. Enhancement of nucleation during hanging drop protein crystallization using HF Treatment of cover glasses. Cryst. Res. Technol. 2010, 45, 158–166. [Google Scholar] [CrossRef]
- Pham, T.; Lai, D.; Ji, D.; Tuntiwechapikul, W.; Friedman, J.M.; Lee, T.R. Well-ordered self-assembled monolayer surfaces can be used to enhance the growth of protein crystals. Colloids Surf. B Biointerfaces 2004, 34, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Ji, D.; Arnold, C.M.; Graupe, M.; Beadle, E.; Dunn, R.V.; Phan, M.N.; Villazana, R.J.; Benson, R.; Colorado, R., Jr; Lee, T.R.; et al. Improved protein crystallization by vapor diffusion from drops in contact with transparent, self-assembled monolayers on gold-coated glass coverslips. J. Cryst. Growth 2000, 218, 390–398. [Google Scholar] [CrossRef]
- Pechkova, E.; Nicolini, C. Accelerated protein crystal growth by protein thin film template. J. Cryst. Growth 2001, 231, 599–602. [Google Scholar] [CrossRef]
- Rothgeb, T.M.; Oldfield, E. Nuclear magnetic resonance of heme protein crystals. General aspects. J. Biol. Chem. 1981, 256, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Gavira, J.A.; Garcı’a-Ruiz, J.M. Effects of a Magnetic Field on Lysozyme Crystal Nucleation and Growth in a Diffusive Environment. Cryst. Growth Des. 2009, 9, 2610–2615. [Google Scholar] [CrossRef]
- Satoa, T.; Yamadaa, Y.; Saijoa, S.; Horia, T.; Hirosea, R.; Tanakaa, N.; Sazakib, G.; Nakajimab, K.; Igarashid, N.; Tanakad, M.; et al. Improvement in diffraction maxima in orthorhombic HEWL crystal grown under high magnetic field. J. Cryst. Growth 2001, 232, 229–236. [Google Scholar] [CrossRef]
- Nakamura, A.; Ohtsuka, J.; Miyazono, K.-i.; Yamamura, A.; Kubota, K.; Hirose, R.; Hirota, N.; Ataka, M.; Sawano, Y.; Tanokura, M. Improvement in Quality of Protein Crystals Grown in a High Magnetic Field Gradient. Cryst. Growth Des. 2012, 12, 1141–1150. [Google Scholar] [CrossRef]
- Sazaki, G.; Yoshida, E.; Komatsu, H.T.; Nakada, S.M.; Watanabe, K. Effects of a magnetic field on the nucleation and growth of protein crystals. J. Cryst. Growth 1997, 173, 231–234. [Google Scholar] [CrossRef]
- Ryu, S.; Oh, I.; Cho, S.; Kim, S.; Song, H. Enhancing Protein Crystallization under a Magnetic Field. Crystals 2020, 10, 821. [Google Scholar] [CrossRef]
- Chin, C.C.; Dence, J.B.; Warren, J.C. Crystallization of human placental estradiol 17beta-dehydrogenase. A new method for crystallizing labile enzymes. J. Biol. Chem. 1976, 251, 3700–3705. [Google Scholar] [CrossRef]
- Moreno, A.; Sazaki, G. The use of a new ad hoc growth cell with parallel electrodes for the nucleation control of lysozyme. J. Cryst. Growth 2004, 264, 438–444. [Google Scholar] [CrossRef]
- Nanev, C.N.; Penkova, A. Nucleation and growth of lysozyme crystals under external electric field. Colloids Surf. A 2002, 209, 139–145. [Google Scholar] [CrossRef]
- Mirkin, N.; Frontana-Uribe, B.A.; Rodríguez-Romero, A.; Hernández-Santoyo, A.; Moreno, A. The influence of an internal electric field upon protein crystallization using the gel-acupuncture method. Acta Crystallogr. D Biol. Crystallogr. 2003, 59 Pt 9, 1533–1538. [Google Scholar] [CrossRef]
- Penkova, A.; Gliko, O.; Dimitrov, I.L.; Hodjaoglu, F.V.; Nanev, C.; Vekilov, P.G. Enhancement and suppression of protein crystal nucleation due to electrically driven convection. J. Cryst. Growth 2005, 275, e1527–e1532. [Google Scholar] [CrossRef]
- Taleb, M.; Didierjean, C.; Jelsch, C.; Mangeot, J.P.; Capelle, B.; Aubry, A. Crystallization of proteins under an external electric field. J. Cryst. Growth 1999, 200, 575–582. [Google Scholar] [CrossRef]
- Sabnis, S.S.; Gogate, P.R. Ultrasound assisted size reduction of DADPS based on recrystallization. Ultrason. Sonochem. 2019, 54, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.H.; Lee, J.; Padilla, S.G.; Martini, S. Altering functional properties of fats using power ultrasound. J. Food Sci. 2010, 75, E208–E214. [Google Scholar] [CrossRef]
- Yu, F.; Mao, Y.; Zhao, H.; Zhang, X.; Wang, T.; Yuan, M.; Ding, S.; Wang, N.; Huang, X.; Hao, H. Enhancement of Continuous Crystallization of Lysozyme through Ultrasound. Org. Process Res. Dev. 2021, 25, 2508–2515. [Google Scholar] [CrossRef]
- Crespo, R.; Martins, P.M.; Gales, L.; Rocha, F.; Damas, A.M. Potential use of ultrasound to promote protein crystallization. J. Appl. Crystallogr. 2010, 43, 1419–1425. [Google Scholar] [CrossRef]
- Mao, Y.; Li, F.; Wang, T.; Cheng, X.; Li, G.; Li, D.; Zhang, X.; Hao, H. Enhancement of lysozyme crystallization under ultrasound field. Ultrason. Sonochem. 2020, 63, 104975. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J. X-ray Crystallography of Biomolecules. In Inventive Geniuses Who Changed the World; Springer: Cham, Switzerland, 2022; pp. 301–312. [Google Scholar]
- Pusey, M.L.; Liu, Z.J.; Tempel, W.; Praissman, J.; Lin, D.; Wang, B.C.; Gavira, J.A.; Ng, J.D. Life in the fast lane for protein crystallization and X-ray crystallography. Prog. Biophys. Mol. Biol. 2005, 88, 359–386. [Google Scholar] [CrossRef]
- Stojanoff, V.; Northrup, P.; Pietri, R.; Zhong, Z. Synchrotron Radiation in Life Sciences. Protein Pept. Lett. 2012, 19, 761–769. [Google Scholar] [CrossRef]
- Kojić-Prodić, B. A century of X-ray crystallography and 2014 international year of X-ray crystallography. Maced. J. Chem. Cheml. Eng. 2015, 34, 19–32. [Google Scholar] [CrossRef]
- Sherwood, D.; Cooper, J. Crystals, X-rays and Proteins: Comprehensive Protein Crystallography; Sherwood, D., Ed.; Oxford University Press: Oxford, UK, 2010. [Google Scholar]
- Yonekura, K.; Kato, K.; Ogasawara, M.; Tomita, M.; Toyoshima, C. Electron crystallography of ultrathin 3D protein crystals: Atomic model with charges. Proc. Natl. Acad. Sci. USA 2015, 112, 3368–3373. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, K.J. Developing a macromolecular crystallography driven CURE. Struct. Dyn. 2021, 8, 020406. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, D.V.; Nizovtseva, I.G. On the theory of crystal growth in metastable systems with biomedical applications: Protein and insulin crystallization. Philos. Trans. A Math. Phys. Eng. Sci. 2019, 377, 20180214. [Google Scholar] [CrossRef]
- Klabunde, T.; Hessler, G. Drug Design Strategies for Targeting G-Protein-Coupled Receptors. Chembiochem 2002, 3, 928–944. [Google Scholar] [CrossRef]
- Reed, J.C. Apoptosis-regulating proteins as targets for drug discovery. Trends Mol. Med. 2001, 7, 314–319. [Google Scholar] [CrossRef]
- Stura, E.A. Protein crystallization for drug design in the last 50 years. Arbor 2015, 191, 8. [Google Scholar] [CrossRef][Green Version]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Zhao, Y.; Sun, J. Heterogeneous Nucleation in Protein Crystallization. Biomimetics 2023, 8, 68. https://doi.org/10.3390/biomimetics8010068
Liu H, Zhao Y, Sun J. Heterogeneous Nucleation in Protein Crystallization. Biomimetics. 2023; 8(1):68. https://doi.org/10.3390/biomimetics8010068
Chicago/Turabian StyleLiu, Hao, Yue Zhao, and Jing Sun. 2023. "Heterogeneous Nucleation in Protein Crystallization" Biomimetics 8, no. 1: 68. https://doi.org/10.3390/biomimetics8010068
APA StyleLiu, H., Zhao, Y., & Sun, J. (2023). Heterogeneous Nucleation in Protein Crystallization. Biomimetics, 8(1), 68. https://doi.org/10.3390/biomimetics8010068







