Macromolecules Absorbed from Influenza Infection-Based Sera Modulate the Cellular Uptake of Polymeric Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Nanoparticle Synthesis, Cargo Encapsulation, and PEG Functionalization
2.2. IAV Infection and Recovery of Sera for Treating HBPE-NPs
2.3. Dynamic Light Scattering (DLS) Analysis and IgG Detection
2.4. Cell Culture
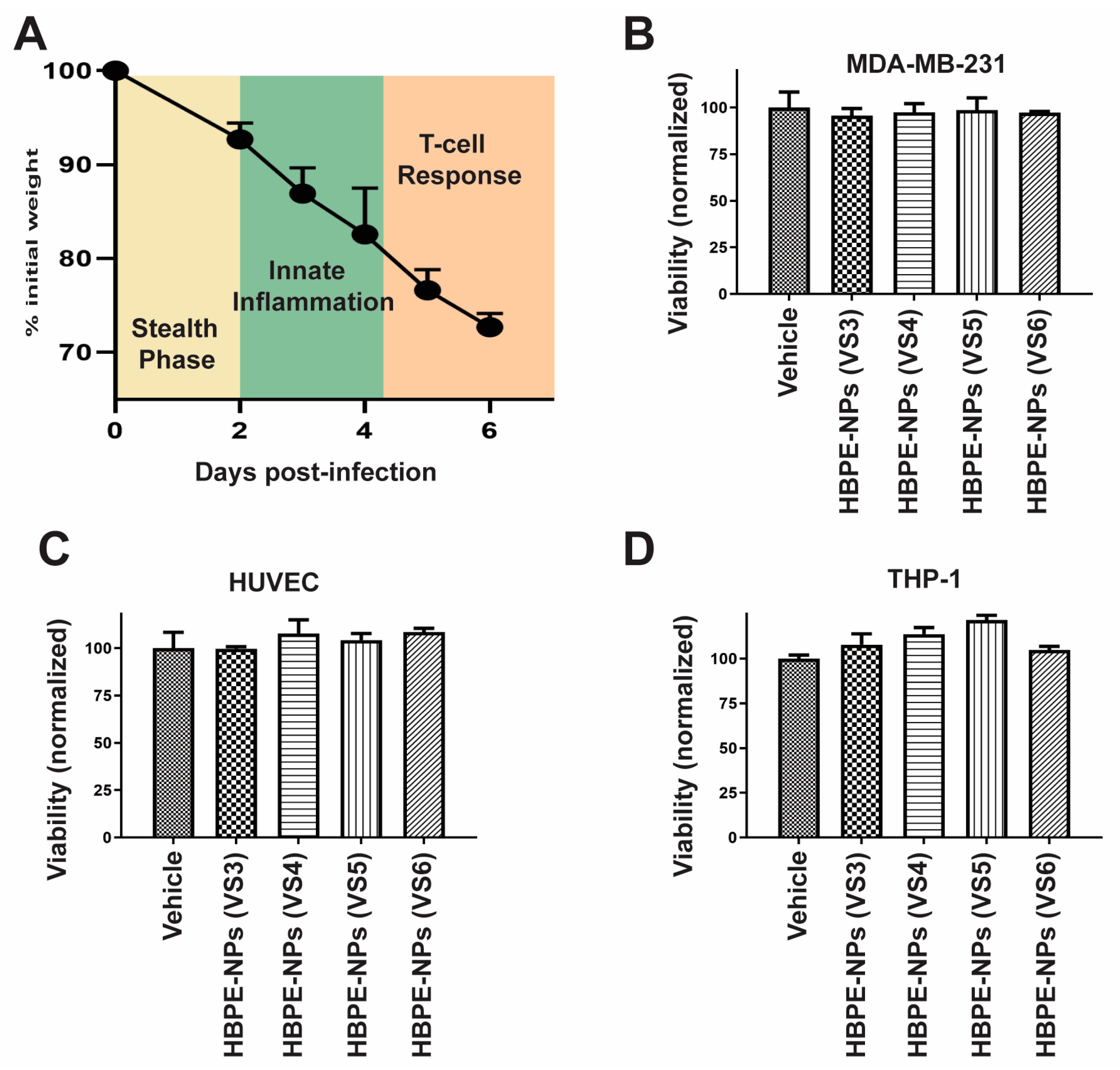
2.5. Viability Assay
2.6. In Vitro Assays and Imaging
2.7. Chemotactic Transwell (CT) Protocol
2.8. Modified Transwell Assay
2.9. Biodistribution Studies
2.10. Gel Electrophoresis and Immunoblotting
2.11. Identification of Proteins Absorbed by NPs Using Mass Spectrometry
2.12. Statistical Analysis and Databases
3. Results
3.1. Formation of Protein Corona on HBPE-NPs Using Sera from IAV-Infected Mice
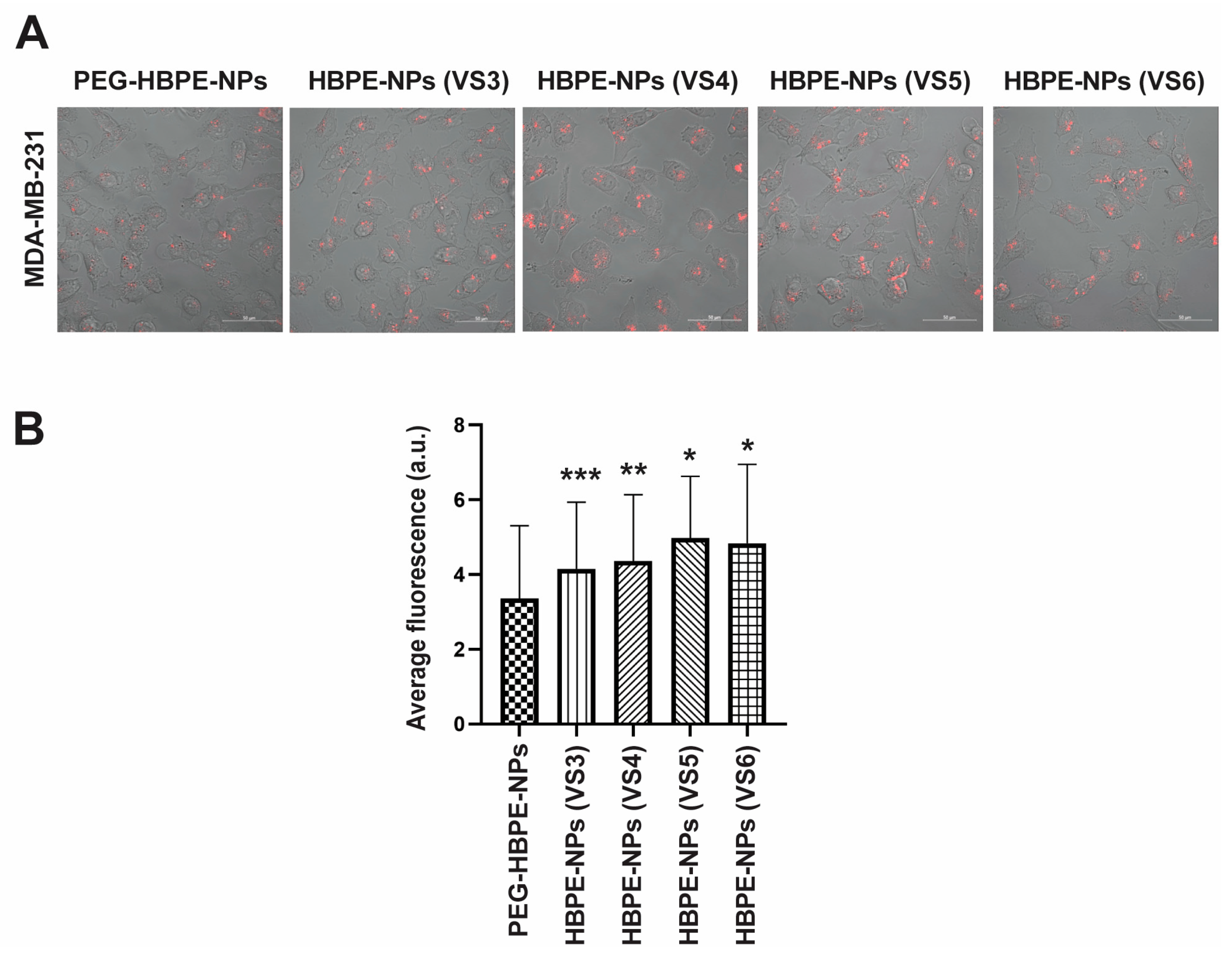
3.2. Direct Uptake of HBPE-NPs Pre-Treated with Sera from IAV-Infected Mice by Monocytic, Endothelial, and Breast Cancer Cells
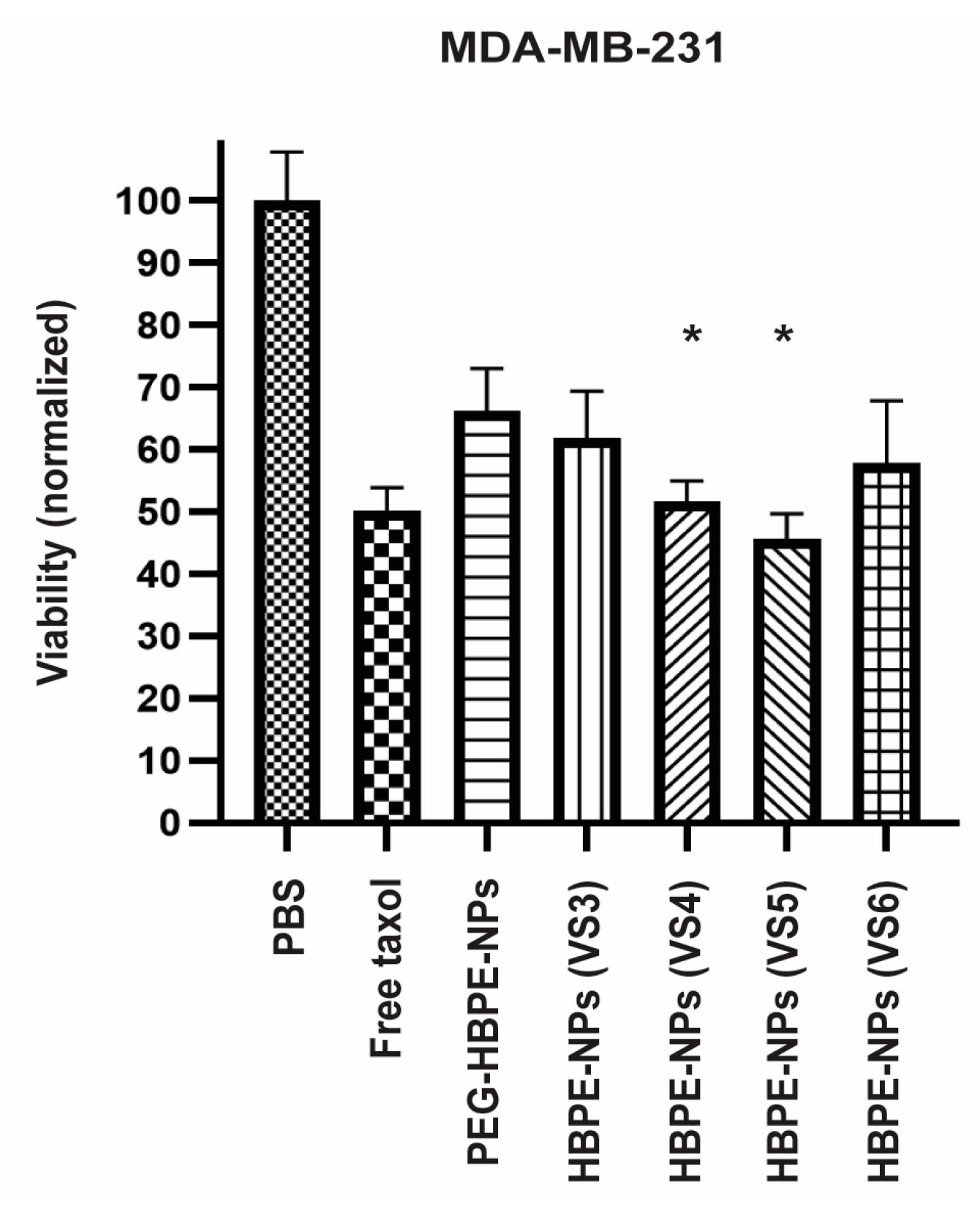
3.3. Drug Delivery to Cancer Cells Using HBPE-NPs Pre-Treated with Sera from IAV-Infected Mice
3.4. Cancer Cell Uptake of HBPE-NPs Pre-Treated with Sera from IAV-Infected Mice after Interaction with an Endothelial Layer
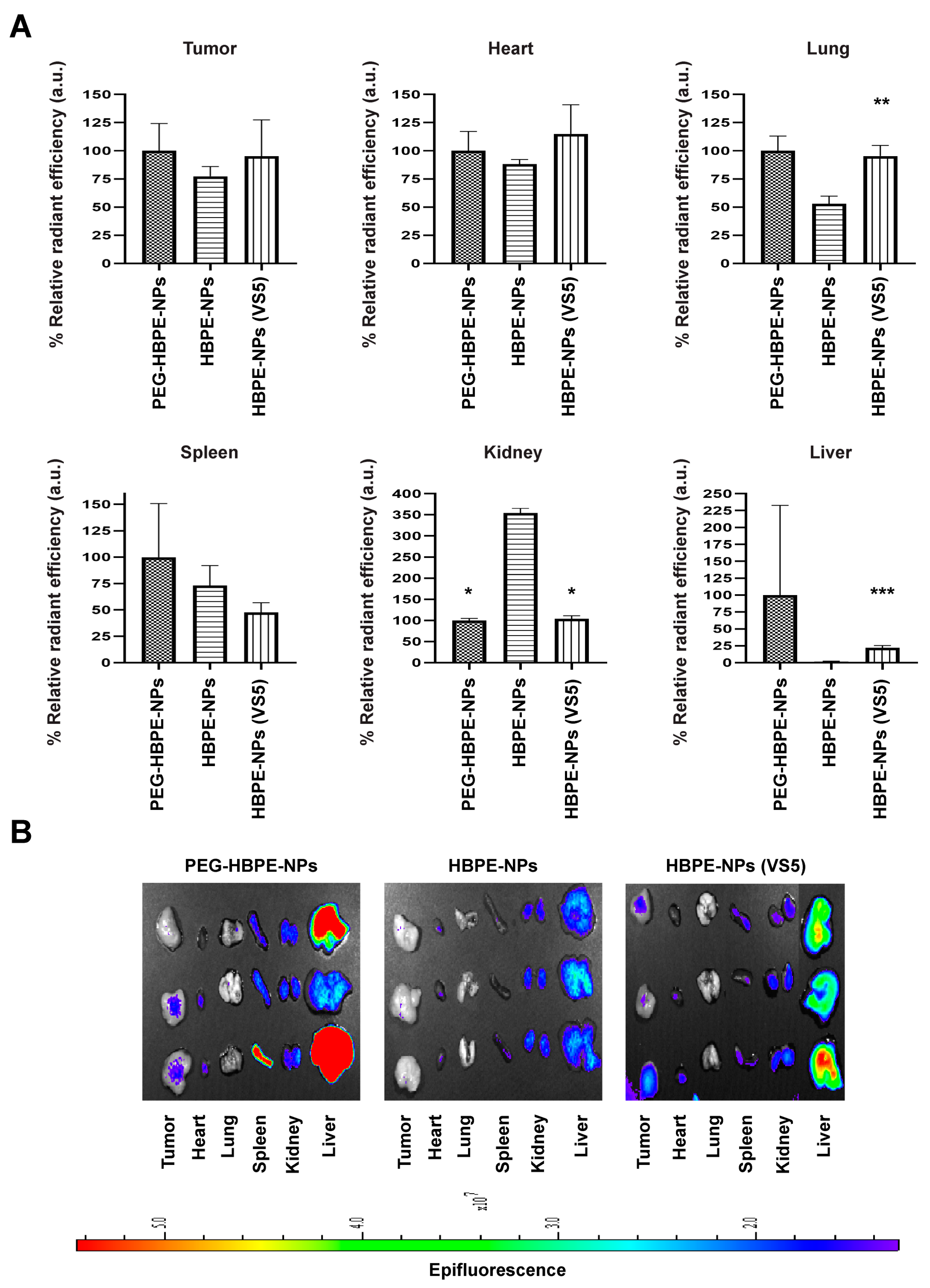
3.5. In Vivo Biodistribution of HBPE-NPs Pre-Treated with VS5
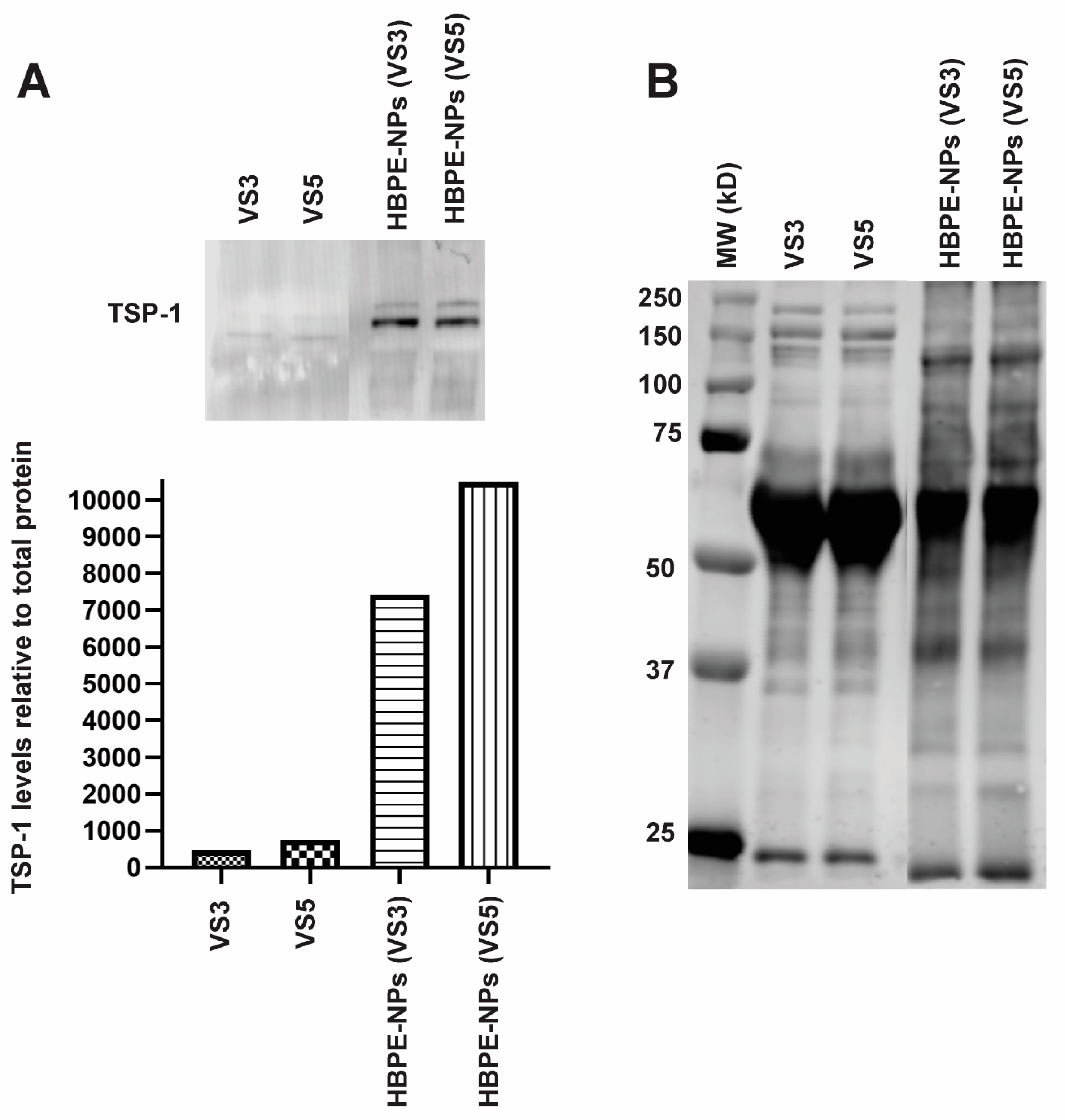
3.6. Identification of Proteins Absorbed by HBPE-NPs Pre-Treated with VS3 and VS5
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, W.; Huang, Z.; Li, Y.; Wang, W.; Shi, J.; Fu, F.; Huang, Y.; Pan, X.; Wu, C. Impact of particle size and pH on protein corona formation of solid lipid nanoparticles: A proof-of-concept study. Acta Pharm. Sin. B 2021, 11, 1030–1046. [Google Scholar] [CrossRef] [PubMed]
- Pustulka, S.M.; Ling, K.; Pish, S.L.; Champion, J.A. Protein Nanoparticle Charge and Hydrophobicity Govern Protein Corona and Macrophage Uptake. ACS Appl. Mater. Interfaces 2020, 12, 48284–48295. [Google Scholar] [CrossRef] [PubMed]
- Madathiparambil Visalakshan, R.; Gonzalez Garcia, L.E.; Benzigar, M.R.; Ghazaryan, A.; Simon, J.; Mierczynska-Vasilev, A.; Michl, T.D.; Vinu, A.; Mailander, V.; Morsbach, S.; et al. The Influence of Nanoparticle Shape on Protein Corona Formation. Small 2020, 16, e2000285. [Google Scholar] [CrossRef] [PubMed]
- Gref, R.; Luck, M.; Quellec, P.; Marchand, M.; Dellacherie, E.; Harnisch, S.; Blunk, T.; Muller, R.H. ‘Stealth’ corona-core nanoparticles surface modified by polyethylene glycol (PEG): Influences of the corona (PEG chain length and surface density) and of the core composition on phagocytic uptake and plasma protein adsorption. Colloids Surf. B Biointerfaces 2000, 18, 301–313. [Google Scholar] [CrossRef]
- Kozma, G.T.; Shimizu, T.; Ishida, T.; Szebeni, J. Anti-PEG antibodies: Properties, formation, testing and role in adverse immune reactions to PEGylated nano-biopharmaceuticals. Adv. Drug Deliv. Rev. 2020, 154–155, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, H.; Akita, H.; Harashima, H. The polyethyleneglycol dilemma: Advantage and disadvantage of PEGylation of liposomes for systemic genes and nucleic acids delivery to tumors. Biol. Pharm. Bull. 2013, 36, 892–899. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, X.; Huang, J.; Wang, J.; Wang, Z. Harnessing Protein Corona for Biomimetic Nanomedicine Design. Biomimetics 2022, 7, 126. [Google Scholar] [CrossRef]
- Simon, J.; Kuhn, G.; Fichter, M.; Gehring, S.; Landfester, K.; Mailander, V. Unraveling the In Vivo Protein Corona. Cells 2021, 10, 132. [Google Scholar] [CrossRef]
- Kokkinopoulou, M.; Simon, J.; Landfester, K.; Mailander, V.; Lieberwirth, I. Visualization of the protein corona: Towards a biomolecular understanding of nanoparticle-cell-interactions. Nanoscale 2017, 9, 8858–8870. [Google Scholar] [CrossRef]
- Kari, O.K.; Ndika, J.; Parkkila, P.; Louna, A.; Lajunen, T.; Puustinen, A.; Viitala, T.; Alenius, H.; Urtti, A. In situ analysis of liposome hard and soft protein corona structure and composition in a single label-free workflow. Nanoscale 2020, 12, 1728–1741. [Google Scholar] [CrossRef]
- Srivastava, I.; Khan, M.S.; Dighe, K.; Alafeef, M.; Wang, Z.; Banerjee, T.; Ghonge, T.; Grove, L.M.; Bashir, R.; Pan, D. On-Chip Electrical Monitoring of Real-Time “Soft” and “Hard” Protein Corona Formation on Carbon Nanoparticles. Small Methods 2020, 4, 2000099. [Google Scholar] [CrossRef]
- Vroman, L. Effect of absorbed proteins on the wettability of hydrophilic and hydrophobic solids. Nature 1962, 196, 476–477. [Google Scholar] [CrossRef] [PubMed]
- Tenzer, S.; Docter, D.; Kuharev, J.; Musyanovych, A.; Fetz, V.; Hecht, R.; Schlenk, F.; Fischer, D.; Kiouptsi, K.; Reinhardt, C.; et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nat. Nanotechnol. 2013, 8, 772–781. [Google Scholar] [CrossRef] [PubMed]
- Schottler, S.; Klein, K.; Landfester, K.; Mailander, V. Protein source and choice of anticoagulant decisively affect nanoparticle protein corona and cellular uptake. Nanoscale 2016, 8, 5526–5536. [Google Scholar] [CrossRef] [PubMed]
- Bonvin, D.; Aschauer, U.; Alexander, D.T.L.; Chiappe, D.; Moniatte, M.; Hofmann, H.; Mionic Ebersold, M. Protein Corona: Impact of Lymph Versus Blood in a Complex In Vitro Environment. Small 2017, 13, 1700409. [Google Scholar] [CrossRef] [PubMed]
- Kianfar, E. Protein nanoparticles in drug delivery: Animal protein, plant proteins and protein cages, albumin nanoparticles. J. Nanobiotechnol. 2021, 19, 159. [Google Scholar] [CrossRef]
- Ngo, W.; Wu, J.L.Y.; Lin, Z.P.; Zhang, Y.; Bussin, B.; Granda Farias, A.; Syed, A.M.; Chan, K.; Habsid, A.; Moffat, J.; et al. Identifying cell receptors for the nanoparticle protein corona using genome screens. Nat. Chem. Biol. 2022, 18, 1023–1031. [Google Scholar] [CrossRef]
- Santra, S.; Kaittanis, C.; Perez, J.M. Aliphatic hyperbranched polyester: A new building block in the construction of multifunctional nanoparticles and nanocomposites. Langmuir 2010, 26, 5364–5373. [Google Scholar] [CrossRef]
- Nierenberg, D.; Flores, O.; Fox, D.; Sip, Y.Y.L.; Finn, C.; Ghozlan, H.; Cox, A.; McKinstry, K.K.; Zhai, L.; Khaled, A.R. Polymeric Nanoparticles with a Sera-Derived Coating for Efficient Cancer Cell Uptake and Killing. ACS Omega 2021, 6, 5591–5606. [Google Scholar] [CrossRef]
- Wen, Y.; Lambrecht, J.; Ju, C.; Tacke, F. Hepatic macrophages in liver homeostasis and diseases-diversity, plasticity and therapeutic opportunities. Cell. Mol. Immunol. 2021, 18, 45–56. [Google Scholar] [CrossRef]
- Steenbrugge, J.; De Jaeghere, E.A.; Meyer, E.; Denys, H.; De Wever, O. Splenic Hematopoietic and Stromal Cells in Cancer Progression. Cancer Res. 2021, 81, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Di Domenico, M.; Pozzi, D.; Palchetti, S.; Digiacomo, L.; Iorio, R.; Astarita, C.; Fiorelli, A.; Pierdiluca, M.; Santini, M.; Barbarino, M.; et al. Nanoparticle-biomolecular corona: A new approach for the early detection of non-small-cell lung cancer. J. Cell. Physiol. 2019, 234, 9378–9386. [Google Scholar] [CrossRef] [PubMed]
- Colapicchioni, V.; Tilio, M.; Digiacomo, L.; Gambini, V.; Palchetti, S.; Marchini, C.; Pozzi, D.; Occhipinti, S.; Amici, A.; Caracciolo, G. Personalized liposome-protein corona in the blood of breast, gastric and pancreatic cancer patients. Int. J. Biochem. Cell Biol. 2016, 75, 180–187. [Google Scholar] [CrossRef]
- Xu, W.; Xu, M.; Xiao, Y.; Yu, L.; Xie, H.; Jiang, X.; Chen, M.; Gao, H.; Wang, L. Changes in target ability of nanoparticles due to protein corona composition and disease state. Asian J. Pharm. Sci. 2022, 17, 401–411. [Google Scholar] [CrossRef]
- Lichtenthaler, S.F.; Lemberg, M.K.; Fluhrer, R. Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments. EMBO J. 2018, 37, e99456. [Google Scholar] [CrossRef]
- Rahman, I.; Collado Sanchez, A.; Davies, J.; Rzeniewicz, K.; Abukscem, S.; Joachim, J.; Hoskins Green, H.L.; Killock, D.; Sanz, M.J.; Charras, G.; et al. L-selectin regulates human neutrophil transendothelial migration. J. Cell Sci. 2021, 134, jcs250340. [Google Scholar] [CrossRef]
- Yang, J.; LeBlanc, M.E.; Cano, I.; Saez-Torres, K.L.; Saint-Geniez, M.; Ng, Y.S.; D’Amore, P.A. ADAM10 and ADAM17 proteases mediate proinflammatory cytokine-induced and constitutive cleavage of endomucin from the endothelial surface. J. Biol. Chem. 2020, 295, 6641–6651. [Google Scholar] [CrossRef]
- Mishra, H.K.; Ma, J.; Walcheck, B. Ectodomain Shedding by ADAM17: Its Role in Neutrophil Recruitment and the Impairment of This Process during Sepsis. Front. Cell. Infect. Microbiol. 2017, 7, 138. [Google Scholar] [CrossRef]
- Zheng, T.; Crews, J.; McGill, J.L.; Dhume, K.; Finn, C.; Strutt, T.; McKinstry, K.K.; Huo, Q. A Single-Step Gold Nanoparticle-Blood Serum Interaction Assay Reveals Humoral Immunity Development and Immune Status of Animals from Neonates to Adults. ACS Infect. Dis. 2019, 5, 228–238. [Google Scholar] [CrossRef]
- Zheng, T.; Pierre-Pierre, N.; Yan, X.; Huo, Q.; Almodovar, A.J.; Valerio, F.; Rivera-Ramirez, I.; Griffith, E.; Decker, D.D.; Chen, S.; et al. Gold nanoparticle-enabled blood test for early stage cancer detection and risk assessment. ACS Appl. Mater. Interfaces 2015, 7, 6819–6827. [Google Scholar] [CrossRef]
- Tien, W.S.; Chen, J.H.; Wu, K.P. SheddomeDB: The ectodomain shedding database for membrane-bound shed markers. BMC Bioinform. 2017, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Sebak, A.A.; Gomaa, I.E.O.; ElMeshad, A.N.; Farag, M.H.; Breitinger, U.; Breitinger, H.G.; AbdelKader, M.H. Distinct Proteins in Protein Corona of Nanoparticles Represent a Promising Venue for Endogenous Targeting—Part II: In vitro and in vivo Kinetics Study. Int. J. Nanomed. 2020, 15, 9539–9556. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Hollenbaugh, J.A.; Zand, M.S.; Holden-Wiltse, J.; Mosmann, T.R.; Perelson, A.S.; Wu, H.; Topham, D.J. Quantifying the early immune response and adaptive immune response kinetics in mice infected with influenza A virus. J. Virol. 2010, 84, 6687–6698. [Google Scholar] [CrossRef] [PubMed]
- Groves, H.T.; McDonald, J.U.; Langat, P.; Kinnear, E.; Kellam, P.; McCauley, J.; Ellis, J.; Thompson, C.; Elderfield, R.; Parker, L.; et al. Mouse Models of Influenza Infection with Circulating Strains to Test Seasonal Vaccine Efficacy. Front. Immunol. 2018, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Li, S.; Fang, J.; Li, F.; Tu, S. Antibody-activated trans-endothelial delivery of mesoporous organosilica nanomedicine augments tumor extravasation and anti-cancer immunotherapy. Bioact. Mater. 2021, 6, 2158–2172. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, T.; Yang, R.; Fu, S.; Guan, L.; Hou, T.; Mu, W.; Pang, X.; Liang, S.; Liu, Y.; et al. Small Morph Nanoparticles for Deep Tumor Penetration via Caveolae-Mediated Transcytosis. ACS Appl. Mater. Interfaces 2020, 12, 38499–38511. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Ren, J.; Ji, Y.; Wang, Y.; Liu, Y.; Chen, Z.; Farhadi Sabet, Z.; Wu, X.; Lynch, I.; Chen, C. Corona of Thorns: The Surface Chemistry-Mediated Protein Corona Perturbs the Recognition and Immune Response of Macrophages. ACS Appl. Mater. Interfaces 2020, 12, 1997–2008. [Google Scholar] [CrossRef] [PubMed]
- Soddu, L.; Trinh, D.N.; Dunne, E.; Kenny, D.; Bernardini, G.; Kokalari, I.; Marucco, A.; Monopoli, M.P.; Fenoglio, I. Identification of physicochemical properties that modulate nanoparticle aggregation in blood. Beilstein J. Nanotechnol. 2020, 11, 550–567. [Google Scholar] [CrossRef]
- Abdelkhaliq, A.; van der Zande, M.; Punt, A.; Helsdingen, R.; Boeren, S.; Vervoort, J.J.M.; Rietjens, I.; Bouwmeester, H. Impact of nanoparticle surface functionalization on the protein corona and cellular adhesion, uptake and transport. J. Nanobiotechnol. 2018, 16, 70. [Google Scholar] [CrossRef]
- Banha, J.; Marques, L.; Oliveira, R.; Martins Mde, F.; Paixao, E.; Pereira, D.; Malho, R.; Penque, D.; Costa, L. Ceruloplasmin expression by human peripheral blood lymphocytes: A new link between immunity and iron metabolism. Free. Radic. Biol. Med. 2008, 44, 483–492. [Google Scholar] [CrossRef]
- Li, S.S.; Ivanoff, A.; Bergstrom, S.E.; Sandstrom, A.; Christensson, B.; van Nerven, J.; Holgersson, J.; Hauzenberger, D.; Arencibia, I.; Sundqvist, K.G. T lymphocyte expression of thrombospondin-1 and adhesion to extracellular matrix components. Eur. J. Immunol. 2002, 32, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- de Castro, C.E.; Panico, K.; Stangherlin, L.M.; Ribeiro, C.A.S.; da Silva, M.C.C.; Carneiro-Ramos, M.S.; Dal-Bo, A.G.; Giacomelli, F.C. The Protein Corona Conundrum: Exploring the Advantages and Drawbacks of its Presence around Amphiphilic Nanoparticles. Bioconjugate Chem. 2020, 31, 2638–2647. [Google Scholar] [CrossRef] [PubMed]
- Partikel, K.; Korte, R.; Stein, N.C.; Mulac, D.; Herrmann, F.C.; Humpf, H.U.; Langer, K. Effect of nanoparticle size and PEGylation on the protein corona of PLGA nanoparticles. Eur. J. Pharm. Biopharm. 2019, 141, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Bewersdorff, T.; Gruber, A.; Eravci, M.; Dumbani, M.; Klinger, D.; Haase, A. Amphiphilic nanogels: Influence of surface hydrophobicity on protein corona, biocompatibility and cellular uptake. Int. J. Nanomed. 2019, 14, 7861–7878. [Google Scholar] [CrossRef] [PubMed]
- Sanita, G.; Armanetti, P.; Silvestri, B.; Carrese, B.; Cali, G.; Pota, G.; Pezzella, A.; d’Ischia, M.; Luciani, G.; Menichetti, L.; et al. Albumin-Modified Melanin-Silica Hybrid Nanoparticles Target Breast Cancer Cells via a SPARC-Dependent Mechanism. Front. Bioeng. Biotechnol. 2020, 8, 765. [Google Scholar] [CrossRef]
- Fritzsche, T.; Schnolzer, M.; Fiedler, S.; Weigand, M.; Wiessler, M.; Frei, E. Isolation and identification of heterogeneous nuclear ribonucleoproteins (hnRNP) from purified plasma membranes of human tumour cell lines as albumin-binding proteins. Biochem. Pharmacol. 2004, 67, 655–665. [Google Scholar] [CrossRef]
- Yang, B.; Kwon, I. Multivalent Albumin-Neonatal Fc Receptor Interactions Mediate a Prominent Extension of the Serum Half-Life of a Therapeutic Protein. Mol. Pharm. 2021, 18, 2397–2405. [Google Scholar] [CrossRef]
- Gianesello, L.; Priante, G.; Ceol, M.; Radu, C.M.; Saleem, M.A.; Simioni, P.; Terrin, L.; Anglani, F.; Del Prete, D. Albumin uptake in human podocytes: A possible role for the cubilin-amnionless (CUBAM) complex. Sci. Rep. 2017, 7, 13705. [Google Scholar] [CrossRef]
- Szweras, M.; Liu, D.; Partridge, E.A.; Pawling, J.; Sukhu, B.; Clokie, C.; Jahnen-Dechent, W.; Tenenbaum, H.C.; Swallow, C.J.; Grynpas, M.D.; et al. alpha 2-HS glycoprotein/fetuin, a transforming growth factor-beta/bone morphogenetic protein antagonist, regulates postnatal bone growth and remodeling. J. Biol. Chem. 2002, 277, 19991–19997. [Google Scholar] [CrossRef]
- Sanchez, M.C.; Chiabrando, G.A.; Vides, M.A. Pregnancy zone protein-tissue-type plasminogen activator complexes bind to low-density lipoprotein receptor-related protein (LRP). Arch. Biochem. Biophys. 2001, 389, 218–222. [Google Scholar] [CrossRef]
- Sand, O.; Folkersen, J.; Westergaard, J.G.; Sottrup-Jensen, L. Characterization of human pregnancy zone protein. Comparison with human alpha 2-macroglobulin. J. Biol. Chem. 1985, 260, 15723–15735. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.M.; Patel, D.D.; Pizzo, S.V. Oxidized alpha2-macroglobulin (alpha2M) differentially regulates receptor binding by cytokines/growth factors: Implications for tissue injury and repair mechanisms in inflammation. J. Immunol. 1998, 161, 4356–4365. [Google Scholar] [PubMed]
- Lin, J.; Jiang, X.; Dong, M.; Liu, X.; Shen, Q.; Huang, Y.; Zhang, H.; Ye, R.; Zhou, H.; Yan, C.; et al. Hepatokine Pregnancy Zone Protein Governs the Diet-Induced Thermogenesis Through Activating Brown Adipose Tissue. Adv. Sci. 2021, 8, e2101991. [Google Scholar] [CrossRef] [PubMed]
- Poon, I.K.; Hulett, M.D.; Parish, C.R. Histidine-rich glycoprotein is a novel plasma pattern recognition molecule that recruits IgG to facilitate necrotic cell clearance via FcgammaRI on phagocytes. Blood 2010, 115, 2473–2482. [Google Scholar] [CrossRef]
- Tiruppathi, C.; Naqvi, T.; Wu, Y.; Vogel, S.M.; Minshall, R.D.; Malik, A.B. Albumin mediates the transcytosis of myeloperoxidase by means of caveolae in endothelial cells. Proc. Natl. Acad. Sci. USA 2004, 101, 7699–7704. [Google Scholar] [CrossRef]
- Sokolov, A.V.; Ageeva, K.V.; Pulina, M.O.; Cherkalina, O.S.; Samygina, V.R.; Vlasova, I.I.; Panasenko, O.M.; Zakharova, E.T.; Vasilyev, V.B. Ceruloplasmin and myeloperoxidase in complex affect the enzymatic properties of each other. Free Radic. Res. 2008, 42, 989–998. [Google Scholar] [CrossRef]
- Larsen, M.T.; Mandrup, O.A.; Schelde, K.K.; Luo, Y.; Sorensen, K.D.; Dagnaes-Hansen, F.; Cameron, J.; Stougaard, M.; Steiniche, T.; Howard, K.A. FcRn overexpression in human cancer drives albumin recycling and cell growth; a mechanistic basis for exploitation in targeted albumin-drug designs. J. Control. Release 2020, 322, 53–63. [Google Scholar] [CrossRef]
- Ramos, D.; Mar, D.; Ishida, M.; Vargas, R.; Gaite, M.; Montgomery, A.; Linder, M.C. Mechanism of Copper Uptake from Blood Plasma Ceruloplasmin by Mammalian Cells. PLoS ONE 2016, 11, e0149516. [Google Scholar] [CrossRef]
- Harned, J.; Ferrell, J.; Nagar, S.; Goralska, M.; Fleisher, L.N.; McGahan, M.C. Ceruloplasmin alters intracellular iron regulated proteins and pathways: Ferritin, transferrin receptor, glutamate and hypoxia-inducible factor-1alpha. Exp. Eye Res. 2012, 97, 90–97. [Google Scholar] [CrossRef]
- Ma, S.; Fu, X.; Liu, L.; Liu, Y.; Feng, H.; Jiang, H.; Liu, X.; Liu, R.; Liang, Z.; Li, M.; et al. Iron-Dependent Autophagic Cell Death Induced by Radiation in MDA-MB-231 Breast Cancer Cells. Front. Cell Dev. Biol. 2021, 9, 723801. [Google Scholar] [CrossRef]
- Olson, G.E.; Winfrey, V.P.; Nagdas, S.K.; Hill, K.E.; Burk, R.F. Apolipoprotein E receptor-2 (ApoER2) mediates selenium uptake from selenoprotein P by the mouse testis. J. Biol. Chem. 2007, 282, 12290–12297. [Google Scholar] [CrossRef] [PubMed]
- Olson, G.E.; Winfrey, V.P.; Hill, K.E.; Burk, R.F. Megalin mediates selenoprotein P uptake by kidney proximal tubule epithelial cells. J. Biol. Chem. 2008, 283, 6854–6860. [Google Scholar] [CrossRef] [PubMed]
- Misu, H.; Takayama, H.; Saito, Y.; Mita, Y.; Kikuchi, A.; Ishii, K.A.; Chikamoto, K.; Kanamori, T.; Tajima, N.; Lan, F.; et al. Deficiency of the hepatokine selenoprotein P increases responsiveness to exercise in mice through upregulation of reactive oxygen species and AMP-activated protein kinase in muscle. Nat. Med. 2017, 23, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, C.; Cui, J.; Song, Q.; Wang, L.; Kang, J.; Li, P.; Hu, X.; Song, H.; Yang, J.; et al. Bioinformatic analysis of the membrane cofactor protein CD46 and microRNA expression in hepatocellular carcinoma. Oncol. Rep. 2014, 31, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Hvidberg, V.; Maniecki, M.B.; Jacobsen, C.; Hojrup, P.; Moller, H.J.; Moestrup, S.K. Identification of the receptor scavenging hemopexin-heme complexes. Blood 2005, 106, 2572–2579. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Herndon, M.E.; Ranganathan, S.; Godyna, S.; Lawler, J.; Argraves, W.S.; Liau, G. Internalization but not binding of thrombospondin-1 to low density lipoprotein receptor-related protein-1 requires heparan sulfate proteoglycans. J. Cell. Biochem. 2004, 91, 766–776. [Google Scholar] [CrossRef]
- Garg, P.; Yang, S.; Liu, A.; Pallero, M.A.; Buchsbaum, D.J.; Mosher, D.F.; Murphy-Ullrich, J.E.; Goldblum, S.E. Thrombospondin-1 opens the paracellular pathway in pulmonary microvascular endothelia through EGFR/ErbB2 activation. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2011, 301, L79–L90. [Google Scholar] [CrossRef]
- Silverstein, R.L.; Nachman, R.L.; Pannell, R.; Gurewich, V.; Harpel, P.C. Thrombospondin forms complexes with single-chain and two-chain forms of urokinase. J. Biol. Chem. 1990, 265, 11289–11294. [Google Scholar] [CrossRef]
- Greenaway, J.; Lawler, J.; Moorehead, R.; Bornstein, P.; Lamarre, J.; Petrik, J. Thrombospondin-1 inhibits VEGF levels in the ovary directly by binding and internalization via the low density lipoprotein receptor-related protein-1 (LRP-1). J. Cell. Physiol. 2007, 210, 807–818. [Google Scholar] [CrossRef]
- Cao, Z.T.; Gan, L.Q.; Jiang, W.; Wang, J.L.; Zhang, H.B.; Zhang, Y.; Wang, Y.; Yang, X.; Xiong, M.; Wang, J. Protein Binding Affinity of Polymeric Nanoparticles as a Direct Indicator of Their Pharmacokinetics. ACS Nano 2020, 14, 3563–3575. [Google Scholar] [CrossRef]
- Chu, Y.; Tang, W.; Zhang, Z.; Li, C.; Qian, J.; Wei, X.; Ying, T.; Lu, W.; Zhan, C. Deciphering Protein Corona by scFv-Based Affinity Chromatography. Nano Lett. 2021, 21, 2124–2131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, J.L.Y.; Lazarovits, J.; Chan, W.C.W. An Analysis of the Binding Function and Structural Organization of the Protein Corona. J. Am. Chem. Soc. 2020, 142, 8827–8836. [Google Scholar] [CrossRef] [PubMed]
- Imperlini, E.; Celia, C.; Cevenini, A.; Mandola, A.; Raia, M.; Fresta, M.; Orru, S.; Di Marzio, L.; Salvatore, F. Nano-bio interface between human plasma and niosomes with different formulations indicates protein corona patterns for nanoparticle cell targeting and uptake. Nanoscale 2021, 13, 5251–5269. [Google Scholar] [CrossRef] [PubMed]
- Rampado, R.; Crotti, S.; Caliceti, P.; Pucciarelli, S.; Agostini, M. Recent Advances in Understanding the Protein Corona of Nanoparticles and in the Formulation of “Stealthy” Nanomaterials. Front. Bioeng. Biotechnol. 2020, 8, 166. [Google Scholar] [CrossRef]
- Spada, A.; Emami, J.; Tuszynski, J.A.; Lavasanifar, A. The Uniqueness of Albumin as a Carrier in Nanodrug Delivery. Mol. Pharm. 2021, 18, 1862–1894. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Liu, X.; Li, T.; Chen, Y.; Chen, Y.; Wang, P.; Zheng, L.; Yang, H.; Wu, C.; Deng, S.; et al. Recent Advancements in Serum Albumin-Based Nanovehicles Toward Potential Cancer Diagnosis and Therapy. Front. Chem. 2021, 9, 746646. [Google Scholar] [CrossRef]
- Tomak, A.; Cesmeli, S.; Hanoglu, B.D.; Winkler, D.; Karakus, C.O. Nanoparticle-protein corona complex: Understanding multiple interactions between environmental factors, corona formation, and biological activity. Nanotoxicology 2021, 15, 1331–1357. [Google Scholar] [CrossRef]
- Vincent, M.P.; Bobbala, S.; Karabin, N.B.; Frey, M.; Liu, Y.; Navidzadeh, J.O.; Stack, T.; Scott, E.A. Surface chemistry-mediated modulation of adsorbed albumin folding state specifies nanocarrier clearance by distinct macrophage subsets. Nat. Commun. 2021, 12, 648. [Google Scholar] [CrossRef]
- Yoneshima, Y.; Morita, S.; Ando, M.; Nakamura, A.; Iwasawa, S.; Yoshioka, H.; Goto, Y.; Takeshita, M.; Harada, T.; Hirano, K.; et al. Phase 3 Trial Comparing Nanoparticle Albumin-Bound Paclitaxel With Docetaxel for Previously Treated Advanced NSCLC. J. Thorac. Oncol. 2021, 16, 1523–1532. [Google Scholar] [CrossRef]
- Li, S.; Li, M.; Huo, S.; Wang, Q.; Chen, J.; Ding, S.; Zeng, Z.; Zhou, W.; Wang, Y.; Wang, J. Voluntary-Opsonization-Enabled Precision Nanomedicines for Inflammation Treatment. Adv. Mater. 2021, 33, e2006160. [Google Scholar] [CrossRef]
- Pinals, R.L.; Yang, D.; Rosenberg, D.J.; Chaudhary, T.; Crothers, A.R.; Iavarone, A.T.; Hammel, M.; Landry, M.P. Quantitative Protein Corona Composition and Dynamics on Carbon Nanotubes in Biological Environments. Angew. Chem. Int. Ed. 2020, 59, 23668–23677. [Google Scholar] [CrossRef] [PubMed]
- Papafilippou, L.; Claxton, A.; Dark, P.; Kostarelos, K.; Hadjidemetriou, M. Protein corona fingerprinting to differentiate sepsis from non-infectious systemic inflammation. Nanoscale 2020, 12, 10240–10253. [Google Scholar] [CrossRef] [PubMed]
- Kumar, Y.; Liang, C.; Limmon, G.V.; Liang, L.; Engelward, B.P.; Ooi, E.E.; Chen, J.; Tannenbaum, S.R. Molecular analysis of serum and bronchoalveolar lavage in a mouse model of influenza reveals markers of disease severity that can be clinically useful in humans. PLoS ONE 2014, 9, e86912. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, A.A.; Tammam, S.N.; Hanafi, R.S.; Rashad, O.; Osama, A.; Abdelnaby, E.; Magdeldin, S.; Mansour, S. Different Serum, Different Protein Corona! The Impact of the Serum Source on Cellular Targeting of Folic Acid-Modified Chitosan-Based Nanoparticles. Mol. Pharm. 2022, 19, 1635–1646. [Google Scholar] [CrossRef]
- Ndumiso, M.; Buchtova, N.; Husselmann, L.; Mohamed, G.; Klein, A.; Aucamp, M.; Canevet, D.; D’Souza, S.; Maphasa, R.E.; Boury, F.; et al. Comparative whole corona fingerprinting and protein adsorption thermodynamics of PLGA and PCL nanoparticles in human serum. Colloids Surf. B Biointerfaces 2020, 188, 110816. [Google Scholar] [CrossRef]
- Mohammad-Beigi, H.; Hayashi, Y.; Zeuthen, C.M.; Eskandari, H.; Scavenius, C.; Juul-Madsen, K.; Vorup-Jensen, T.; Enghild, J.J.; Sutherland, D.S. Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 2020, 11, 4535. [Google Scholar] [CrossRef]
- Vanderbeke, L.; Van Mol, P.; Van Herck, Y.; De Smet, F.; Humblet-Baron, S.; Martinod, K.; Antoranz, A.; Arijs, I.; Boeckx, B.; Bosisio, F.M.; et al. Monocyte-driven atypical cytokine storm and aberrant neutrophil activation as key mediators of COVID-19 disease severity. Nat. Commun. 2021, 12, 4117. [Google Scholar] [CrossRef]
- Sudan, K.; Vijayan, V.; Madyaningrana, K.; Gueler, F.; Igarashi, K.; Foresti, R.; Motterlini, R.; Immenschuh, S. TLR4 activation alters labile heme levels to regulate BACH1 and heme oxygenase-1 expression in macrophages. Free. Radic. Biol. Med. 2019, 137, 131–142. [Google Scholar] [CrossRef]
- Bowman, E.R.; Cameron, C.M.; Richardson, B.; Kulkarni, M.; Gabriel, J.; Cichon, M.J.; Riedl, K.M.; Mustafa, Y.; Cartwright, M.; Snyder, B.; et al. Macrophage maturation from blood monocytes is altered in people with HIV, and is linked to serum lipid profiles and activation indices: A model for studying atherogenic mechanisms. PLoS Pathog. 2020, 16, e1008869. [Google Scholar] [CrossRef]
- Guo, C.; Zhou, H.X. Fatty Acids Compete with Abeta in Binding to Serum Albumin by Quenching Its Conformational Flexibility. Biophys. J. 2019, 116, 248–257. [Google Scholar] [CrossRef]
- Li, Q.; Yang, W.Y.; Qu, L.L.; Qi, H.Y.; Huang, Y.; Zhang, Z. Interaction of Warfarin with Human Serum Albumin and Effect of Ferulic Acid on the Binding. J. Spectrosc. 2014, 2014, 8345501. [Google Scholar] [CrossRef]
- Stenken, J.A.; Poschenrieder, A.J. Bioanalytical chemistry of cytokines—A review. Anal. Chim. Acta 2015, 853, 95–115. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Roth, D.; Isoe, Y.; Hayashi, K.; Mochizuki, C.; Kalkum, M.; Nakamura, M. Protein corona components of polyethylene glycol-conjugated organosilica nanoparticles modulates macrophage uptake. Colloids Surf. B Biointerfaces 2021, 199, 111527. [Google Scholar] [CrossRef] [PubMed]
- Papini, E.; Tavano, R.; Mancin, F. Opsonins and Dysopsonins of Nanoparticles: Facts, Concepts, and Methodological Guidelines. Front. Immunol. 2020, 11, 567365. [Google Scholar] [CrossRef]
- de Oliveira, F.A.; Albuquerque, L.J.C.; Castro, C.E.; Riske, K.A.; Bellettini, I.C.; Giacomelli, F.C. Reduced cytotoxicity of nanomaterials driven by nano-bio interactions: Case study of single protein coronas enveloping polymersomes. Colloids Surf. B Biointerfaces 2022, 213, 112387. [Google Scholar] [CrossRef]
- Gonzalez-Garcia, L.E.; MacGregor, M.N.; Visalakshan, R.M.; Lazarian, A.; Cavallaro, A.A.; Morsbach, S.; Mierczynska-Vasilev, A.; Mailander, V.; Landfester, K.; Vasilev, K. Nanoparticles Surface Chemistry Influence on Protein Corona Composition and Inflammatory Responses. Nanomaterials 2022, 12, 682. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, M.; Li, M.; Zhang, L.; Zhao, J.; Cao, J.; Wang, W. Controlled Recognition and Corona Formation by Cascade Micellar Nanoprobes: For Boosting Glioma Theranostics. Anal. Chem. 2022, 94, 11118–11123. [Google Scholar] [CrossRef]
- Sofias, A.M.; Toner, Y.C.; Meerwaldt, A.E.; van Leent, M.M.T.; Soultanidis, G.; Elschot, M.; Gonai, H.; Grendstad, K.; Flobak, A.; Neckmann, U.; et al. Tumor Targeting by alphavbeta3-Integrin-Specific Lipid Nanoparticles Occurs via Phagocyte Hitchhiking. ACS Nano 2020, 14, 7832–7846. [Google Scholar] [CrossRef]
- Mao, H.L.; Qian, F.; Li, S.; Shen, J.W.; Ye, C.K.; Hua, L.; Zhang, L.Z.; Wu, D.M.; Lu, J.; Yu, R.T.; et al. Delivery of Doxorubicin from Hyaluronic Acid-Modified Glutathione-Responsive Ferrocene Micelles for Combination Cancer Therapy. Mol. Pharm. 2019, 16, 987–994. [Google Scholar] [CrossRef] [PubMed]

| Nanoparticle | Particle Size (nm ± σ) | PDI ± σ | ζ-Potential (mV ± σ) |
|---|---|---|---|
| HBPE-NPs | 159.0 ± 2.7 | 0.147 ± 0.040 | −32.6 ± 3.8 |
| HBPE-NPs + VS3 | 145.9 ± 5.2 | 0.154 ± 0.060 | −24.0 ± 0.9 |
| HBPE-NP + VS3 + anti-IgG | 177.9 ± 5.7 | 0.102 ± 0.011 | ND |
| HBPE-NPs + VS4 | 148.4 ± 4.4 | 0.183 ± 0.026 | −32.8 ± 1.4 |
| HBPE-NPs + VS4 + anti-IgG | 166.6 ± 3.9 | 0.190 ± 0.012 | ND |
| HBPE-NPs + VS5 | 147.9 ± 6.8 | 0.179 ± 0.028 | −28.0 ± 2.2 |
| HBPE-NPs + VS5 + anti-IgG | 157.3 ± 2.3 | 0.183 ± 0.041 | ND |
| HBPE-NPs + VS6 | 143.7 ± 2.0 | 0.174 ± 0.036 | −30.4 ± 1.3 |
| HBPE-NPs + VS6 + anti-IgG | 159.5 ± 5.1 | 0.157 ± 0.017 | ND |
| Protein Abundance (VS3 > VS5) | Protein Abundance (VS5 > VS3) |
|---|---|
| Complement C3 | Albumin |
| Alpha-2-HS-glycoprotein | Fibronectin |
| Complement factor B | Apolipoprotein B-100 |
| Vitronectin | Complement factor H |
| Clusterin | Haptoglobin |
| Inhibitor of carbonic anhydrase | Immunoglobulin heavy constant mu |
| H-2 class I histocompatibility antigen, Q10 alpha chain | Complement component C8 beta chain |
| Complement C5 | Ig gamma-2B chain C region |
| Carboxypeptidase N subunit 2 | Protein AMBP |
| Plasma protease C1 inhibitor | Ig gamma-1 chain C region, membrane-bound form |
| Alpha-1-acid glycoprotein 1 | Complement component C8 gamma chain |
| Alpha-2-antiplasmin | Alpha-1-acid glycoprotein 2 |
| Complement component C8 alpha chain | Immunoglobulin kappa constant |
| Complement component C9 | Mannose-binding protein C |
| Serum amyloid A-1 protein | Beta-2-microglobulin |
| Complement factor D | Serum amyloid P-component |
| Serum amyloid A-2 protein | Complement C1s-B subcomponent |
| Ig-like domain-containing protein | Transthyretin |
| Complement C1s-A subcomponent | |
| N-acetylmuramoyl-L-alanine amidase | |
| Carboxypeptidase N catalytic chain | |
| Complement C2 | |
| Complement component 7 | |
| Mannan-binding lectin serine protease 2 | |
| Ficolin-1 | |
| Complement C1r-A subcomponent | |
| Vitamin K-dependent protein S | |
| Mannan-binding lectin serine protease 1 | |
| Glyceraldehyde-3-phosphate dehydrogenase | |
| Vitamin K-dependent protein C | |
| Interleukin-1 receptor accessory protein |
| Protein Abundance (VS3 > VS5) | Protein Abundance (VS5 > VS3) | ||
|---|---|---|---|
| Protein | Potential Cancer Interaction | Protein | Potential Cancer Interaction |
| Inter alpha-trypsin inhibitor, heavy chain 4 | Hyaluronan | Albumin | SPARC, hnRNPs, calreticulin, FcRn, Cubilin [45,46,47,48] |
| Alpha-2-HS-glycoprotein | Transforming growth factor-beta [49] | Pregnancy zone protein | LRP1, IL-1, GRP78 [50,51,52,53] |
| Inter-alpha-trypsin inhibitor heavy chain H2 | Hyaluronan | Apolipoprotein B-100 | Lipids, lipid receptors |
| Clusterin | Low-density lipoprotein receptor | Beta-2-glycoprotein 1 | Phospholipids |
| Histidine-rich glycoprotein | Phospholipids [54] | Ceruloplasmin | Albumin (FcRn), Ctr1, ferritin, ferroportin [55,56,57,58,59,60] |
| Afamin | Fatty acids, vitamin E receptors | Serum paraoxonase/arylesterase 1 | High-density lipoprotein |
| Carboxypeptidase N subunit 2 | Kinin receptors | Glutathione peroxidase 3 | Selenium (ApoER2 + LRP1 + LRP2) [61,62,63] |
| Apolipoprotein A-II | Lipids, lipid receptors | Insulin-like growth factor-binding protein complex acid labile subunit | Insulin-like growth factor 1 receptors |
| Corticosteroid-binding globulin | Glucocorticoid receptors | Beta-2-glycoprotein 1 | Phospholipids |
| Flavin reductase (NADPH) | Riboflavin receptors | Apolipoprotein C-III | Lipids, lipid receptors |
| Identified Protein | Accession # | Molecular Weight (kD) | Potential Cancer Interaction (MDA-MB-231) |
|---|---|---|---|
| Albumin | ALBU | 69 | SPARC, hnRNPs, calreticulin, FcRn, Cubilin [45,46,47,48] |
| Alpha-2-HS-glycoprotein | FETUA | 37 | TGF-beta [49] |
| Apolipoprotein A-I | APOA1 | 31 | Lipids, lipid receptors |
| Apolipoprotein A-IV | APOA4 | 45 | Lipids, lipid receptors |
| Apolipoprotein B-100 | APOB | 509 | Lipids, lipid receptors |
| Apolipoprotein E | APOE | 36 | Lipids, lipid receptors |
| Beta-2-glycoprotein 1 | APOH | 39 | Phospholipids |
| Ceruloplasmin | CERU | 121 | Albumin (FcRn), Ctr1, ferritin, ferroportin [55,56,57,59,60] |
| Clusterin | CLUS | 52 | Low-density lipoprotein receptor |
| Complement factor I | CFAI | 67 | CD46 [64] |
| Hemopexin | HEMO | 51 | LRP1 [65] |
| Histidine-rich glycoprotein | HRG | 59 | Phospholipids [54] |
| Inter alpha-trypsin inhibitor, heavy chain 4 | ITIH4 | 105 | Hyaluronan |
| Plasminogen | PLMN | 91 | Apolipoproteins, thrombospondin |
| Pregnancy zone protein | PZP | 166 | LRP1, interleukin-1, Glycoprotein 78 [50,51,52,53] |
| Thrombospondin-1 | TSP1 | 130 | Integrins, LRP1, EGFR, TGF-beta, uPA, VEGF-A [66,67,68,69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nierenberg, D.; Flores, O.; Fox, D.; Sip, Y.Y.L.; Finn, C.M.; Ghozlan, H.; Cox, A.; Coathup, M.; McKinstry, K.K.; Zhai, L.; et al. Macromolecules Absorbed from Influenza Infection-Based Sera Modulate the Cellular Uptake of Polymeric Nanoparticles. Biomimetics 2022, 7, 219. https://doi.org/10.3390/biomimetics7040219
Nierenberg D, Flores O, Fox D, Sip YYL, Finn CM, Ghozlan H, Cox A, Coathup M, McKinstry KK, Zhai L, et al. Macromolecules Absorbed from Influenza Infection-Based Sera Modulate the Cellular Uptake of Polymeric Nanoparticles. Biomimetics. 2022; 7(4):219. https://doi.org/10.3390/biomimetics7040219
Chicago/Turabian StyleNierenberg, Daniel, Orielyz Flores, David Fox, Yuen Yee Li Sip, Caroline M. Finn, Heba Ghozlan, Amanda Cox, Melanie Coathup, Karl Kai McKinstry, Lei Zhai, and et al. 2022. "Macromolecules Absorbed from Influenza Infection-Based Sera Modulate the Cellular Uptake of Polymeric Nanoparticles" Biomimetics 7, no. 4: 219. https://doi.org/10.3390/biomimetics7040219
APA StyleNierenberg, D., Flores, O., Fox, D., Sip, Y. Y. L., Finn, C. M., Ghozlan, H., Cox, A., Coathup, M., McKinstry, K. K., Zhai, L., & Khaled, A. R. (2022). Macromolecules Absorbed from Influenza Infection-Based Sera Modulate the Cellular Uptake of Polymeric Nanoparticles. Biomimetics, 7(4), 219. https://doi.org/10.3390/biomimetics7040219







