Advances in Biomimetic Systems for Molecular Recognition and Biosensing
Abstract
1. Introduction
2. Why Are Biomimetic Systems Required?
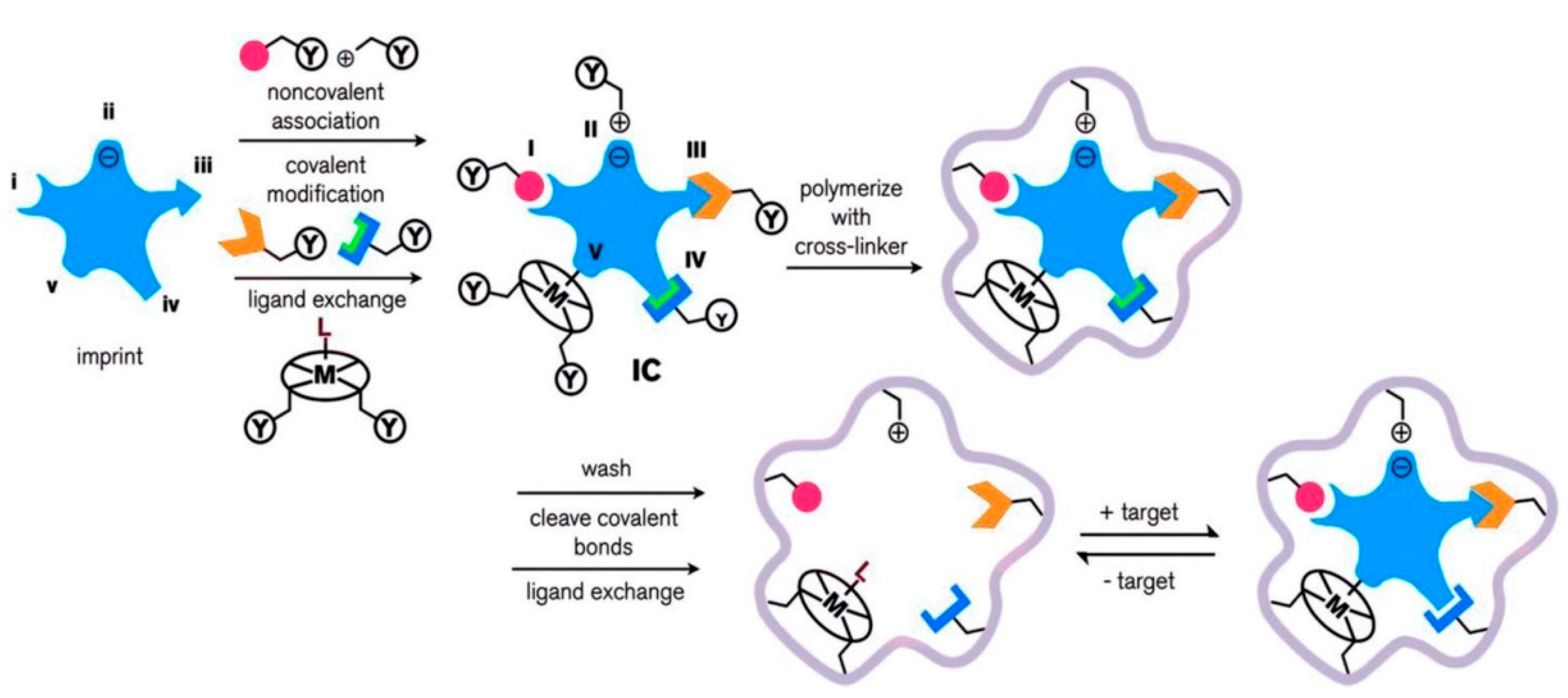
3. Molecular Imprinting Method
4. Molecularly Imprinted Polymers-Integrated Systems
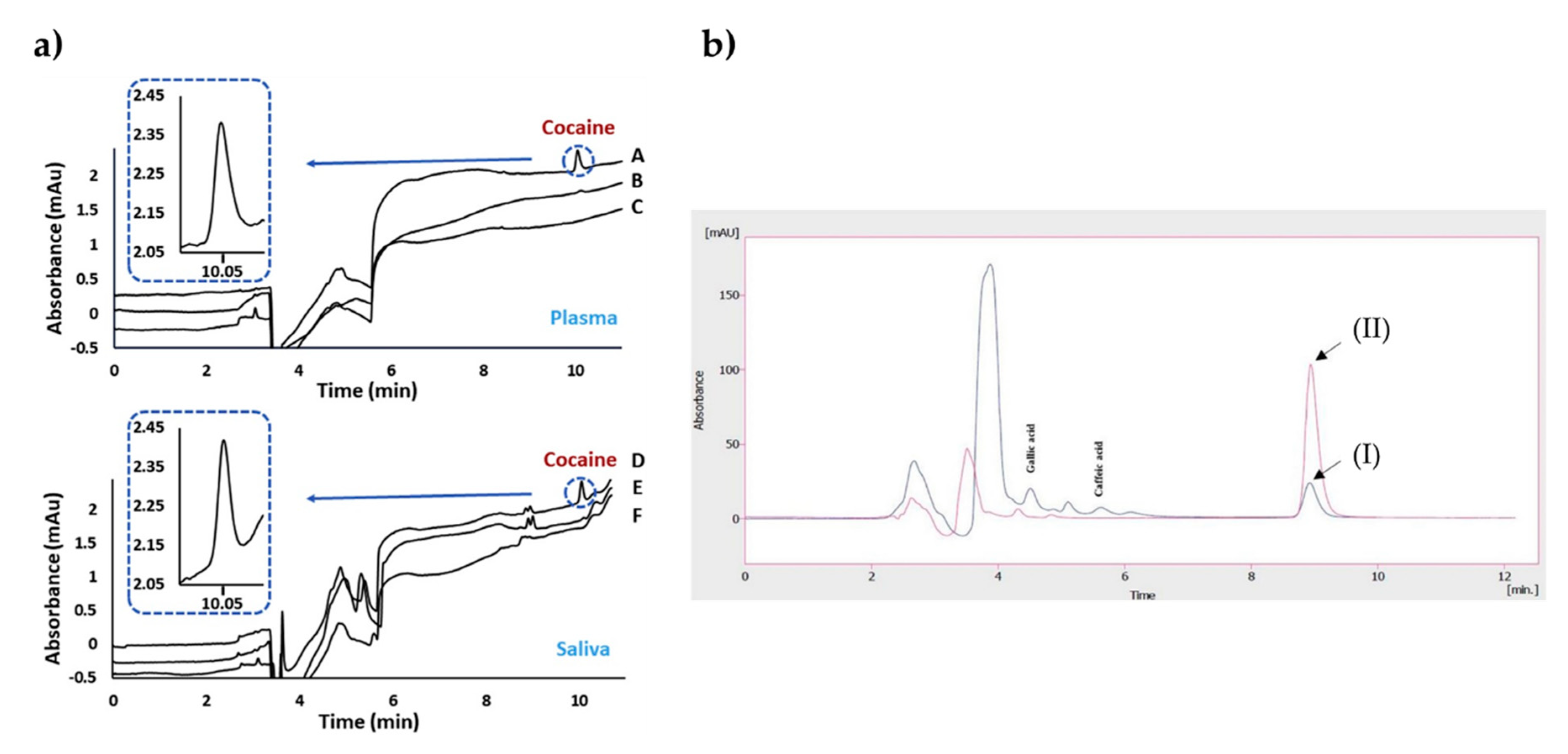
4.1. Chromatographic Systems
4.2. Sensor Systems
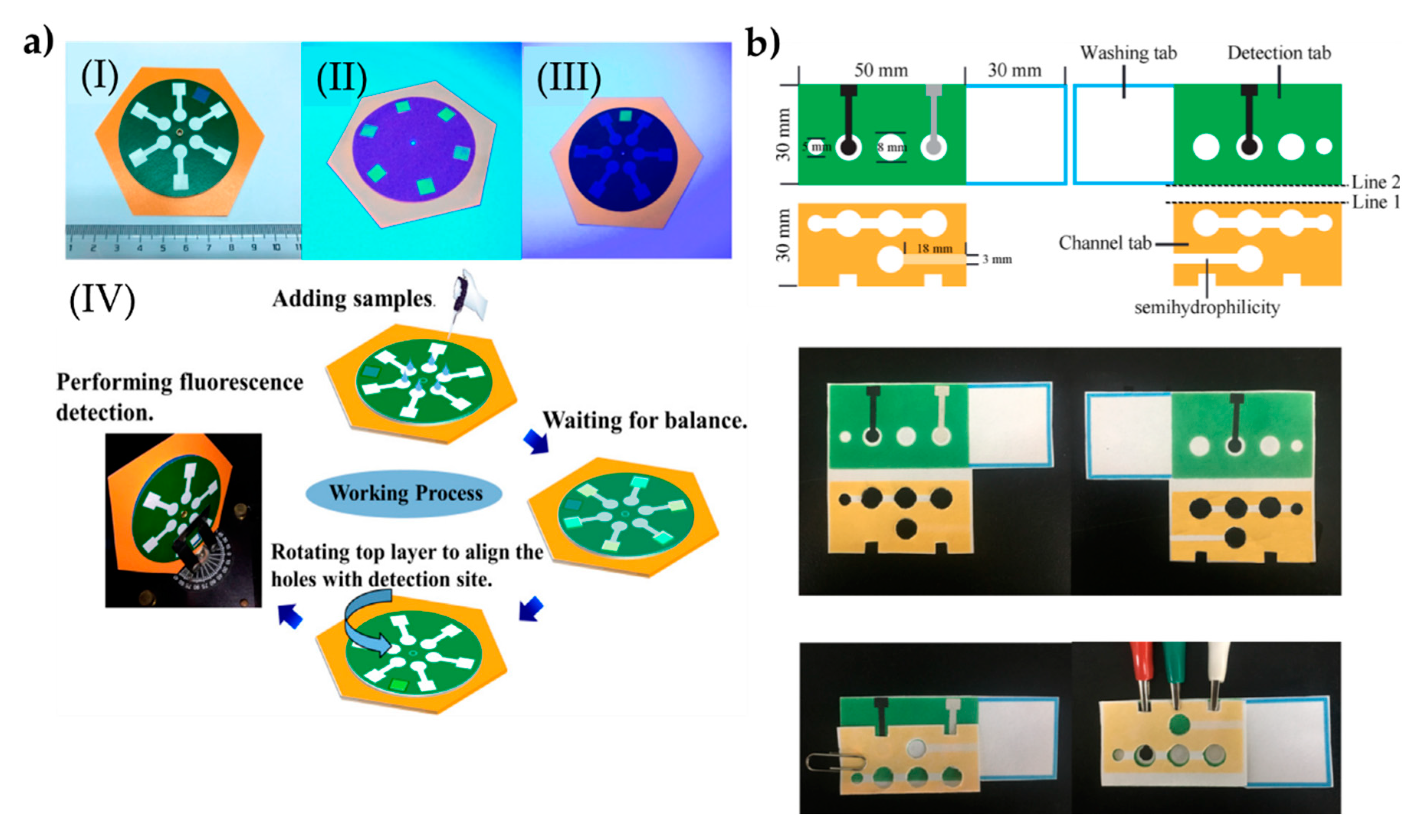
4.3. Lab-on-a-Chip Systems
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Fisher, R.J. Biomimetic systems. Tissue Eng. Artif. Organs 2016, 51–64. [Google Scholar]
- Bandyopadhyay, A.; Chowdhury, S.K.; Dey, S.; Moses, J.C.; Mandal, B.B. Silk: A promising biomaterial opening new vistas towards affordable healthcare solutions. J. Indian Inst. Sci. 2019, 99, 445–487. [Google Scholar] [CrossRef]
- Eiben, S.; Koch, C.; Altintoprak, K.; Southan, A.; Tovar, G.; Laschat, S.; Weiss, I.M.; Wege, C. Plant virus-based materials for biomedical applications: Trends and prospects. Adv. Drug Deliv. Rev. 2019, 145, 96–118. [Google Scholar] [CrossRef]
- Sunderland, K.S.; Yang, M.; Mao, C. Phage-enabled nanomedicine: From probes to therapeutics in precision medicine. Angew. Chemie Int. Ed. 2017, 56, 1964–1992. [Google Scholar] [CrossRef]
- Tromans, R.A.; Carter, T.S.; Chabanne, L.; Crump, M.P.; Li, H.; Matlock, J.V.; Orchard, M.G.; Davis, A.P. A biomimetic receptor for glucose. Nat. Chem. 2019, 11, 52–56. [Google Scholar] [CrossRef]
- Persch, E.; Dumele, O.; Diederich, F. Molecular recognition in chemical and biological systems. Angew. Chem. Int. Ed. 2015, 54, 3290–3327. [Google Scholar] [CrossRef]
- Naik, R.R.; Singamaneni, S. Introduction: Bioinspired and biomimetic materials. Chem. Rev. 2017, 117, 12581–12583. [Google Scholar] [CrossRef]
- Jia, Y.; Li, J. Reconstitution of FoF1-ATPase-based biomimetic systems. Nat. Rev. Chem. 2019, 3, 361–374. [Google Scholar] [CrossRef]
- van Leeuwen, T.; Lubbe, A.S.; Štacko, P.; Wezenberg, S.J.; Feringa, B.L. Dynamic control of function by light-driven molecular motors. Nat. Rev. Chem. 2017, 1, 1–7. [Google Scholar] [CrossRef]
- Pezzato, C.; Cheng, C.; Stoddart, J.F.; Astumian, R.D. Mastering the non-equilibrium assembly and operation of molecular machines. Chem. Soc. Rev. 2017, 46, 5491–5507. [Google Scholar] [CrossRef]
- Smith, B.D.; Anslyn, E.; Gokel, G.; Kubik, S.; Wang, B.; Hong, J.-I.; Hargrove, A.E.; Schmuck, C.; Rotello, D.; Gale, P. Synthetic Receptors for Biomolecules: Design Principles and Applications; Royal Society of Chemistry: Cambridge, UK, 2015. [Google Scholar]
- Kolesnichenko, I.V.; Anslyn, E. V Practical applications of supramolecular chemistry. Chem. Soc. Rev. 2017, 46, 2385–2390. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, Y. Biomedical applications of supramolecular systems based on host–guest interactions. Chem. Rev. 2015, 115, 7794–7839. [Google Scholar] [CrossRef]
- Kataev, E.A.; Müller, C. Recent advances in molecular recognition in water: Artificial receptors and supramolecular catalysis. Tetrahedron 2014, 70, 137–167. [Google Scholar] [CrossRef]
- Razzak, M.; De Brabander, J.K. Lessons and revelations from biomimetic syntheses. Nat. Chem. Biol. 2011, 7, 865–875. [Google Scholar] [CrossRef]
- Kramer, W. Transporters, Trojan horses and therapeutics: Suitability of bile acid and peptide transporters for drug delivery. Biol. Chem. 2011, 392, 77–94. [Google Scholar] [CrossRef]
- Alvarez-Lorenzo, C.; Concheiro, A. Bioinspired drug delivery systems. Curr. Opin. Biotechnol. 2013, 24, 1167–1173. [Google Scholar] [CrossRef]
- Sabu, C.; Rejo, C.; Kotta, S.; Pramod, K. Bioinspired and biomimetic systems for advanced drug and gene delivery. J. Control. Release 2018, 287, 142–155. [Google Scholar] [CrossRef]
- Saylan, Y.; Denizli, A. Molecular fingerprints of hemoglobin on a nanofilm chip. Sensors 2018, 18, 3016. [Google Scholar] [CrossRef]
- Luan, J.; Liu, K.-K.; Tadepalli, S.; Jiang, Q.; Morrissey, J.J.; Kharasch, E.D.; Singamaneni, S. PEGylated artificial antibodies: Plasmonic biosensors with improved selectivity. ACS Appl. Mater. Interfaces 2016, 8, 23509–23516. [Google Scholar] [CrossRef]
- Saylan, Y.; Erdem, Ö.; Cihangir, N.; Denizli, A. Detecting fingerprints of waterborne bacteria on a sensor. Chemosensors 2019, 7, 33. [Google Scholar] [CrossRef]
- Huynh, T.-P.; KC, C.B.; Lisowski, W.; D’Souza, F.; Kutner, W. Molecularly imprinted polymer of bis (2, 2′-bithienyl) methanes for selective determination of adrenaline. Bioelectrochemistry 2013, 93, 37–45. [Google Scholar] [CrossRef]
- Tang, X.; Liu, W.; Chen, J.; Jia, J.; Ma, Z.; Shi, Q.; Gao, Y.; Wang, X.; Xu, S.; Wang, K. Preparation and evaluation of polydopamine imprinting layer coated multi-walled carbon nanotubes for the determination of testosterone in prostate cancer LNcap cells. Anal. Methods 2015, 7, 8326–8334. [Google Scholar] [CrossRef]
- Assavapanumat, S.; Ketkaew, M.; Kuhn, A.; Wattanakit, C. Synthesis, characterization, and electrochemical applications of chiral imprinted mesoporous Ni surfaces. J. Am. Chem. Soc. 2019, 141, 18870–18876. [Google Scholar] [CrossRef]
- Ozcelikay, G.; Kurbanoglu, S.; Zhang, X.; Kosak Soz, C.; Wollenberger, U.; Ozkan, S.A.; Yarman, A.; Scheller, F.W. Electrochemical MIP sensor for butyrylcholinesterase. Polymers 2019, 11, 1970. [Google Scholar] [CrossRef]
- Yang, K.; Li, S.; Liu, L.; Chen, Y.; Zhou, W.; Pei, J.; Liang, Z.; Zhang, L.; Zhang, Y. Epitope imprinting technology: Progress, applications, and perspectives toward artificial antibodies. Adv. Mater. 2019, 31, 1902048. [Google Scholar] [CrossRef]
- Hwang, J.; Jeong, Y.; Park, J.M.; Lee, K.H.; Hong, J.W.; Choi, J. Biomimetics: Forecasting the future of science, engineering, and medicine. Int. J. Nanomed. 2015, 10, 5701–5713. [Google Scholar]
- Bar-Cohen, Y. Biomimetics: Nature-Based Innovation; CRC press: Boca Raton, FL, USA, 2011; ISBN 1439834768. [Google Scholar]
- Zhao, X.; Lin, C.-W.; Wang, J.; Oh, D.H. Advances in rapid detection methods for foodborne pathogens. J. Microbiol. Biotechnol. 2014, 24, 297–312. [Google Scholar] [CrossRef]
- Yang, Y.; Song, X.; Li, X.; Chen, Z.; Zhou, C.; Zhou, Q.; Chen, Y. Recent progress in biomimetic additive manufacturing technology: From materials to functional structures. Adv. Mater. 2018, 30, 1706539. [Google Scholar] [CrossRef]
- Cui, J.; Drotlef, D.; Larraza, I.; Fernández-Blázquez, J.P.; Boesel, L.F.; Ohm, C.; Mezger, M.; Zentel, R.; del Campo, A. Bioinspired actuated adhesive patterns of liquid crystalline elastomers. Adv. Mater. 2012, 24, 4601–4604. [Google Scholar] [CrossRef]
- Steven, C.R.; Busby, G.A.; Mather, C.; Tariq, B.; Briuglia, M.L.; Lamprou, D.A.; Urquhart, A.J.; Grant, M.H.; Patwardhan, S. V Bioinspired silica as drug delivery systems and their biocompatibility. J. Mater. Chem. B 2014, 2, 5028–5042. [Google Scholar] [CrossRef]
- Mason, T.O.; Shimanovich, U. Fibrous protein self-assembly in biomimetic materials. Adv. Mater. 2018, 30, 1–11. [Google Scholar] [CrossRef]
- Xu, Q.; Wan, Y.; Hu, T.S.; Liu, T.X.; Tao, D.; Niewiarowski, P.H.; Tian, Y.; Liu, Y.; Dai, L.; Yang, Y. Robust self-cleaning and micromanipulation capabilities of gecko spatulae and their bio-mimics. Nat. Commun. 2015, 6, 1–9. [Google Scholar] [CrossRef]
- Lissandrello, C.; Inci, F.; Francom, M.; Paul, M.R.; Demirci, U.; Ekinci, K.L. Nanomechanical motion of Escherichia coli adhered to a surface. Appl. Phys. Lett. 2014, 105, 113701. [Google Scholar] [CrossRef]
- Glaser, D.E.; Viney, C. Biomimetic Materials, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2013; ISBN 9780123746269. [Google Scholar]
- Chen, H.; Yada, R. Nanotechnologies in agriculture: New tools for sustainable development. Trends Food Sci. Technol. 2011, 22, 585–594. [Google Scholar] [CrossRef]
- Sekhon, B.S. Nanotechnology in agri-food production: An overview. Nanotechnol. Sci. Appl. 2014, 7, 31. [Google Scholar] [CrossRef]
- Pendergast, M.M.; Hoek, E.M. V A review of water treatment membrane nanotechnologies. Energy Environ. Sci. 2011, 4, 1946–1971. [Google Scholar] [CrossRef]
- Inci, F.; Filippini, C.; Baday, M.; Ozen, M.O.; Calamak, S.; Durmus, N.G.; Wang, S.; Hanhauser, E.; Hobbs, K.S.; Juillard, F. Multitarget, quantitative nanoplasmonic electrical field-enhanced resonating device (NE2RD) for diagnostics. Proc. Natl. Acad. Sci. USA 2015, 112, E4354–E4363. [Google Scholar] [CrossRef]
- Inci, F.; Celik, U.; Turken, B.; Özer, H.Ö.; Kok, F.N. Construction of P-glycoprotein incorporated tethered lipid bilayer membranes. Biochem. Biophys. Rep. 2015, 2, 115–122. [Google Scholar] [CrossRef]
- Cunha, C.; Panseri, S.; Antonini, S. Emerging nanotechnology approaches in tissue engineering for peripheral nerve regeneration. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 50–59. [Google Scholar] [CrossRef]
- Suri, K.; Wolfram, J.; Shen, H.; Ferrari, M. Advances in nanotechnology-based drug delivery platforms and novel drug delivery systems. In Novel Approaches and Strategies for Biologics, Vaccines and Cancer Therapies; Elsevier: Holly Springs, NC, USA, 2015; pp. 41–58. [Google Scholar]
- Schroeder, A.; Heller, D.A.; Winslow, M.M.; Dahlman, J.E.; Pratt, G.W.; Langer, R.; Jacks, T.; Anderson, D.G. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 2012, 12, 39–50. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Hunter, A.C.; Andresen, T.L. Factors controlling nanoparticle pharmacokinetics: An integrated analysis and perspective. Annu. Rev. Pharmacol. Toxicol. 2012, 52, 481–503. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.K.; Treetong, A.; Suedee, R. Biomimetic insulin-imprinted polymer nanoparticles as a potential oral drug delivery system. Acta Pharm. 2017, 67, 149–168. [Google Scholar] [CrossRef] [PubMed]
- Suedee, R.; Naklua, W.; Laengchokshoi, S.; Thepkaue, K.; Pathaburee, P.; Nuanplub, M. Investigation of a self-assembling microgel containing an (S)-propranolol molecularly imprinted polymer in a native tissue microenvironment: Part I. Preparation and characterization. Process Biochem. 2015, 50, 517–544. [Google Scholar] [CrossRef]
- Ko, E.; Cho, S.-W. Biomimetic polymer scaffolds to promote stem cell-mediated osteogenesis. Int. J. Stem Cells 2013, 6, 87. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Saylan, Y.; Yilmaz, F.; Özgür, E.; Derazshamshir, A.; Yavuz, H.; Denizli, A. Molecular imprinting of macromolecules for sensor applications. Sensors 2017, 19, 1279. [Google Scholar] [CrossRef]
- He, Y.; Yang, T.; Mo, H.; Chen, T.; Feng, J.; Zhang, W. Low-cost potentiometric sensor based on a molecularly imprinted polymer for the rapid determination of matrine in herbal medicines. Instrum. Sci. Technol. 2019, 47, 581–596. [Google Scholar] [CrossRef]
- Saylan, Y.; Denizli, A. Molecularly imprinted polymer-based microfluidic systems for point-of-care applications. Micromachines 2019, 10, 766. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Zeng, C.; Qu, Q.; Wang, M.; Qi, W.; Su, R.; He, Z. A sandwich-like sensor for the highly specific and reproducible detection of rhodamine 6G on a surface-enhanced Raman scattering platform. ACS Appl. Mater. Interfaces 2020, 12, 4699–4706. [Google Scholar] [CrossRef]
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-based impedimetric sensor for detecting dengue fever biomarker. Appl. Biochem. Biotechnol. 2020, 1–11. [Google Scholar] [CrossRef]
- Uzun, L.; Turner, A.P.F. Molecularly-imprinted polymer sensors: Realising their potential. Biosens. Bioelectron. 2016, 76, 131–144. [Google Scholar] [CrossRef]
- Regasa, M.B.; Refera Soreta, T.; Femi, O.E.; C. Ramamurthy, P. Development of molecularly imprinted conducting polymer composite film-based electrochemical sensor for melamine detection in infant formula. ACS Omega 2020, 5, 4090–4099. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.A. Molecular imprinting: A useful approach for drug delivery. Mater. Sci. Energy Technol. 2020, 3, 72–77. [Google Scholar] [CrossRef]
- Bodoki, A.E.; Iacob, B.-C.; Bodoki, E. Perspectives of molecularly imprinted polymer-based drug delivery systems in cancer therapy. Polymers 2019, 11, 2085. [Google Scholar] [CrossRef]
- Pirzada, M.; Altintas, Z. Nanomaterials for healthcare biosensing applications. Sensors 2019, 19, 5311. [Google Scholar] [CrossRef] [PubMed]
- Sharifi, M.; Attar, F.; Saboury, A.A.; Akhtari, K.; Hooshmand, N.; Hasan, A.; El-Sayed, M.A.; Falahati, M. Plasmonic gold nanoparticles: Optical manipulation, imaging, drug delivery and therapy. J. Control. Release 2019, 311–312, 170–189. [Google Scholar] [CrossRef]
- Khan, I.M.; Niazi, S.; Khan, M.K.I.; Pasha, I.; Mohsin, A.; Haider, J.; Iqbal, M.W.; Rehman, A.; Yue, L.; Wang, Z. Recent advances and perspectives of AIE as an emerging platform for detection and bioimaging. TrAC Trends Anal. Chem. 2019, 119, 115637. [Google Scholar] [CrossRef]
- Ansari, S. Application of magnetic molecularly imprinted polymer as a versatile and highly selective tool in food and environmental analysis: Recent developments and trends. TrAC Trends Anal. Chem. 2017, 90, 89–106. [Google Scholar] [CrossRef]
- Rutkowska, M.; Płotka-Wasylka, J.; Morrison, C.; Wieczorek, P.P.; Namieśnik, J.; Marć, M. Application of molecularly imprinted polymers in analytical chiral separations and analysis. TrAC Trends Anal. Chem. 2018, 102, 91–102. [Google Scholar] [CrossRef]
- Pardeshi, S.; Singh, S.K. Precipitation polymerization: A versatile tool for preparing molecularly imprinted polymer beads for chromatography applications. Rsc Adv. 2016, 6, 23525–23536. [Google Scholar] [CrossRef]
- Wackerlig, J.; Lieberzeit, P.A. Molecularly imprinted polymer nanoparticles in chemical sensing—Synthesis, characterisation and application. Sens. Actuators B Chem. 2015, 207, 144–157. [Google Scholar] [CrossRef]
- Matos, M.J.B.; Pina, A.S.; Roque, A.C.A. Rational design of affinity ligands for bioseparation. J. Chromatogr. A 2020, 460871. [Google Scholar] [CrossRef]
- Andaç, M.; Galaev, I.Y.; Denizli, A. Affinity based and molecularly imprinted cryogels: Applications in biomacromolecule purification. J. Chromatogr. B 2016, 1021, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.M.; Lin, D.Q.; Yao, S.J. Review on biomimetic affinity chromatography with short peptide ligands and its application to protein purification. J. Chromatogr. A 2018, 1571, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhou, T.; Kettisen, K.; Ye, L.; Bülow, L. Chromatographic separation of hemoglobin variants using robust molecularly imprinted polymers. Talanta 2019, 199, 27–31. [Google Scholar] [CrossRef]
- Hudson, A.D.; Ueta, J.T.; Battell, W.; Jamieson, O.; Dunbar, T.; Maciá, B.; Peeters, M. Synthesis of optimized molecularly imprinted polymers for the isolation and detection of antidepressants via HPLC. Biomimetics 2019, 4, 18. [Google Scholar] [CrossRef]
- Razym, G.; Bakhshpour, M.; Yavuz, H.; Kip, Ç.; Tuncel, A.; Denizli, A. Surface-imprinted silica particles for Concanavalin A purification from Canavalia ensiformis. J. Chromatogr. B. 2020, 1136, 121852. [Google Scholar] [CrossRef]
- Bouvarel, T.; Delaunay, N.; Pichon, V. Selective extraction of cocaine from biological samples with a miniaturized monolithic molecularly imprinted polymer and on-line analysis in nano-liquid chromatography. Anal. Chim. Acta 2020, 1096, 89–99. [Google Scholar] [CrossRef]
- Rahimi, M.; Bahar, S.; Heydari, R.; Amininasab, S.M. Determination of quercetin using a molecularly imprinted polymer as solid-phase microextraction sorbent and high-performance liquid chromatography. Microchem. J. 2019, 148, 433–441. [Google Scholar] [CrossRef]
- Lu, W.; Liu, J.; Li, J.; Wang, X.; Lv, M.; Cui, R.; Chen, L. Dual-template molecularly imprinted polymers for dispersive solid-phase extraction of fluoroquinolones in water samples coupled with high performance liquid chromatography. Analyst 2019, 144, 1292–1302. [Google Scholar] [CrossRef]
- Ma, X.-H.; Li, J.-P.; Wang, C.; Xu, G.-B. A Review on bio-macromolecular imprinted sensors and their applications. Chinese J. Anal. Chem. 2016, 44, 152–159. [Google Scholar] [CrossRef]
- Anantha-Iyengar, G.; Shanmugasundaram, K.; Nallal, M.; Lee, K.P.; Whitcombe, M.J.; Lakshmi, D.; Sai-Anand, G. Functionalized conjugated polymers for sensing and molecular imprinting applications. Prog. Polym. Sci. 2019, 88, 1–129. [Google Scholar] [CrossRef]
- Ahmad, O.S.; Bedwell, T.S.; Esen, C.; Garcia-Cruz, A.; Piletsky, S.A. Molecularly imprinted polymers in electrochemical and optical sensors. Trends Biotechnol. 2019, 37, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Li, J.; Ma, X.; Liu, C.; Pang, C.; Luo, J. Highly selective molecular imprinting electrochemiluminescence switch sensor for biotoxin L-canavanine measurement. Microchem. J. 2019, 148, 397–403. [Google Scholar] [CrossRef]
- Nawaz, T.; Ahmad, M.; Yu, J.; Wang, S.; Wei, T. The biomimetic detection of progesterone by novel bifunctional group monomer based molecularly imprinted polymers prepared in UV. New J. Chem. 2020, 44, 6992–7000. [Google Scholar] [CrossRef]
- Phonklam, K.; Wannapob, R.; Sriwimol, W.; Thavarungkul, P.; Phairatana, T. A novel molecularly imprinted polymer PMB/MWCNTs sensor for highly-sensitive cardiac troponin T detection. Sens. Actuators B Chem. 2020, 308, 127630. [Google Scholar] [CrossRef]
- Pandey, H.; Khare, P.; Singh, S.; Singh, S.P. Carbon nanomaterials integrated molecularly imprinted polymers for biological sample analysis: A critical review. Mater. Chem. Phys. 2020, 239, 121966. [Google Scholar] [CrossRef]
- Cennamo, N.; Maniglio, D.; Tatti, R.; Zeni, L.; Bossi, A.M. Deformable molecularly imprinted nanogels permit sensitivity-gain in plasmonic sensing. Biosens. Bioelectron. 2020, 156, 112126. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015, 478, 107–120. [Google Scholar] [CrossRef]
- Turner, A.P.F. Biosensors: Sense and sensibility. Chem. Soc. Rev. 2013, 42, 3184–3196. [Google Scholar] [CrossRef]
- Luo, L.; Zhang, F.; Chen, C.; Cai, C. Visual simultaneous detection of hepatitis a and b viruses based on a multifunctional molecularly imprinted fluorescence sensor. Anal. Chem. 2019, 91, 15748–15756. [Google Scholar] [CrossRef]
- Erdem, Ö.; Saylan, Y.; Cihangir, N.; Denizli, A. Molecularly imprinted nanoparticles based plasmonic sensors for real-time Enterococcus faecalis detection. Biosens. Bioelectron. 2019, 126, 608–614. [Google Scholar] [CrossRef] [PubMed]
- Sergeyeva, T.; Yarynka, D.; Piletska, E.; Linnik, R.; Zaporozhets, O.; Brovko, O.; Piletsky, S.; El’skaya, A. Development of a smartphone-based biomimetic sensor for aflatoxin B1 detection using molecularly imprinted polymer membranes. Talanta 2019, 201, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Ayankojo, A.G.; Reut, J.; Ciocan, V.; Öpik, A.; Syritski, V. Molecularly imprinted polymer-based sensor for electrochemical detection of erythromycin. Talanta 2020, 209, 120502. [Google Scholar] [CrossRef] [PubMed]
- Kidakova, A.; Boroznjak, R.; Reut, J.; Öpik, A.; Saarma, M.; Syritski, V. Molecularly imprinted polymer-based SAW sensor for label-free detection of cerebral dopamine neurotrophic factor protein. Sens. Actuators B Chem. 2020, 308, 127708. [Google Scholar] [CrossRef]
- Özgür, E.; Topçu, A.A.; Yılmaz, E.; Denizli, A. Surface plasmon resonance based biomimetic sensor for urinary tract infections. Talanta 2020, 212, 120778. [Google Scholar] [CrossRef]
- Feng, J.; Tao, Y.; Shen, X.; Jin, H.; Zhou, T.; Zhou, Y.; Hu, L.; Luo, D.; Mei, S.; Lee, Y.I. Highly sensitive and selective fluorescent sensor for tetrabromobisphenol-A in electronic waste samples using molecularly imprinted polymer coated quantum dots. Microchem. J. 2019, 144, 93–101. [Google Scholar] [CrossRef]
- Akgönüllü, S.; Battal, D.; Yalcin, M.S.; Yavuz, H.; Denizli, A. Rapid and sensitive detection of synthetic cannabinoids JWH-018, JWH-073 and their metabolites using molecularly imprinted polymer-coated QCM nanosensor in artificial saliva. Microchem. J. 2019, 153, 104454. [Google Scholar] [CrossRef]
- Inci, F.; Saylan, Y.; Kojouri, A.M.; Ogut, M.G.; Denizli, A.; Demirci, U. A disposable microfluidic-integrated hand-held plasmonic platform for protein detection. Appl. Mater. Today 2020, 18, 100478. [Google Scholar] [CrossRef]
- Zhong, Q.; Ding, H.; Gao, B.; He, Z.; Gu, Z. Advances of Microfluidics in Biomedical Engineering. Adv. Mater. Technol. 2019, 4, 1800663. [Google Scholar] [CrossRef]
- Hong, C.-C.; Lin, C.-C.; Hong, C.-L.; Chang, P.-H. Enhanced anesthetic propofol biochips by modifying molecularly imprinted nanocavities of biosensors. Biomed. Microdevices 2012, 14, 435–441. [Google Scholar] [CrossRef]
- Krupadam, R.J.; Korde, B.A.; Ashokkumar, M.; Kolev, S.D. Novel molecularly imprinted polymeric microspheres for preconcentration and preservation of polycyclic aromatic hydrocarbons from environmental samples. Anal. Bioanal. Chem. 2014, 406, 5313–5321. [Google Scholar] [CrossRef]
- Janfaza, S.; Kim, E.; O’Brien, A.; Najjaran, H.; Nikkhah, M.; Alizadeh, T.; Hoorfar, M. A nanostructured microfluidic artificial olfaction for organic vapors recognition. Sci. Rep. 2019, 9, 19051. [Google Scholar] [CrossRef] [PubMed]
- Wagner, S.; Bell, J.; Biyikal, M.; Gawlitza, K.; Rurack, K. Integrating fluorescent molecularly imprinted polymer (MIP) sensor particles with a modular microfluidic platform for nanomolar small-molecule detection directly in aqueous samples. Biosens. Bioelectron. 2018, 99, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, B.; Wang, X.; Zhang, Z.; Wang, Z.; Han, J.; Chen, L. Three-dimensional paper-based microfluidic chip device for multiplexed fluorescence detection of Cu2+ and Hg2+ ions based on ion imprinting technology. Sens. Actuators B Chem. 2017, 251, 224–233. [Google Scholar] [CrossRef]
- Qi, J.; Li, B.; Wang, X.; Fu, L.; Luo, L.; Chen, L. Rotational paper-based microfluidic-chip device for multiplexed and simultaneous fluorescence detection of phenolic pollutants based on a molecular-imprinting technique. Anal. Chem. 2018, 90, 11827–11834. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Jian, Y.; Wang, H.; Ge, S.; Yan, M.; Yu, J. Ultrasensitive microfluidic paper-based electrochemical biosensor based on molecularly imprinted film and boronate affinity sandwich assay for glycoprotein detection. ACS Appl. Mater. Interfaces 2019, 11, 16198–16206. [Google Scholar] [CrossRef] [PubMed]

| Polymerization | General Advantages and Disadvantages |
|---|---|
| Bulk | • Simple and universal type of polymerization. • No need for sophisticated instrumentation. • Obtaining spherical materials. • Providing reproducible results • Allowing a large-scale examination of products. • Requiring lengthy procedures. • Resulting in irregularity in size and shape. • Low performance. |
| Precipitation | • Providing uniform size and high yields of imprinted materials • Creating homogeneous binding sites. • One of the easiest and well-suited type with a high dilution factor. • Requirement for a polymerization mixture in the presence of a much higher amount of porogen maker. • The growing polymer chains are unable to occupy the entire volume. |
| Suspension | • An organic-based medium is mixed with an excess of water and the amount of suspension stabilizer. • Two phases are mixed by stirring to form a suspension of organic droplets in the aqueous phase. • The imprinted materials are scarce because water might disrupt non-covalent interactions between the template molecule and the monomers. |
| Multi-step swelling | • Producing mono-disperse and outstanding materials with controlled diameter. • The size of the imprinted materials might be controlled by changing the polymerization conditions. • Requiring complex and long polymerization conditions. • Requiring laborious procedure and aqueous emulsions. |
| Surface | • Producing mono-disperse materials and thin imprinted layers. • Creating more accessible binding sites. • Allowing rapid binding and high desorption rates. • Providing more effective ability to recognize the template molecules. • Providing a large specific surface area for the particles, hence leading to excellent affinity and selectivity. • Requireing a complicated system and time-consuming procedure. |
| In-situ | • Requiring a single-step preparation strategy. • Beinga cost-friendly fashion. • Providing a, well-porous structure. • Requiring a comprehensive and lengthy optimization procedure that needs to be optimized for every template molecules systems. |
| Application | Template Molecule | Polymerization Type | Dynamic Range | Adsorption Capacity | Reference |
|---|---|---|---|---|---|
| HPLC-UV | Fluoxetine | Bulk | 0–1.5 mM | 800 µmol/g | [69] |
| SDS-PAGE | Concanavalin A | Surface | 0–2.0 mg/mL | 305.2 mg/g | [70] |
| NanoLC-UV | Cocaine | In-situ | 100–2000 ng/mL | Not available | [71] |
| HPLC-UV | Quercetin | Sol-gel | 0.05–100 μg/mL | 19.98 ng/g | [72] |
| HPLC | Norfloxacin | Precipitation | 1.0–200 μg/L | 32 mg/g | [73] |
| Sensor Type | Template Molecule | Polymerization Type | Dynamic Range | Limit of Detection | Reference |
|---|---|---|---|---|---|
| Optical | Enterococcus faecalis | Emulsion | 2 × 104–1 × 108 cfu/mL | 1.05 × 102 cfu/mL | [85] |
| Optical | Aflatoxin B1 | In-situ | 20–100 ng/mL | 20 ng/mL | [86] |
| Electrochemical | Erythromycin | Electro-polymerization | 12.8 nM–40 μM | 0.1 nM | [87] |
| Surface acoustic wave | Cerebral dopamine neurotrophic factor protein | Surface | 5.0–300 ng/mL | 0.1 pg/mL | [88] |
| Optical | Escherichia coli | Micro-contact | 101–106 cfu/mL | 0.57 cfu/mL | [89] |
| Fluorescent | Tetrabromobisphenol-A | Sol-gel | 1.0–60 ng/mL | 3.6 ng/g | [90] |
| Piezoelectric | Cannabinoids | Emulsion | 0.0005–1.0 ng/mL | 0.28 ng/mL | [91] |
| Combination | Template Molecule | Polymerization Type | Dynamic Range | Limit of Detection | Reference |
|---|---|---|---|---|---|
| Fluorescent sensor | 2,4-dichloro phenoxyacetic acid | RAFT | 20 nM–5 μM | 20 nM | [97] |
| Fluorescent sensor | Cu2+ | Surface | 0.11–58 μg/L | 0.035 μg/L | [98] |
| Fluorescent sensor | 4-nitrophenol | Surface | 0.5–20 mg/L | 0.097 mg/L | [99] |
| Electrochemical sensor | Ovalbumin | In-situ | 1 pg/mL–1000 ng/mL | 0.87 pg/mL | [100] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saylan, Y.; Erdem, Ö.; Inci, F.; Denizli, A. Advances in Biomimetic Systems for Molecular Recognition and Biosensing. Biomimetics 2020, 5, 20. https://doi.org/10.3390/biomimetics5020020
Saylan Y, Erdem Ö, Inci F, Denizli A. Advances in Biomimetic Systems for Molecular Recognition and Biosensing. Biomimetics. 2020; 5(2):20. https://doi.org/10.3390/biomimetics5020020
Chicago/Turabian StyleSaylan, Yeşeren, Özgecan Erdem, Fatih Inci, and Adil Denizli. 2020. "Advances in Biomimetic Systems for Molecular Recognition and Biosensing" Biomimetics 5, no. 2: 20. https://doi.org/10.3390/biomimetics5020020
APA StyleSaylan, Y., Erdem, Ö., Inci, F., & Denizli, A. (2020). Advances in Biomimetic Systems for Molecular Recognition and Biosensing. Biomimetics, 5(2), 20. https://doi.org/10.3390/biomimetics5020020








