Diagnosis of Schizophrenia Using Feature Extraction from EEG Signals Based on Markov Transition Fields and Deep Learning
Abstract
1. Introduction
- We are the first to integrate Markov Transition Fields (MTF) with deep feature extraction (VGG-16) on EEG data for schizophrenia detection.
- We implemented a traditional Support Vector Machine (SVM) and a Neural Network (NN) integrated with an autoencoder for dimensionality reduction, ensuring both accuracy and computational efficiency.
- We have performed a comprehensive evaluation of the model performance, including accuracy, precision, recall, specificity, F1-score, and AUC to validate the reliability of the proposed architecture.
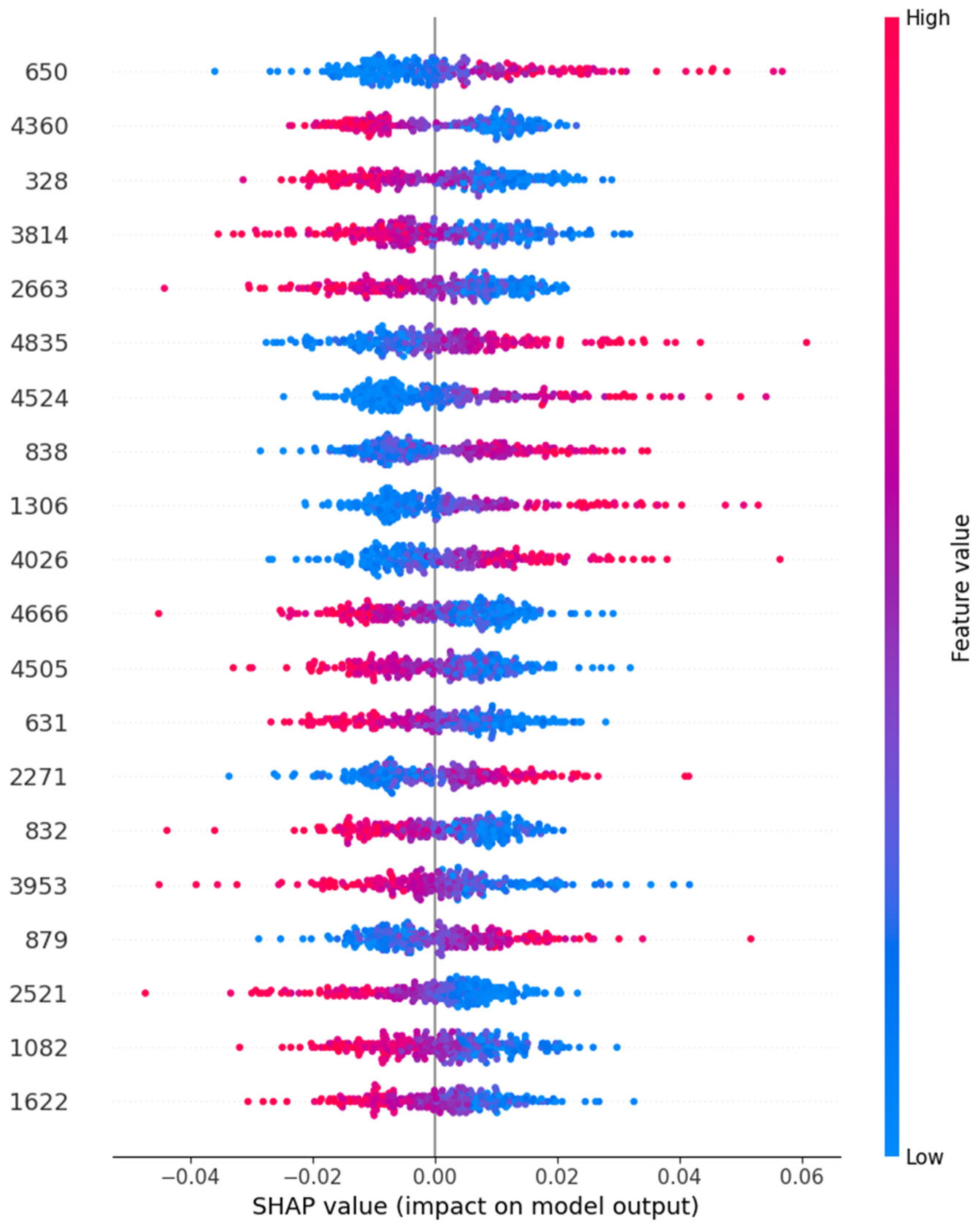
- We have demonstrated the model’s explainability and generalizability by plotting a SHAP (SHapley Additive exPlanations) plot for our autoencoder with a neural network model, positioning it as a scalable tool for aiding clinical decision-making in mental health diagnostics.
2. Related Work
3. Methodology
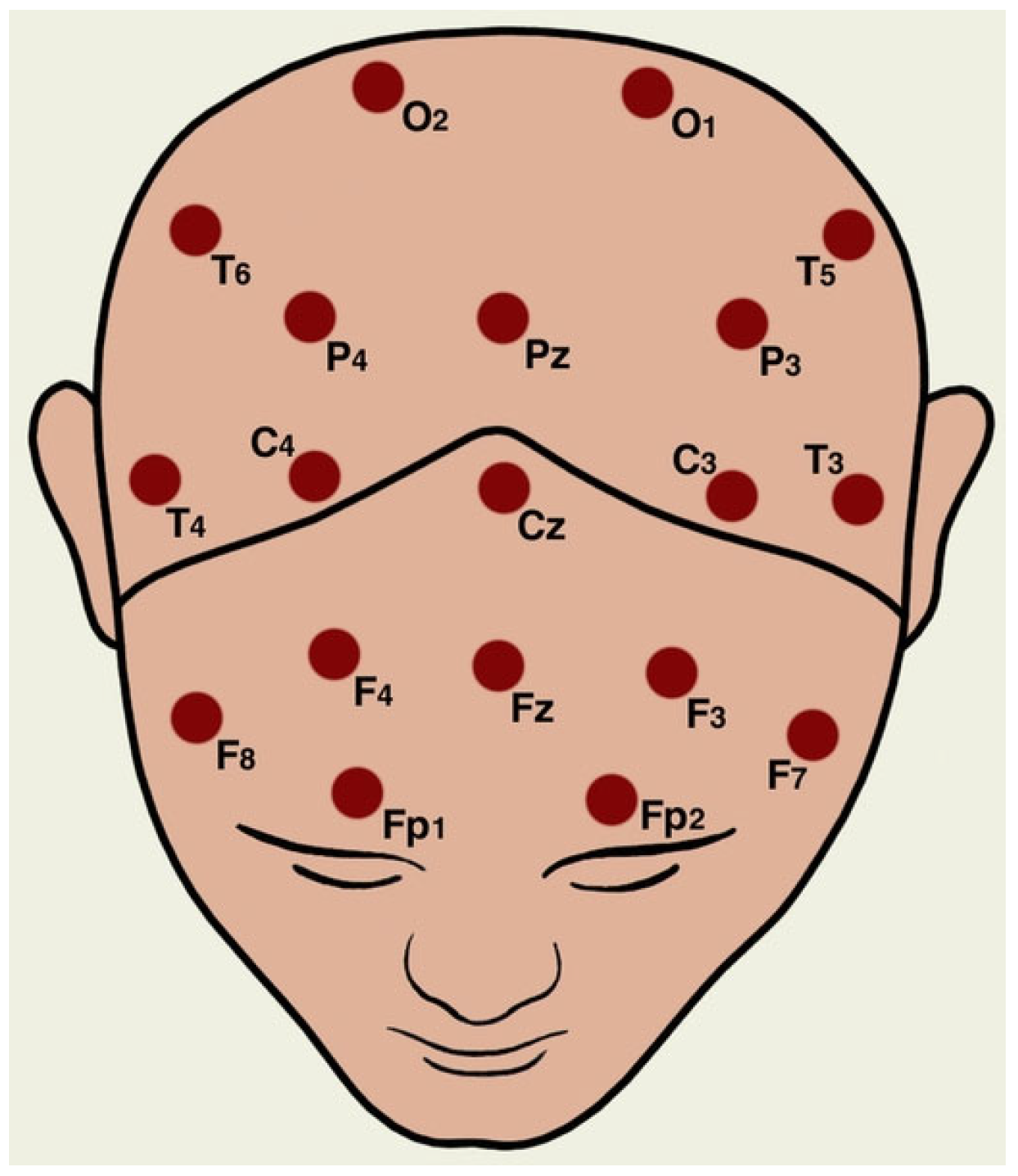
3.1. Dataset
3.2. Preprocessing
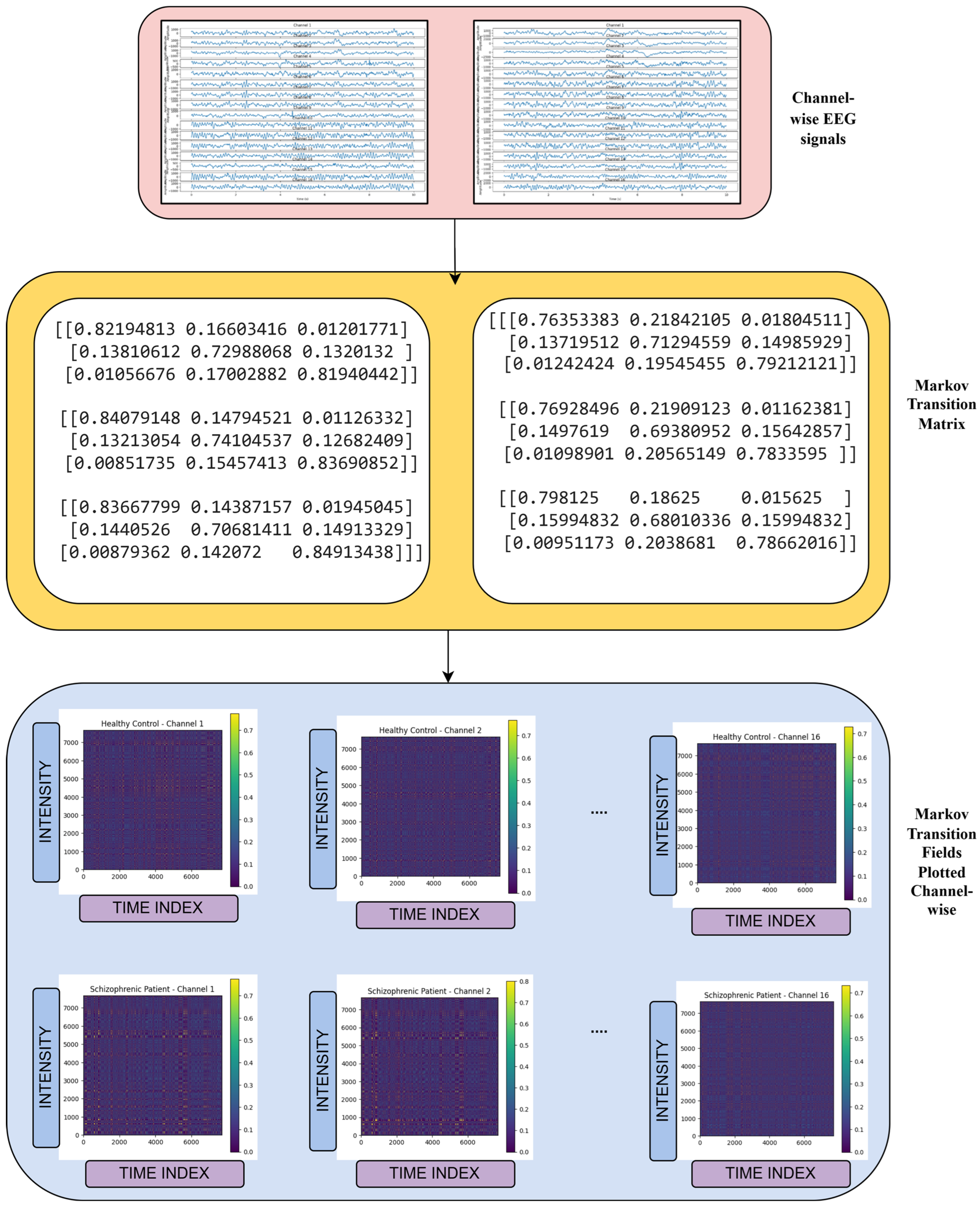
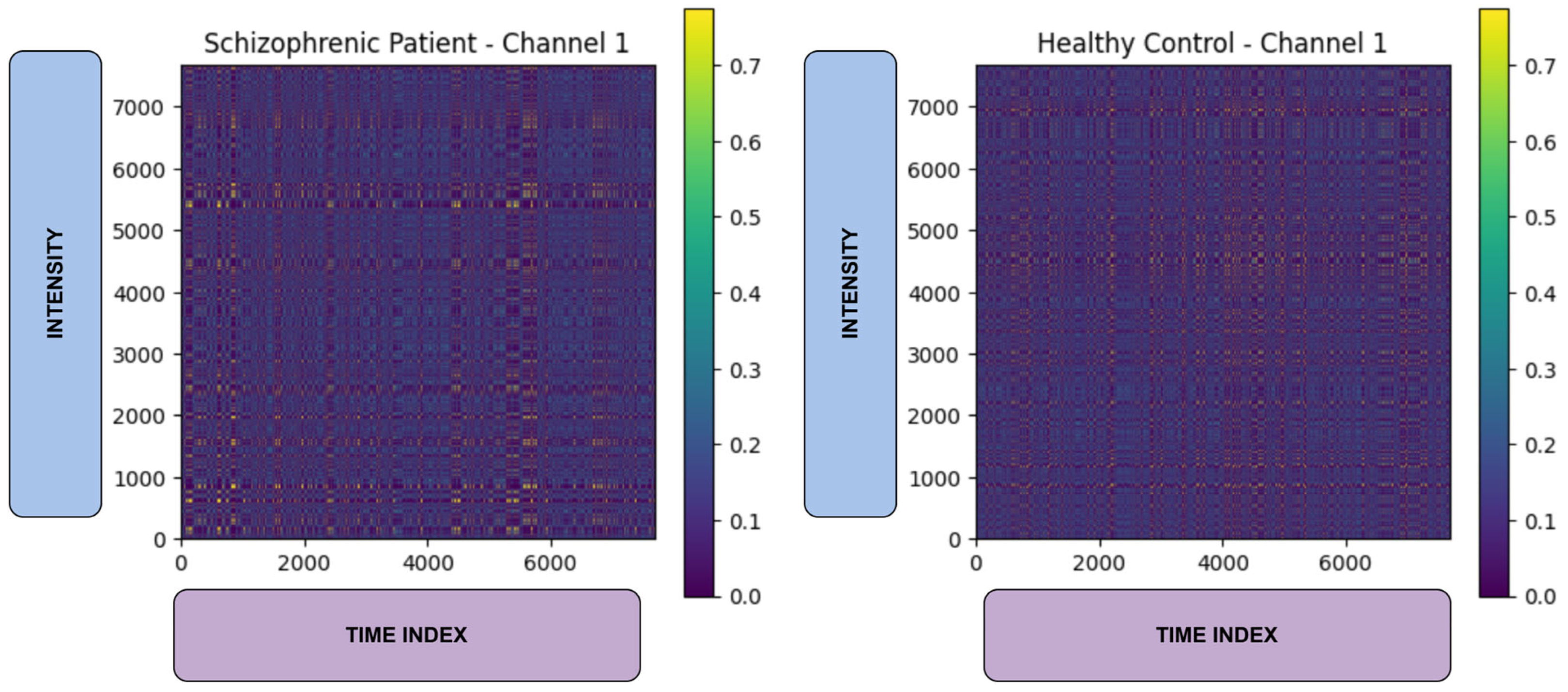
3.3. Markov Transition Field Transformation
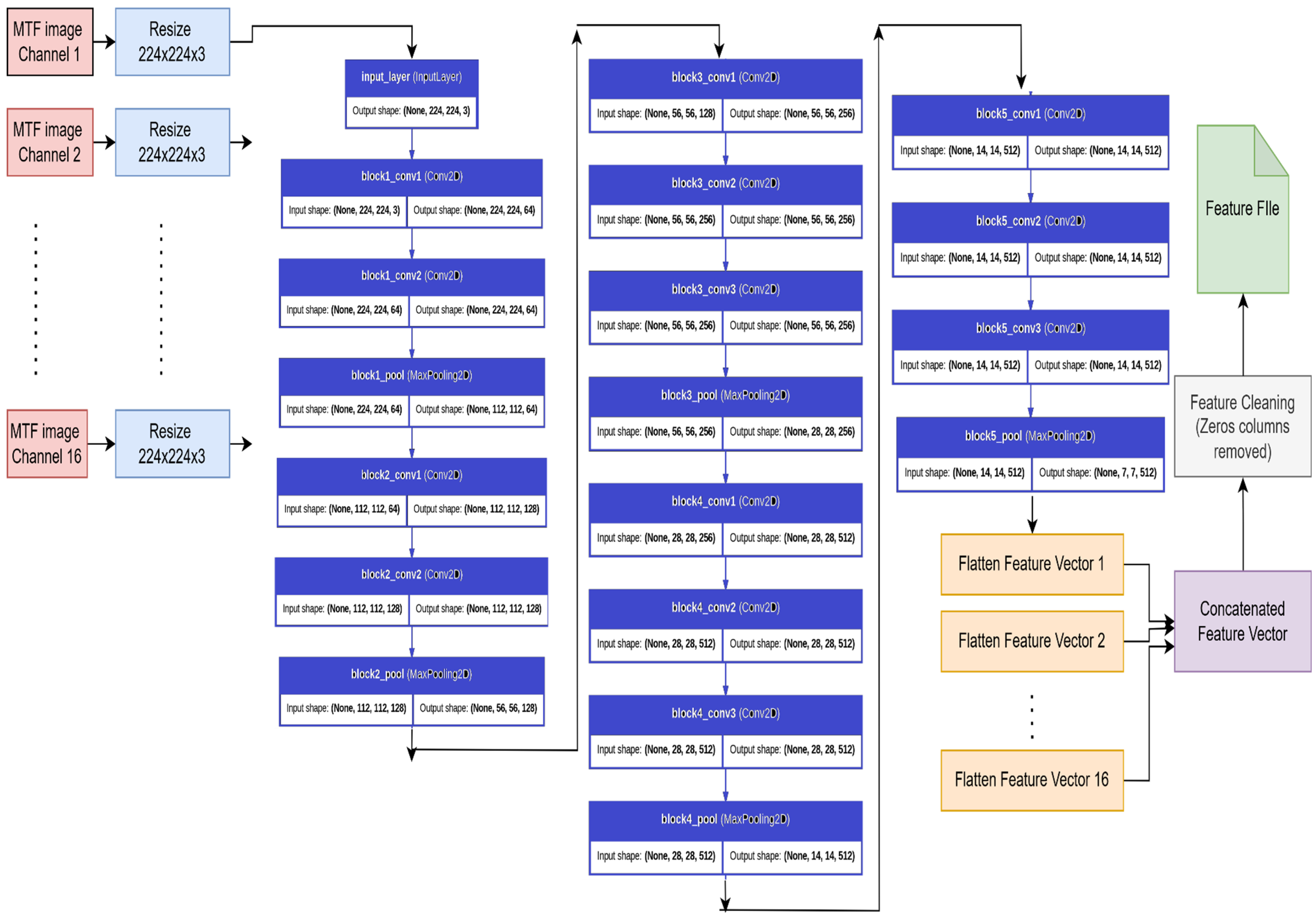
3.4. Feature Extraction Using Pre-Trained VGG-16 Network
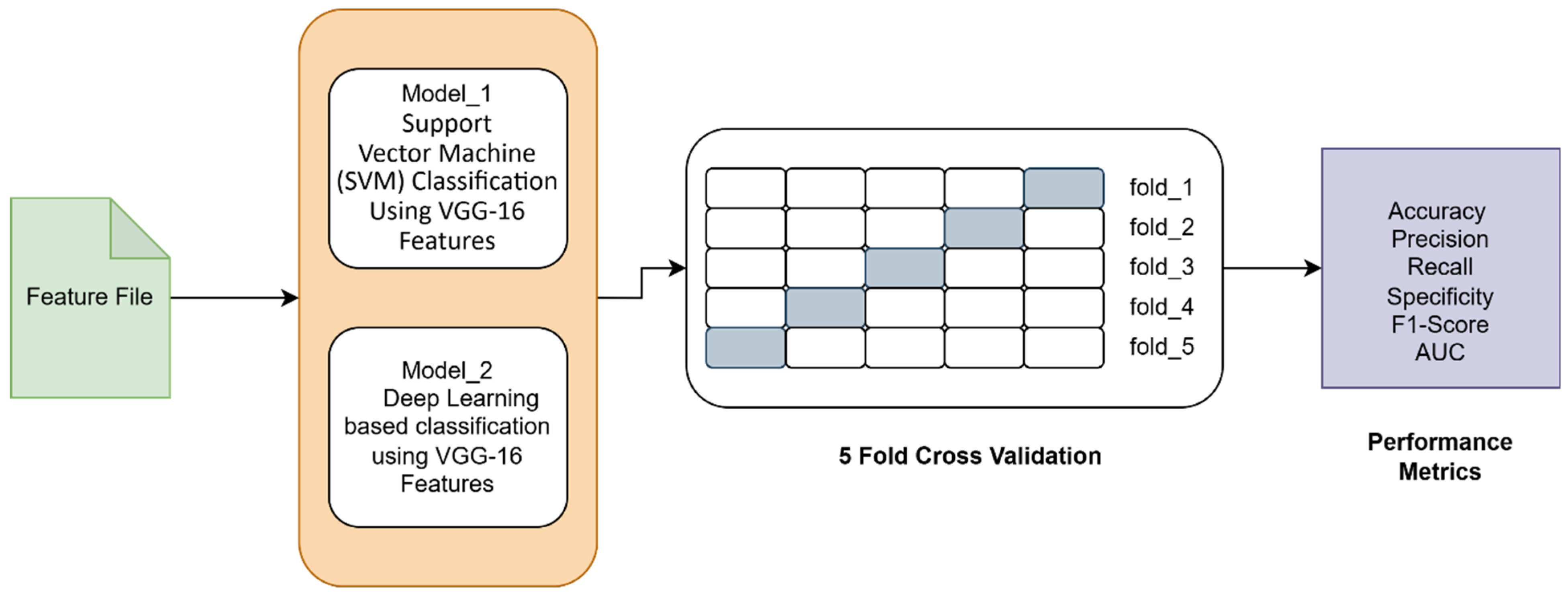
3.4.1. Model_1: Support Vector Machine (SVM) Classification Using VGG-16 Features
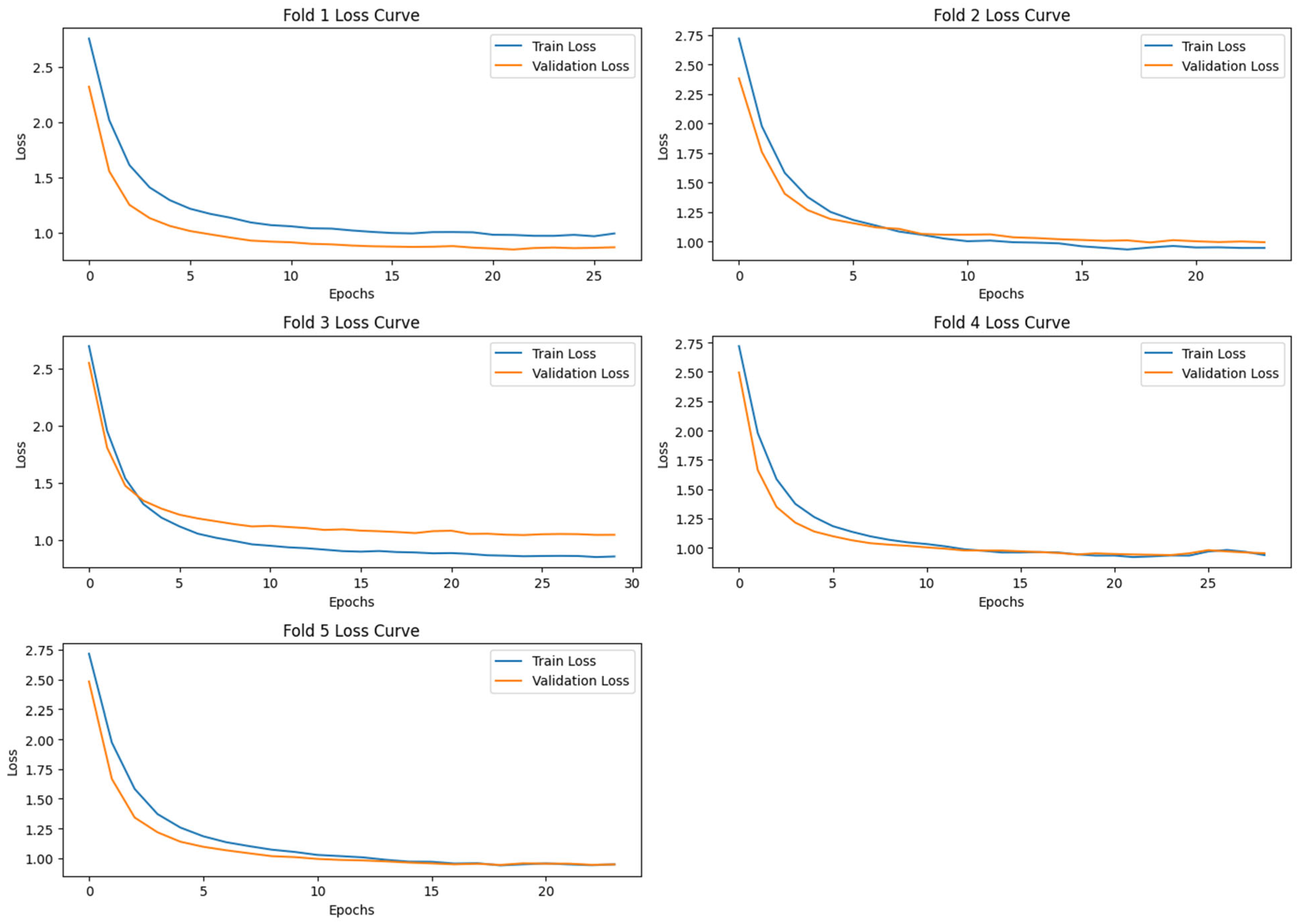
3.4.2. Model_2: Deep Learning Based Classification Using VGG-16 Features
- Autoencoder for Feature Compression: A symmetrical autoencoder was designed with gaussian noise injection [17] at the input to enhance generalizability. It is random noise sampled from a gaussian distribution with a mean and a standard deviation. It simulates real-world EEG-variability, forcing the autoencoder to learn noise-invariant features. The encoder comprised two hidden layers with 512 and 256 neurons, respectively, activated using ELU and regularized using L2 weight decay and dropout layers. The encoded latent representation was size 512. The decoder mirrored the encoder architecture in reverse order to reconstruct the input features. The model was trained to minimize mean squared error (MSE) loss. The hyperparameters that are used in the model are batch_size = 64, learning_rate = 0.001, encoding_dim = 512, epochs = 30, and droupout = 0.4.
- Classification with Fully Connected Neural Network: The encoded features were then fed into a Fully Connected Neural Network classifier comprising two hidden layers (128 and 64 neurons) with ReLU activation, batch normalization, L2 regularization, and dropout. The output layer consisted of a single sigmoid unit for binary classification. The model was compiled using the Adam optimizer with a binary cross-entropy loss function. The hyperparameters that are used in the model are batch_size = 64, learning_rate = 0.001, epochs = 30, and droupout = 0.5.
3.5. Hardware and Software
3.6. Evaluation Metrics
4. Results and Analysis
5. Discussion
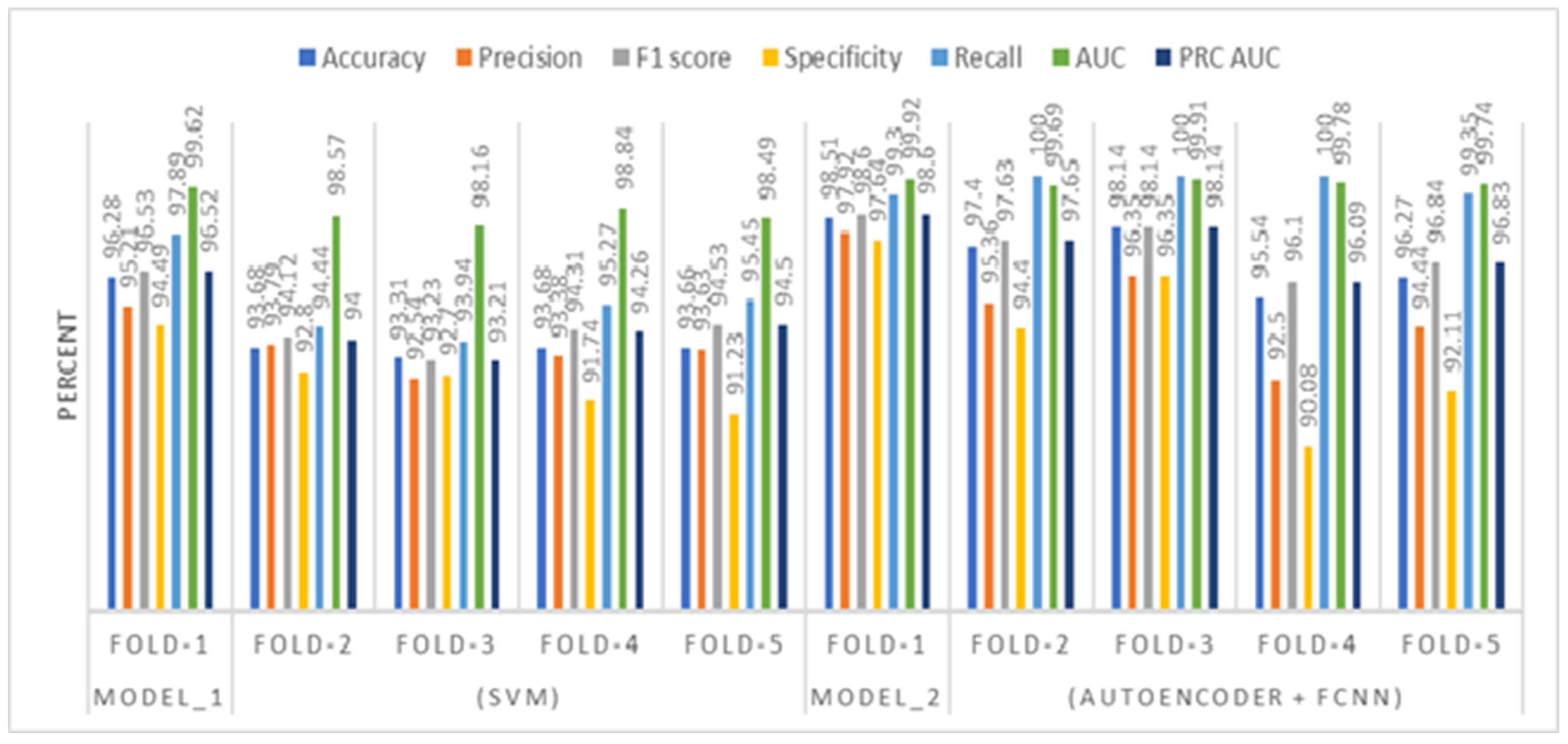
5.1. Comparative Analysis of SVM and Neural Network Model
5.2. Model Explainability Using SHAP
5.3. Advantages of the Proposed Model
- High Accuracy and Robust Performance: The model achieved exceptionally high classification performance (98.51% accuracy, 100% recall), indicating strong potential for reliable schizophrenia detection.
- Effective Feature Representation: By transforming EEG signals into Markov Transition Fields (MTFs) and extracting deep features through VGG-16, the model captures complex spatial–temporal patterns associated with brain activity.
- Dimensionality Reduction: The use of an autoencoder significantly reduces feature dimensionality while retaining critical information, leading to a more efficient and noise-resilient model.
- Model Explainability: SHAP (SHapley Additive exPlanations) analysis adds a layer of explainability, making it easier to understand which features contribute most to the classification outcome—a crucial step toward clinical adoption.
- Comparative Versatility: The framework is flexible, as demonstrated by the competitive performance of both deep learning (Autoencoder + NN) and classical (SVM) classifiers.
5.4. Limitations of the Proposed Model
- Binary Classification Scope: The model currently addresses only binary classification (schizophrenia vs. healthy), which limits its applicability in diagnosing other psychiatric or neurological disorders.
- Dataset Constraints: Results are based on a single dataset with a limited number of subjects. The model’s performance and generalizability across diverse populations or recording settings remain to be tested.
- Computational Requirements: Deep learning models, particularly those involving VGG-16 and autoencoders, require substantial computational resources and may not be feasible in all clinical environments.
- Black-Box Nature: Despite SHAP-based insights, deep learning models are still relatively opaque compared to traditional statistical methods, which may hinder clinician trust and adoption.
5.5. Practical Applications
- Clinical Decision Support: The proposed system can serve as a valuable tool to assist psychiatrists in early and objective diagnosis of schizophrenia, reducing reliance on subjective assessments.
- Screening in Remote or Resource-Limited Settings: With appropriate optimization, this model could enable preliminary screening using portable EEG devices in underserved regions.
- Neuropsychiatric Research: The framework can aid in studying neural biomarkers of schizophrenia, helping identify key EEG patterns linked to the disorder.
5.6. Challenges in Real-World Deployment
- Standardization of EEG Acquisition: EEG signals are sensitive to noise, electrode placement, and individual variability. Standardizing data collection protocols is essential for reliable results.
- Regulatory and Ethical Concerns: Deployment in clinical settings would require regulatory approval and careful handling of patient data to ensure privacy and ethical use.
- Model Adaptability: Models need to adapt to individual variability, comorbid conditions, and medication effects, which can alter EEG patterns.
- Clinician Acceptance: To be integrated into routine practice, the model must be transparent, easy to use, and complement (not replace) clinical expertise.
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- The Natural History of Schizophrenia in the Long Term | GHDx. [Online]. Available online: https://ghdx.healthdata.org/record/natural-history-schizophrenia-long-term (accessed on 20 June 2025).
- Oh, S.L.; Vicnesh, J.; Ciaccio, E.J.; Yuvaraj, R.; Acharya, U.R. Deep convolutional neural network model for automated diagnosis of Schizophrenia using EEG signals. Appl. Sci. 2019, 9, 2870. [Google Scholar] [CrossRef]
- Laursen, T.M.; Nordentoft, M.; Mortensen, P.B. Excess Early Mortality in Schizophrenia. Annu. Rev. Clin. Psychol. 2014, 10, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Harvey, P.D.; Heaton, R.K.; Carpenter, W.T.; Green, M.F.; Gold, J.M.; Schoenbaum, M. Diagnosis of schizophrenia: Consistency across information sources and stability of the condition. Schizophr. Res. 2012, 140, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Aslan, Z.; Akin, M. A deep learning approach in automated detection of schizophrenia using scalogram images of EEG signals. Phys. Eng. Sci. Med. 2022, 45, 83–96. [Google Scholar] [CrossRef]
- Miller, D.D.; Brown, E.W. Artificial Intelligence in Medical Practice: The Question to the Answer? Am. J. Med. 2018, 131, 129–133. [Google Scholar] [CrossRef]
- Gosala, B.; Singh, A.R.; Tiwari, H.; Gupta, M. GCN-LSTM: A hybrid graph convolutional network model for schizophrenia classification. Biomed. Signal Process Control 2025, 105, 107657. [Google Scholar] [CrossRef]
- Gosala, B.; Kapgate, P.D.; Jain, P.; Chaurasia, R.N.; Gupta, M. Wavelet transforms for feature engineering in EEG data processing: An application on Schizophrenia. Biomed. Signal Process Control 2023, 85, 104811. [Google Scholar] [CrossRef]
- Agarwal, M.; Singhal, A. Fusion of pattern-based and statistical features for Schizophrenia detection from EEG signals. Med. Eng. Phys. 2023, 112, 103949. [Google Scholar] [CrossRef] [PubMed]
- Aslan, Z.; Akin, M. Automatic Detection of Schizophrenia by Applying Deep Learning over Spectrogram Images of EEG Signals. Trait. Signal 2020, 37, 235–244. [Google Scholar] [CrossRef]
- Ko, D.-W.; Yang, J.-J. EEG-Based Schizophrenia Diagnosis through Time Series Image Conversion and Deep Learning. Electronics 2022, 11, 2265. [Google Scholar] [CrossRef]
- EEG Database—Schizophrenia. [Online]. Available online: http://brain.bio.msu.ru/eeg_schizophrenia.htm (accessed on 26 March 2025).
- Homan, R.W.; Herman, J.; Purdy, P. Cerebral location of international 10–20 system electrode placement. Electroencephalogr. Clin. Neurophysiol. 1987, 66, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-R.; Yen, C.-T. Markov Transition Field and Convolutional Long Short-Term Memory Neural Network for Manufacturing Quality Prediction. In Proceedings of the 2020 IEEE International Conference on Consumer Electronics—Taiwan (ICCE-Taiwan), Taoyuan, Taiwan, 28–30 September 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–2. [Google Scholar] [CrossRef]
- Ji, L.; Wei, Z.; Hao, J.; Wang, C. An intelligent diagnostic method of ECG signal based on Markov transition field and a ResNet. Comput. Methods Programs Biomed. 2023, 242, 107784. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F. Adaptive Gaussian Noise Injection Regularization for Neural Networks. In International Symposium on Neural Networks; Springer International Publishing: Cham, Switzerland, 2020; pp. 176–189. [Google Scholar] [CrossRef]
- Bisong, E. Google Colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform; Apress: Berkeley, CA, USA, 2019; pp. 59–64. [Google Scholar] [CrossRef]
- Faouzi, J. pyts: A Python Package for Time Series Classification. 2020. Available online: https://github.com/johannfaouzi/pyts (accessed on 20 June 2025).
- Ari, N.; Ustazhanov, M. Matplotlib in python. In Proceedings of the 2014 11th International Conference on Electronics, Computer and Computation (ICECCO), Abuja, Nigeria, 29 September–1 October 2014; IEEE: Piscataway, NJ, USA, 2014; pp. 1–6. [Google Scholar] [CrossRef]


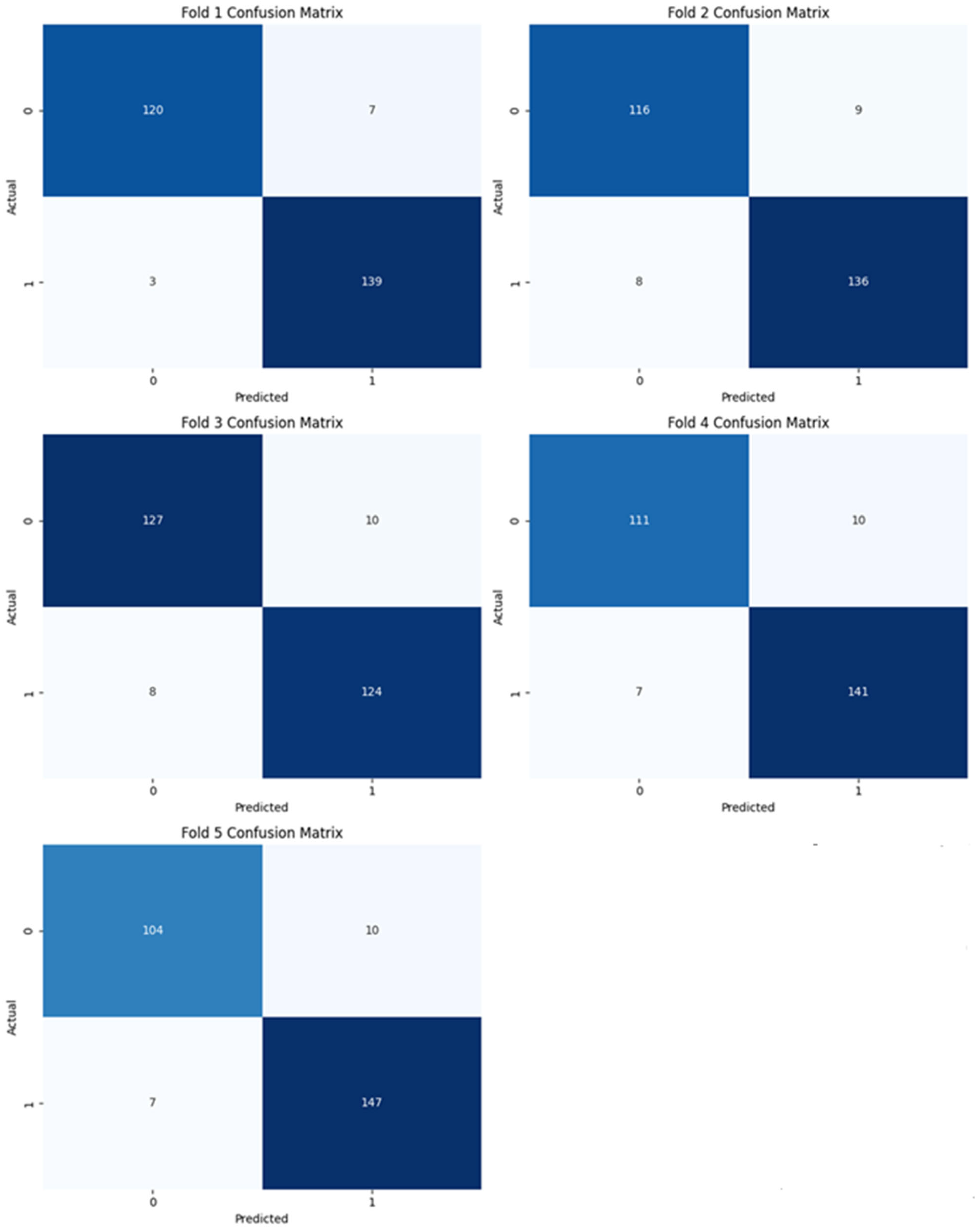
| Predicted Positive | Predicted Negative | |
|---|---|---|
| Actual Positive | True Positive (TP) | False Negative (FN) |
| Actual Negative | False Positive (FP) | True Negative (TN) |
| Performance Metrics | Accuracy | Precision | F1 Score | Specificity | Recall | AUC | PRC AUC | |
|---|---|---|---|---|---|---|---|---|
| Model_1 (SVM) | Fold-1 Fold-2 Fold-3 Fold-4 Fold-5 | 96.28 93.68 93.31 93.68 93.66 | 95.21 93.79 92.54 93.38 93.63 | 96.53 94.12 93.23 94.31 94.53 | 94.49 92.80 92.70 91.74 91.23 | 97.89 94.44 93.94 95.27 95.45 | 99.62 98.57 98.16 98.84 98.49 | 96.52 94.00 93.21 94.26 94.50 |
| Model_2 (Autoencoder + FCNN) | Fold-1 Fold-2 Fold-3 Fold-4 Fold-5 | 98.51 97.40 98.14 95.54 96.27 | 97.92 95.36 96.35 92.50 94.44 | 98.60 97.63 98.14 96.10 96.84 | 97.64 94.40 96.35 90.08 92.11 | 99.30 100.00 100.00 100.00 99.35 | 99.92 99.69 99.91 99.78 99.74 | 98.60 97.65 98.14 96.09 96.83 |
| Authors | Dataset | Classification Algorithm | Performance Metrics (%) |
|---|---|---|---|
| Aslan et al., 2020 [10] | Open-access Schizophrenia EEG database provided by MV Lomonosov Moscow State University and the Institute of Psychiatry and Neurology, Warsaw, Poland | Deep features were extracted from STFT images using a pre-trained VGG-16 convolutional neural network model | Accuracy (Dataset1) = 95.00 Accuracy (Dataset 2) = 97.00 |
| Proposed model | Open-access Schizophrenia EEG database provided by MV Lomonosov Moscow State University | The approach combined Markov Transition Field (MTF) transformation with deep feature extraction using VGG-16, followed by dimensionality reduction through an autoencoder and final classification using a neural network (NN) (Model_2). A Support Vector Machine (SVM) (Model_1) classifier was also used for comparison. | Model_1: Accuracy = 96.28, precision = 95.21, Specificity = 94.49 recall = 97.89, F1-score = 96.53, and AUC = 99.62 Model_2: Accuracy = 98.51, precision = 97.92, Specificity = 97.64 recall = 100.00, F1-score = 98.60, and AUC = 99.92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jalan, A.; Mishra, D.; Marisha; Gupta, M. Diagnosis of Schizophrenia Using Feature Extraction from EEG Signals Based on Markov Transition Fields and Deep Learning. Biomimetics 2025, 10, 449. https://doi.org/10.3390/biomimetics10070449
Jalan A, Mishra D, Marisha, Gupta M. Diagnosis of Schizophrenia Using Feature Extraction from EEG Signals Based on Markov Transition Fields and Deep Learning. Biomimetics. 2025; 10(7):449. https://doi.org/10.3390/biomimetics10070449
Chicago/Turabian StyleJalan, Alka, Deepti Mishra, Marisha, and Manjari Gupta. 2025. "Diagnosis of Schizophrenia Using Feature Extraction from EEG Signals Based on Markov Transition Fields and Deep Learning" Biomimetics 10, no. 7: 449. https://doi.org/10.3390/biomimetics10070449
APA StyleJalan, A., Mishra, D., Marisha, & Gupta, M. (2025). Diagnosis of Schizophrenia Using Feature Extraction from EEG Signals Based on Markov Transition Fields and Deep Learning. Biomimetics, 10(7), 449. https://doi.org/10.3390/biomimetics10070449










