1. Introduction
The increasing importance of feature selection arises from the complexities introduced by high-dimensional datasets [
1]. In such datasets, irrelevant or redundant features can obscure meaningful patterns, compromise model performance, and escalate computational demands [
2]. By concentrating on the identification of a subset of features that maintains or enhances a model’s predictive power, feature selection boosts the efficiency and efficacy of machine learning workflows [
3].
Feature selection methods can be broadly categorized into three main types: filter methods, embedded methods, and wrapper methods, each distinguished by their underlying principles and inherent trade-offs [
4]. Filter methods employ statistical measures to assess and rank features independent of any specific predictive model. Widely used techniques encompass correlation coefficients [
5], mutual information [
6], and variance thresholds [
7]. Although computationally efficient, filter methods often overlook xinteractions among features, limiting their effectiveness in more complex situations. Embedded methods, conversely, integrate the feature selection process directly within the model training phase. Examples include Lasso regression [
8], which introduces a penalty term to shrink the coefficients of less relevant features to zero and where feature importance is derived from split criteria. These methods generally provide improved performance by aligning feature selection with the model’s objectives but are restricted by the choice of the base algorithm. Wrapper methods take a more comprehensive and iterative approach, evaluating feature subsets using a predictive model [
9]. Despite their computational overheads, they are effective at addressing feature interactions and customizing the selected subset for a particular problem. Techniques like forward selection, backward elimination, and recursive feature elimination illustrate this category, underscoring its ability to effectively optimize feature sets. The very nature of wrapper-based feature selection represents a global optimization problem, where the search for the optimal subset within an exponentially growing number of combinations necessitates the use of efficient algorithms. Formally, this problem can be defined as follows:
Let denote the complete set of features, and let be a binary vector where indicates whether the -th feature is selected () or not (). The goal of wrapper-based feature selection is to identify the optimal subset of features that maximizes (or minimizes) a predefined objective function , which typically evaluates the performance of a predictive model trained on the selected features. The search space for this problem is combinatorial in nature, with a total of possible feature subsets.
Traditional methods, such as exhaustive search or greedy algorithms, often struggle with the curse of dimensionality, thereby prompting the adoption of metaheuristic approaches [
10].
Metaheuristic algorithms have emerged as effective tools for addressing challenging optimization problems, particularly in high-dimensional, multimodal, and non-convex search spaces [
11]. These algorithms can be generally categorized into two main types: evolutionary algorithms and swarm intelligence algorithms. Evolutionary algorithms, drawing inspiration from natural selection, encompass techniques such as genetic algorithms (GA) [
12] and differential evolution (DE) [
13], which emulate biological evolutionary processes. Conversely, swarm intelligence algorithms, inspired by the collective behaviors of animal groups, include methods like particle swarm optimization (PSO) [
14] and ant colony optimization (ACO) [
15]. While both categories emphasize the importance of balancing exploration and exploitation, they diverge in their foundational principles and operational mechanisms.
Over the past few years, metaheuristic algorithms, particularly swarm intelligence-based approaches, have shown significant promise in addressing FS challenges. Several studies have explored improved versions of established metaheuristic algorithms for FS. For instance, Gao et al. [
16] introduced clustering probabilistic particle swarm optimization (CPPSO), which enhances traditional PSO with probabilistic velocity representation and a K-means clustering strategy to improve both exploration and exploitation for high-dimensional data. Similarly, hybrid approaches have gained traction, such as the particle swarm-guided bald eagle search (PS-BES) by Kwakye et al. [
17], which combines the speed of PSO to guide bald eagle search, introducing an attack–retreat–surrender mechanism to better balance diversification and intensification. These studies showcase the effectiveness of leveraging different search mechanisms for improved performance on benchmark datasets and real-world problems. Other variations of metaheuristics have explored improved exploration strategies, such as a modified version of the forensic-based investigation algorithm (DCFBI) proposed by Hu et al. [
18], incorporating dynamic individual selection and crisscross mechanisms for improved convergence and avoidance of local optima. Furthermore, Askr et al. [
19] proposed binary-enhanced golden jackal optimization (BEGJO), using copula entropy for dimensionality reduction while integrating enhancement strategies to improve exploration and exploitation capabilities. Beyond the improved variations of metaheuristic algorithms, novel algorithms have also emerged. Lian et al. [
20] presented the parrot optimizer (PO), inspired by parrot behaviors, integrating stochasticity to enhance population diversity and avoid local optima. Likewise, Singh et al. [
21] explored combining emperor penguin optimization, bacterial foraging optimization, and their hybrid to optimize feature selection for glaucoma classification. These studies indicate the emergence of diverse metaheuristic strategies to balance exploration and exploitation for FS.
While metaheuristic approaches have shown considerable success in feature selection, the no free lunch (NFL) theorem highlights their inherent limitations [
22]. The NFL theorem asserts that no single optimization algorithm can consistently outperform all others across all problem instances. This necessitates ongoing innovation and adaptation of metaheuristic strategies to address diverse feature selection challenges. Researchers are thus motivated to refine existing algorithms or explore the combination of multiple techniques, such as hybridizing algorithms or incorporating adaptive mechanisms, to enhance their generalizability and robustness [
23,
24]. Informed by these considerations and the need to overcome the constraints of current metaheuristic algorithms, this study introduces an innovative approach to enhance existing algorithms, aiming to advance their applicability to both feature selection and global optimization tasks.
The polar lights optimization (PLO) algorithm, a recent metaheuristic optimization approach proposed by Yuan et al. in 2024 [
25], draws its inspiration from the natural phenomenon of the aurora. PLO emulates the movement of high-energy particles as they are affected by the Earth’s magnetic field and atmosphere, incorporating three fundamental mechanisms: gyration motion for local exploitation, aurora oval walk for global exploration, and particle collision to facilitate an escape from local optima. A key advantage of PLO lies in its ability to balance local and global search through the use of adaptive weights. However, similar to other metaheuristic algorithms, PLO’s performance can be susceptible to parameter settings, and its convergence may be challenged by high-dimensional problems. Therefore, further research is warranted to investigate parameter-tuning strategies and assess PLO’s performance across diverse real-world applications to validate its robustness and practical utility.
This paper introduces CPLODE, an enhanced version of the polar lights optimization (PLO) algorithm, designed to improve its search capabilities through the integration of a cryptobiosis mechanism and differential evolution (DE) operators. Specifically, the cryptobiosis mechanism refines the greedy selection process within PLO, allowing the algorithm to retain and reuse historically effective search directions. Moreover, the original particle collision strategy in PLO is replaced by DE’s mutation and crossover operators, which provide a more effective means for global exploration and employ a dynamic crossover rate to promote improved convergence. These modifications collectively contribute to the enhanced performance of CPLODE. The key contributions of this paper can be summarized as follows:
A novel enhanced polar lights optimization algorithm, CPLODE, is proposed by integrating a cryptobiosis mechanism and differential evolution operators to enhance the search effectiveness.
The DE mutation and crossover operators replace the original particle collision strategy and use a dynamic and adaptive crossover rate to enable better solution convergence.
The cryptobiosis mechanism replaces the greedy selection approach and allows for the preservation and reuse of historically successful solutions to improve the overall performance.
The performance of CPLODE is validated through comprehensive experiments, demonstrating its efficacy in solving complex optimization problems.
The remainder of this paper is organized as follows:
Section 1 introduces the research background, motivation, and key contributions.
Section 2 describes the fundamentals of the original PLO algorithm.
Section 3 presents the proposed CPLODE algorithm, including detailed explanations of the cryptobiosis mechanism and the DE operators.
Section 4 covers the experimental setup, results, and their analysis to evaluate CPLODE’s performance.
Section 5 explores the application of the proposed CPLODE algorithm in feature selection. Finally,
Section 6 concludes the paper by summarizing the key findings and outlining potential future work.
2. The Original PLO
Polar lights optimization (PLO), introduced by Yuan et al. [
25] in 2024, is a novel metaheuristic algorithm that mimics the movement of high-energy particles interacting with the Earth’s geomagnetic field and atmosphere, inspired by the natural phenomenon of the aurora. The algorithm solves optimization problems by modeling this particle motion, which is divided into three core phases: gyration motion, aurora oval walk, and particle collision.
1. Gyration motion: Inspired by the spiraling trajectory of high-energy particles under Lorentz force and atmospheric damping, gyration motion facilitates local exploitation. Mathematically, this is represented by the following equation:
where
represents the particle’s velocity at time
,
is a constant,
represents the particle’s charge,
is the strength of Earth’s magnetic field, α represents the atmospheric damping factor, and m is the mass of the particle. In the PLO algorithm,
,
, and
are set to 1 for simplicity, and
m is set to 100. The damping factor α is a random value within the range [1, 1.5]. The fitness evaluation process of the current particle represents the time (
) to model the decaying spiraling trajectories, enabling fine-grained local searches.
2. Aurora Oval Walk: The aurora oval walk emulates the dynamic movement of energetic particles along the auroral oval, facilitating global exploration. This movement is influenced by a Levy flight distribution, the average population position, and a random search component. The aurora oval walk of each particle is calculated using the following equation:
where
represents the movement of a particle in the auroral oval walk,
represents the
-th individual and ranges from 1 to
(population size), and
j represents the
-th dimension, ranging from 1 to
(problem dimension).
is the Levy distribution,
is the step size,
represents the direction the particles tend to move toward the average location,
is the lower bound of the search space,
is the upper bound of the search space, and
is a random number [0, 1].
This auroral oval walk enables rapid exploration of the solution space through a seemingly random walk. To integrate both gyration motion and the auroral oval walk, the updated position of each particle (
) is computed as follows:
where
is the current particle position, and
introduces randomness, taking values between 0 and 1.
and
are adaptive weights that balance exploration and exploitation. They are updated in each iteration as follows:
where
is the current iteration, and
is the maximum number of iterations.
increases over time, giving more weight to gyration motion and
decreases, giving less weight to the auroral oval walk, shifting from global search towards local exploitation.
3. Particle Collision: Inspired by the violent particle collisions in Earth’s magnetic field, which result in energy transfer and changes in particle directions, this strategy enables particles to escape local optima. In PLO, each particle may collide with any other particle in the swarm and is modeled mathematically with:
where
is the new position of particle
in dimension
,
is the current position of particle
in dimension
,
is the position of a randomly selected particle in the population, and
,
, and
are random numbers from [0, 1]. The sine function introduces a variable direction of movement after the collision. The collision probability,
, increases with iterations as follows:
The PLO algorithm iteratively updates the particle positions by combining gyration motion and the aurora oval walk by Equations (2) and (3). These motion patterns are balanced by adaptive weights that gradually shift emphasis from global exploration to local exploitation. The particle collision behavior occurs stochastically, allowing particles to escape local optima. This process continues until a maximum number of iterations is reached, resulting in a near-optimal solution. The core strength of PLO lies in the combination of these inspired physical behaviors to enable an effective search and a specific mechanism to avoid local optima.
Figure 1 shows the flowchart of PLO.
5. Application to Feature Selection
This section explores the application of the proposed CPLODE algorithm to feature selection problems. Feature selection is a critical task in machine learning, aimed at identifying the most relevant features from a dataset, thereby reducing dimensionality and improving model performance. To apply CPLODE, a method designed for continuous domains, to this discrete problem, we employed a binary encoding strategy. Specifically, we constrained the problem’s upper and lower bounds to the interval [0, 1] and utilized Equation (11) to determine the selection status of each feature. This approach enables CPLODE, which is inherently suited for continuous domains, to operate effectively within the binary search space of feature selection.
To transition from the continuous search space of CPLODE to the binary feature selection space, each dimension is converted using a threshold of 0.5. Specifically, if a dimension’s value is greater than or equal to 0.5, the corresponding feature is selected; otherwise, it is not selected. This process maps the continuous space to a binary selection space.
In this study, we use the K-nearest neighbors (KNN) classifier to evaluate the quality of the selected feature subsets and use the following fitness function, which seeks to simultaneously minimize the classification error and the number of selected features:
where
represents the classification error rate,
l is the number of selected features,
L is the total number of features, and
μ is a constant between 0 and 1 controlling the trade-off between the error rate and the number of selected features. Because we primarily focus on the accuracy of the selected feature subset, we set
μ to 0.05 to prioritize error minimization. This setting gives more weight to the classification error and less weight to the number of features selected.
5.1. Detailed Description of Datasets
To evaluate the performance of the proposed CPLODE algorithm for feature selection, experiments were conducted on ten datasets selected from the UCI Machine Learning Repository. These datasets represent a range of complexity with varying numbers of samples, features, and classes. The number of samples in these datasets ranges from 72 to 2310, while the number of features varies from 8 to 7130. These variations ensure that the performance of the algorithm is tested across diverse feature selection scenarios, from low-dimensional to high-dimensional data and with different class distributions.
Table 4 provides a detailed description of each dataset used in this study, including the dataset name, number of samples, number of features, and number of classes.
5.2. Feature Selection Results and Discussion
This subsection presents the experimental results of CPLODE for feature selection, compared with several other well-known binary metaheuristic algorithms as follows: BPSO [
35], BGSA [
36], BALO [
37], BBA [
38], and BSSA [
39]. These experiments were conducted on the ten real-world datasets described in
Section 5.1. All algorithms were run with a population size of 30 and a maximum of 1000 iterations. To ensure robustness and avoid bias, a 10-fold cross-validation technique was used in all experiments. The detailed experimental results are shown in
Table 5 and
Table 6.
Table 5 presents the average classification error rates, with standard deviations in parentheses, obtained by each algorithm on each dataset. From the table, it can be observed that CPLODE achieves the lowest error rates on the majority of datasets, demonstrating its superior performance in feature selection. Notably, CPLODE achieves significantly lower error rates on datasets such as “Hepatitis_full_data” and “Segment”, where the error rates of CPLODE are well below other binary metaheuristic algorithms. While some algorithms, such as BPSO and BSSA, perform well on datasets like “Leukemia”, their results are not consistently better than CPLODE across all datasets. On the “Heart” dataset, CPLODE, BGSA, and BSSA exhibited similar performances, but all of the algorithms outperformed BBA, which had the worst performance. These results indicate that CPLODE achieves better accuracy on many of the datasets presented.
Table 6 presents the average number of selected features, with standard deviations in parentheses, achieved by each algorithm on each dataset. From the table, it can be observed that while CPLODE shows a competitive ability in selecting the relevant features, it does not consistently select the shortest feature subsets. For example, the BGSA algorithm achieves lower feature selections on the “Hepatitis_full_data” and “Leukemia” datasets. However, this lower feature count comes at the expense of higher error rates, as shown in
Table 5. Overall, CPLODE tends to select feature subsets that strike a good balance between classification performance and feature reduction, although not always selecting the absolute minimum number of features.
In summary, the experimental results demonstrate that the proposed CPLODE algorithm provides a highly competitive performance for feature selection. The proposed CPLODE demonstrates superior performance on most of the datasets used in this study, achieving a better classification performance. These superior results can be attributed to the integration of effective global search mechanisms through DE while employing the cryptobiosis mechanism for population quality control and the gyration motion strategy of PLO for local exploitation.
6. Conclusions
In this work, we have introduced CPLODE, a novel enhancement of the PLO algorithm, achieved through the integration of a cryptobiosis mechanism and DE operators. These modifications were designed to improve the original PLO’s search capabilities. Specifically, we replaced the original particle collision strategy with DE’s mutation and crossover operators, which enables more effective global exploration while also employing a dynamic crossover rate to enhance convergence. Furthermore, the cryptobiosis mechanism was incorporated to refine the greedy selection approach by recording and reusing historically successful solutions.
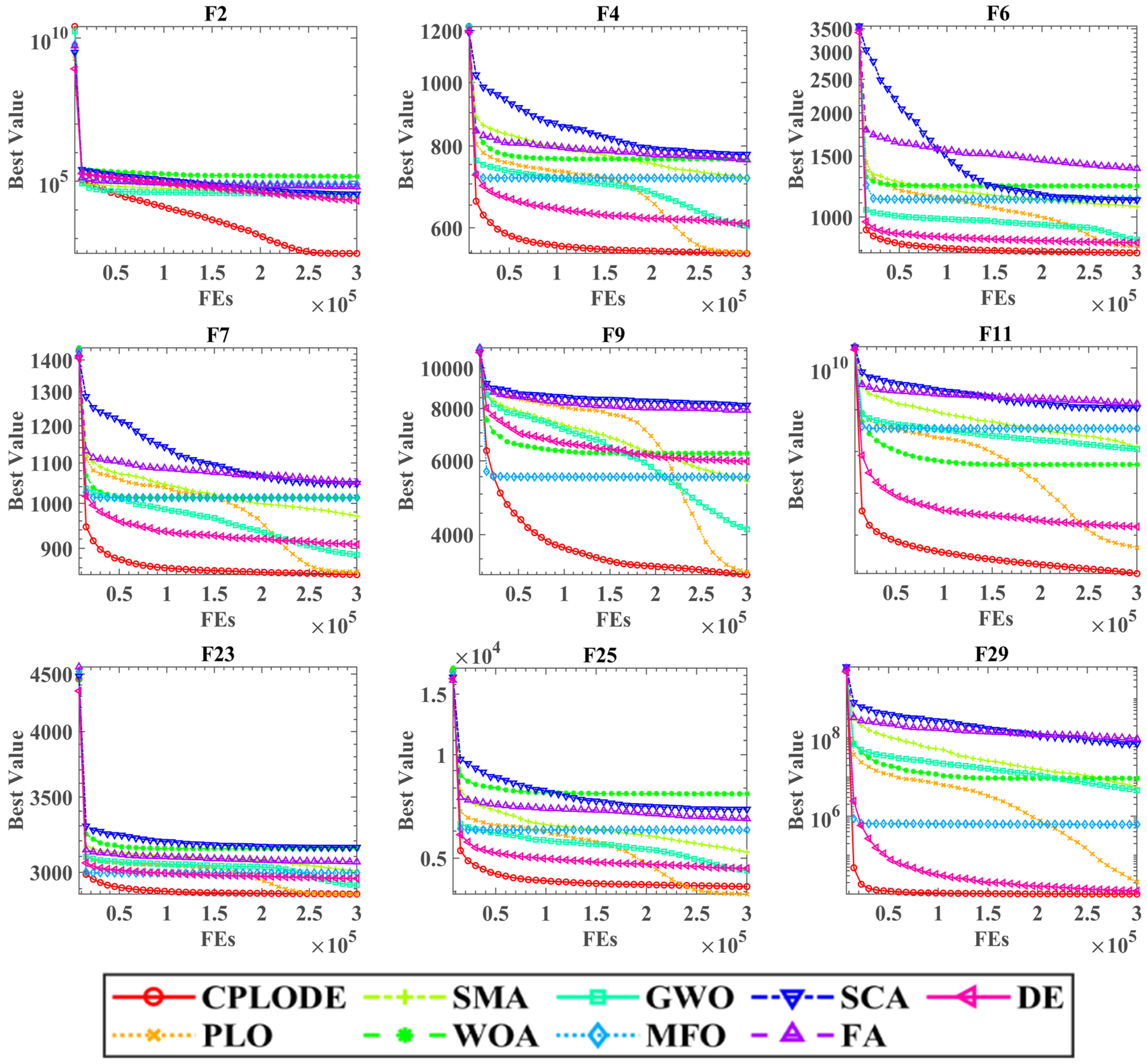
The performance of CPLODE was assessed on 29 benchmark functions from the CEC 2017 test suite, demonstrating superior performance across diverse fitness landscapes when compared to eight classical optimization algorithms. CPLODE achieved a higher average rank and statistically significant improvements, according to the Wilcoxon signed-rank test, particularly on more complex functions. Convergence curves further validated its enhanced convergence rate and optimal function values. These results emphasize the effectiveness of the integrated cryptobiosis mechanism and DE operators within CPLODE, confirming its improved search capability.
Moreover, CPLODE was applied to ten real-world datasets for feature selection, showcasing competitive performance by outperforming several well-known binary metaheuristic algorithms on most datasets and achieving a good balance between classification accuracy and feature reduction.
Future research will focus on further refining CPLODE with advanced adaptive mechanisms and exploring its application to a wider range of real-world optimization and feature selection tasks, including comparisons with traditional methods. We also intend to investigate the integration of machine learning and reinforcement learning techniques for more intelligent optimization strategies.