Gender, Smoking History, and Age Prediction from Laryngeal Images
Abstract
1. Introduction
- We performed the first study on predicting the gender, age, and smoking status of the patient purely based on laryngeal images from laryngoscopy.
- We created a dataset of 33,906 laryngeal image frames captured from 398 patients. The dataset is annotated with the clinical diagnosis, the pathologic diagnosis for the lesion, and the patient’s demographic information. This is the first large laryngoscopic image set for machine learning studies.
- We implemented and benchmarked the performance of eight classical deep learning models and achieved very promising results.
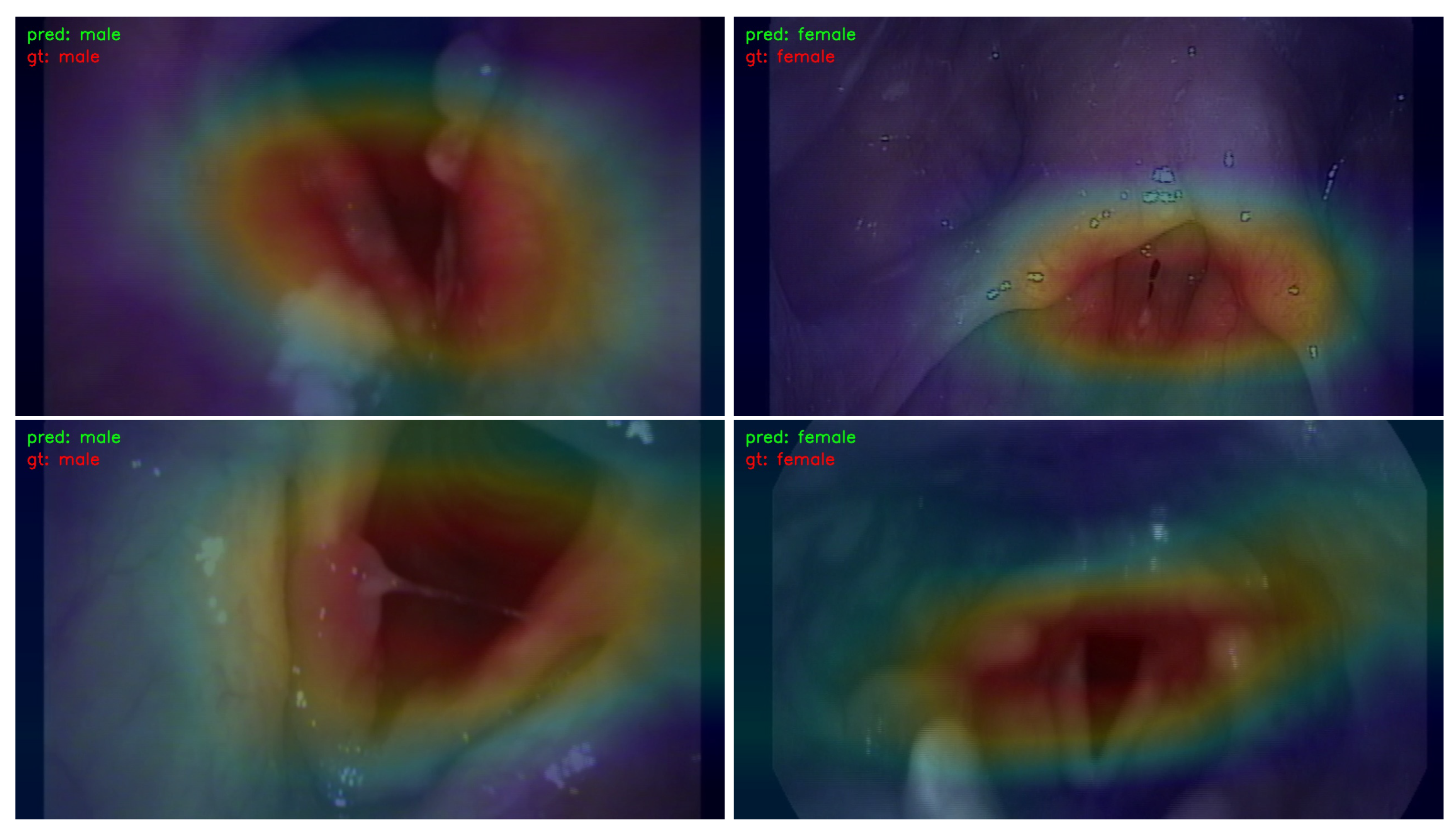
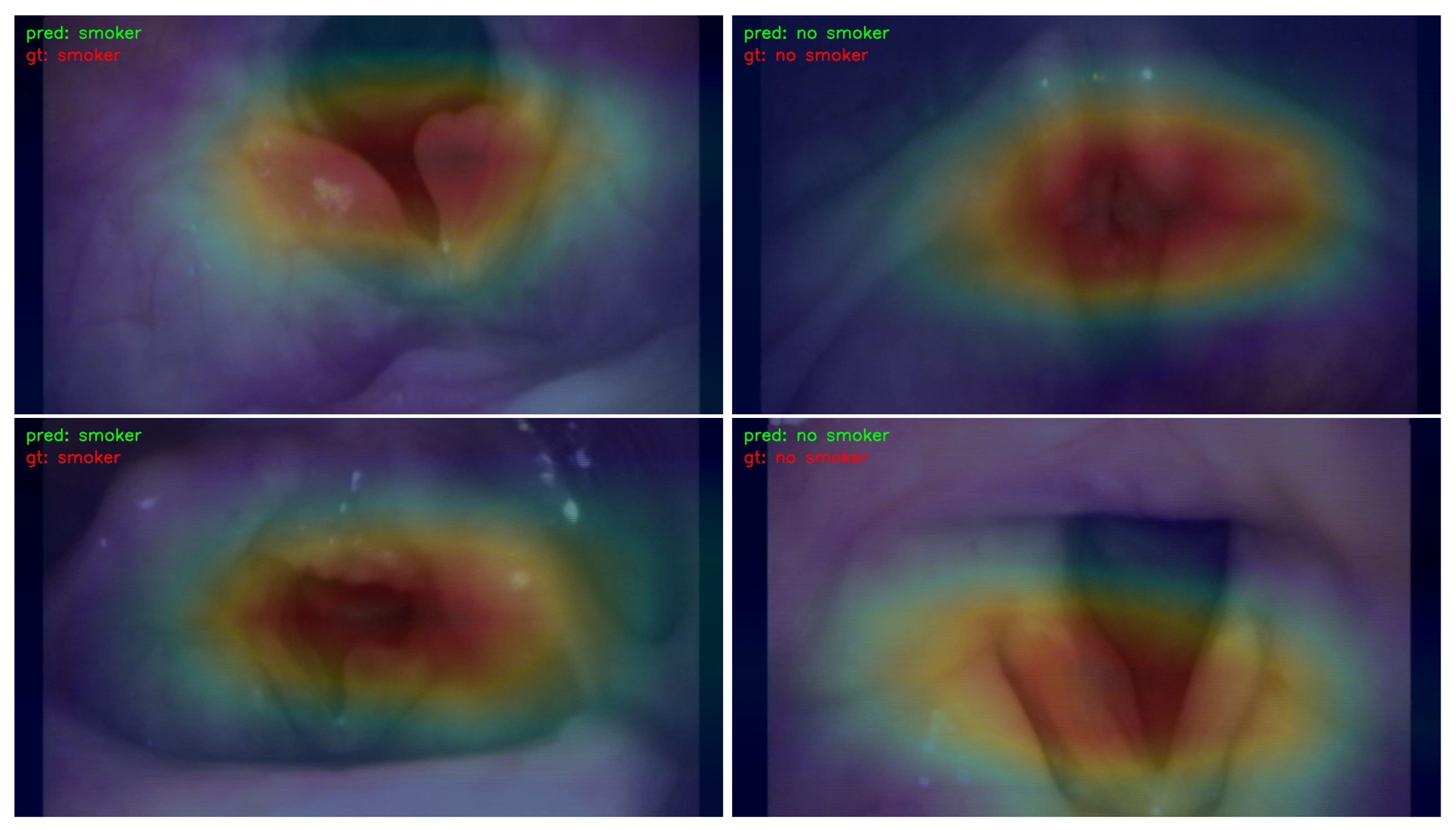
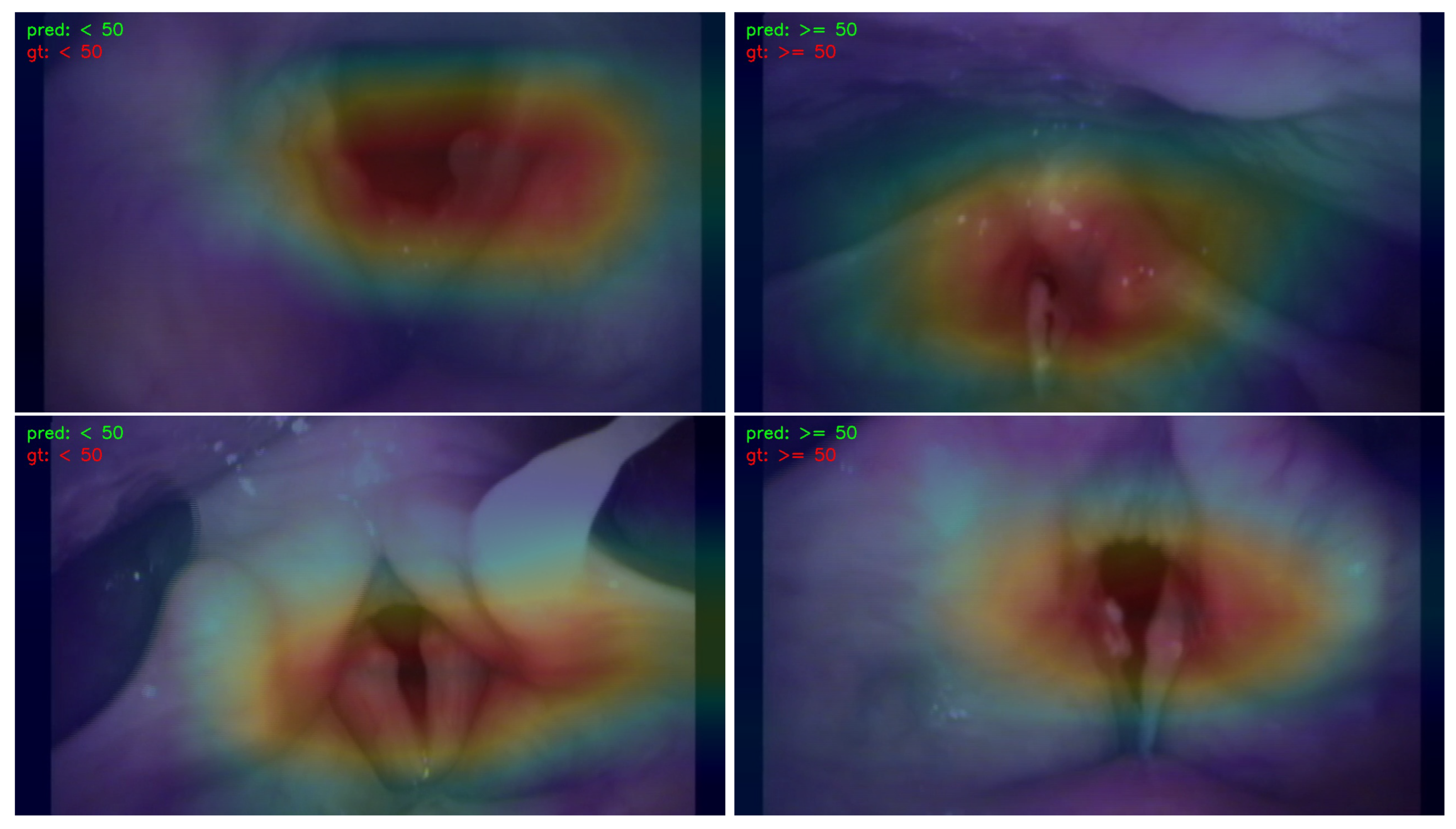
- We employed the Classification Activation Map (CAM) to visualize and analyze the regions of interest in the image. This approach contributes to the explainability of the learning models by providing insights into which specific areas of the image influenced the decision-making process.
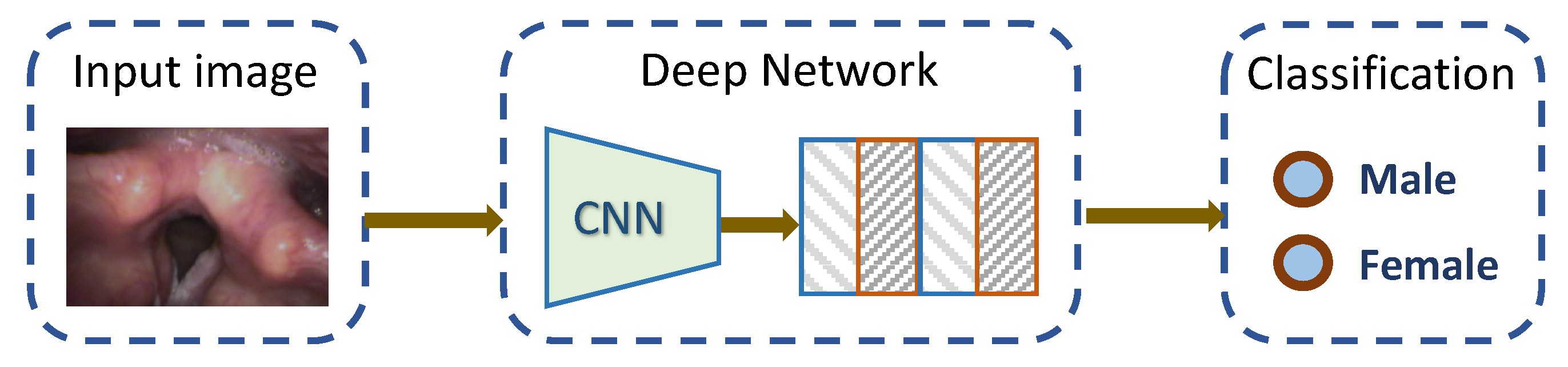
2. Materials and Methods
2.1. Dataset
2.2. Deep Learning Models
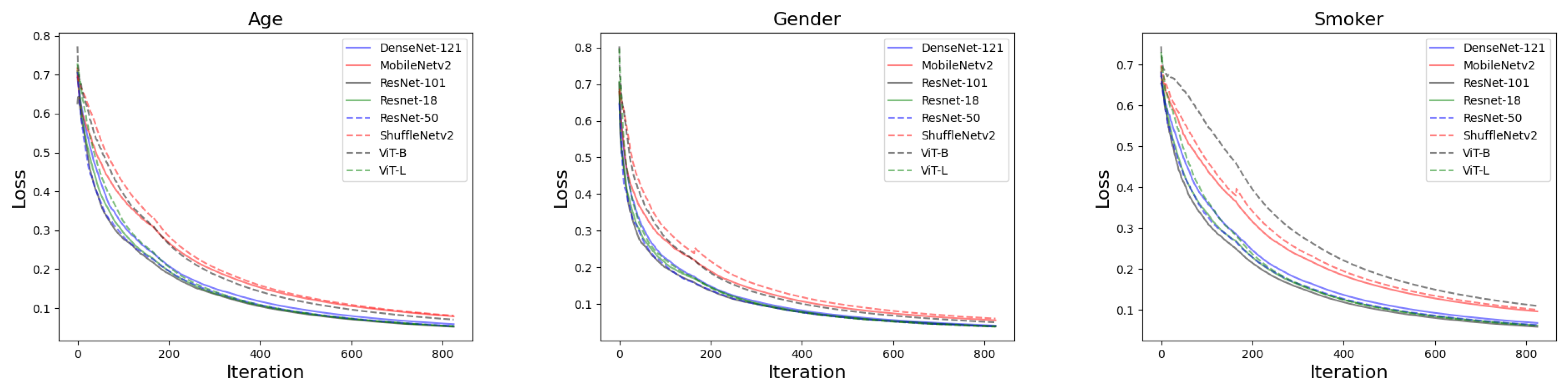
2.3. Training Settings
2.4. The Metrics for Evaluation
3. Results
3.1. The Performance of Deep Learning Models at Image Level
3.2. Overall Performance Based on Patients
3.3. Visualization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Leipzig, B.; Zellmer, J.E.; Klug, D. The Role of Endoscopy in Evaluating Patients With Head and Neck Cancer: A Multi-Institutional Prospective Study. Arch. Otolaryngol. Neck Surg. 1985, 111, 589–594. [Google Scholar] [CrossRef]
- Ebisumoto, K.; Sakai, A.; Maki, D.; Robinson, K.; Murakami, T.; Iijima, H.; Yamauchi, M.; Saito, K.; Watanabe, T.; Okami, K. Tumor detection with transoral use of flexible endoscopy for unknown primary head and neck cancer. Laryngoscope Investig. Otolaryngol. 2021, 6, 1037–1043. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Lin, P.; Yu, J.G.; Ye, J.; Xiao, L.; Tao, Y.; Jiang, Z.; Lin, W.; Liu, M.; Xu, J.; et al. Computer-aided diagnosis of laryngeal cancer via deep learning based on laryngoscopic images. EBioMedicine 2019, 48, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Halicek, M.; Lu, G.; Little, J.V.; Wang, X.; Patel, M.; Griffith, C.C.; El-Deiry, M.W.; Chen, A.Y.; Fei, B. Deep convolutional neural networks for classifying head and neck cancer using hyperspectral imaging. J. Biomed. Opt. 2017, 22, 060503. [Google Scholar] [CrossRef] [PubMed]
- Azam, M.A.; Sampieri, C.; Ioppi, A.; Africano, S.; Vallin, A.; Mocellin, D.; Fragale, M.; Guastini, L.; Moccia, S.; Piazza, C.; et al. Deep Learning Applied to White Light and Narrow Band Imaging Videolaryngoscopy: Toward Real-Time Laryngeal Cancer Detection. Laryngoscope 2022, 132, 1798–1806. [Google Scholar] [CrossRef] [PubMed]
- Takiyama, H.; Ozawa, T.; Ishihara, S.; Fujishiro, M.; Shichijo, S.; Nomura, S.; Miura, M.; Tada, T. Automatic anatomical classification of esophagogastroduodenoscopy images using deep convolutional neural networks. Sci. Rep. 2018, 8, 7497. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, Y.; Chen, S.; Zhong, Q.; Zhang, K. Hierarchical dynamic convolutional neural network for laryngeal disease classification. Sci. Rep. 2022, 12, 13914. [Google Scholar] [CrossRef]
- Ren, J.; Jing, X.; Wang, J.; Ren, X.; Xu, Y.; Yang, Q.; Ma, L.; Sun, Y.; Xu, W.; Yang, N.; et al. Automatic recognition of laryngoscopic images using a deep-learning technique. Laryngoscope 2020, 130, E686–E693. [Google Scholar] [CrossRef]
- Wilson, B.S.; Tucci, D.L.; Moses, D.A.; Chang, E.F.; Young, N.M.; Zeng, F.G.; Lesica, N.A.; Bur, A.M.; Kavookjian, H.; Mussatto, C.; et al. Harnessing the Power of Artificial Intelligence in Otolaryngology and the Communication Sciences. J. Assoc. Res. Otolaryngol. 2022, 23, 319–349. [Google Scholar] [CrossRef]
- Li, K.; Fathan, M.I.; Patel, K.; Zhang, T.; Zhong, C.; Bansal, A.; Rastogi, A.; Wang, J.S.; Wang, G. Colonoscopy polyp detection and classification: Dataset creation and comparative evaluations. PLoS ONE 2021, 16, e0255809. [Google Scholar] [CrossRef]
- Patel, K.B.; Li, F.; Wang, G. FuzzyNet: A Fuzzy Attention Module for Polyp Segmentation. In Proceedings of the NeurIPS’22 Workshop on All Things Attention: Bridging Different Perspectives on Attention, New Orleans, LO, USA, 2 December 2022. [Google Scholar]
- Patel, K.; Bur, A.M.; Wang, G. Enhanced u-net: A feature enhancement network for polyp segmentation. In Proceedings of the 2021 18th Conference on Robots and Vision (CRV), Burnaby, BC, Canada, 26–28 May 2021; pp. 181–188. [Google Scholar]
- Patel, K.; Li, K.; Tao, K.; Wang, Q.; Bansal, A.; Rastogi, A.; Wang, G. A comparative study on polyp classification using convolutional neural networks. PLoS ONE 2020, 15, e0236452. [Google Scholar] [CrossRef]
- Militello, C.; Prinzi, F.; Sollami, G.; Rundo, L.; La Grutta, L.; Vitabile, S. CT radiomic features and clinical biomarkers for predicting coronary artery disease. Cogn. Comput. 2023, 15, 238–253. [Google Scholar] [CrossRef]
- Gu, R.; Wang, G.; Song, T.; Huang, R.; Aertsen, M.; Deprest, J.; Ourselin, S.; Vercauteren, T.; Zhang, S. CA-Net: Comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans. Med. Imaging 2020, 40, 699–711. [Google Scholar] [CrossRef]
- Van der Velden, B.H.; Kuijf, H.J.; Gilhuijs, K.G.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Med. Image Anal. 2022, 79, 102470. [Google Scholar] [CrossRef] [PubMed]
- Prinzi, F.; Orlando, A.; Gaglio, S.; Midiri, M.; Vitabile, S. ML-Based Radiomics Analysis for Breast Cancer Classification in DCE-MRI. In Proceedings of the Applied Intelligence and Informatics: Second International Conference, AII 2022, Reggio Calabria, Italy, 1–3 September 2022; Springer: Berlin/Heidelberg, Germany, 2023; pp. 144–158. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Ma, N.; Zhang, X.; Zheng, H.T.; Sun, J. Shufflenet v2: Practical guidelines for efficient cnn architecture design. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 116–131. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 6848–6856. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. arXiv 2017, arXiv:1706.03762. [Google Scholar]
- Chen, X.; Hu, Q.; Li, K.; Zhong, C.; Wang, G. Accumulated Trivial Attention Matters in Vision Transformers on Small Datasets. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 2–7 January 2023; pp. 3984–3992. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-end object detection with transformers. In Proceedings of the European Conference on Computer Vision, Glasgow, UK, 23–28 August 2020; pp. 213–229. [Google Scholar]
- Ma, W.; Zhang, T.; Wang, G. Miti-detr: Object detection based on transformers with mitigatory self-attention convergence. arXiv 2022, arXiv:2112.13310. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. Pytorch: An imperative style, high-performance deep learning library. arXiv 2019, arXiv:1912.01703. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Conti, V.; Militello, C.; Rundo, L.; Vitabile, S. A novel bio-inspired approach for high-performance management in service-oriented networks. IEEE Trans. Emerg. Top. Comput. 2020, 9, 1709–1722. [Google Scholar] [CrossRef]
| DL Models | Gender | Smoking History | Age | |||
|---|---|---|---|---|---|---|
| Male | Female | Smoker | Non-Smoker | 50 | 50 | |
| ResNet-18 | 93.9 | 73.4 | 65.3 | 63.3 | 41.6 | 88.6 |
| ResNet-50 | 94.7 | 70.9 | 64.2 | 62.8 | 44.5 | 87.8 |
| ResNet-101 | 92.4 | 70.9 | 66.7 | 64.0 | 45.7 | 89.7 |
| DenseNet-121 | 94.6 | 72.3 | 62.0 | 60.0 | 44.3 | 89.5 |
| MobileNetv2 | 93.6 | 66.1 | 64.1 | 61.5 | 43.3 | 89.5 |
| ShuffleNetv2 | 92.0 | 66.8 | 64.6 | 63.1 | 38.5 | 88.0 |
| ViT-L | 93.5 | 66.4 | 66.6 | 63.1 | 47.7 | 89.1 |
| ViT-B | 94.3 | 69.4 | 64.7 | 63.6 | 46.6 | 88.4 |
| Mean | 93.6 | 69.5 | 64.8 | 62.7 | 44.0 | 88.8 |
| Std | 0.99 | 2.81 | 1.50 | 1.30 | 2.93 | 0.73 |
| DL Models | Gender | Smoking History | Age | |||
|---|---|---|---|---|---|---|
| Male | Female | Smoker | Non-Smoker | 50 | 50 | |
| ResNet-18 | 83.5 | 89.3 | 59.6 | 68.7 | 68.7 | 71.7 |
| ResNet-50 | 81.1 | 91.0 | 59.8 | 67.1 | 63.6 | 76.7 |
| ResNet-101 | 81.9 | 86.8 | 59.8 | 70.6 | 70.6 | 75.4 |
| DenseNet-121 | 82.3 | 90.8 | 54.8 | 66.8 | 70.5 | 74.0 |
| MobileNetv2 | 76.7 | 89.6 | 56.3 | 68.9 | 71.1 | 72.7 |
| ShuffleNetv2 | 78.1 | 86.6 | 59.9 | 67.6 | 68.4 | 67.9 |
| ViT-L | 76.9 | 89.5 | 57.7 | 71.4 | 67.3 | 78.4 |
| ViT-B | 79.8 | 90.5 | 61.2 | 67.0 | 65.0 | 78.2 |
| Mean | 80.0 | 89.3 | 58.6 | 68.5 | 68.2 | 74.4 |
| Std | 2.58 | 1.70 | 2.17 | 1.73 | 2.72 | 3.57 |
| DL Models | Gender | Smoking History | Age | |||
|---|---|---|---|---|---|---|
| Male | Female | Smoker | Non-Smoker | 50 | 50 | |
| ResNet-18 | 88.4 | 80.6 | 62.3 | 65.9 | 51.8 | 79.3 |
| ResNet-50 | 87.3 | 79.7 | 61.9 | 64.9 | 52.4 | 81.9 |
| ResNet-101 | 86.9 | 78.1 | 63.1 | 67.1 | 55.5 | 82.0 |
| DenseNet-121 | 88.0 | 80.5 | 58.2 | 63.2 | 54.4 | 81.0 |
| MobileNetv2 | 84.3 | 76.1 | 60.0 | 65.0 | 53.8 | 80.2 |
| ShuffleNetv2 | 84.5 | 75.4 | 62.2 | 65.2 | 49.3 | 76.7 |
| ViT-L | 84.4 | 76.2 | 61.8 | 67.0 | 55.9 | 83.4 |
| ViT-B | 86.4 | 78.6 | 62.9 | 65.3 | 54.3 | 83.0 |
| Mean | 86.3 | 78.2 | 61.6 | 65.5 | 53.4 | 80.9 |
| Std | 1.67 | 2.06 | 1.65 | 1.25 | 2.17 | 2.19 |
| DL Models | Gender | Smoking History | Age |
|---|---|---|---|
| ResNet-18 | 85.5 | 64.2 | 71.0 |
| ResNet-50 | 84.4 | 63.5 | 73.7 |
| ResNet-101 | 83.6 | 65.2 | 74.3 |
| DenseNet-121 | 85.2 | 60.9 | 73.2 |
| MobileNetv2 | 81.0 | 62.6 | 72.3 |
| ShuffleNetv2 | 81.0 | 63.8 | 68.0 |
| ViT-L | 81.2 | 64.6 | 75.9 |
| ViT-B | 83.4 | 64.1 | 75.2 |
| Mean | 83.2 | 63.6 | 73.0 |
| Std | 1.87 | 1.34 | 2.53 |
| DL Models | Gender | Smoking History | Age | |||
|---|---|---|---|---|---|---|
| Majority | Prob | Majority | Prob | Majority | Prob | |
| ResNet-18 | 90.7 | 88.9 | 66.9 | 62.5 | 77.5 | 73.1 |
| ResNet-50 | 88.9 | 86.5 | 62.1 | 59.1 | 81.0 | 78.2 |
| ResNet-101 | 84.5 | 84.7 | 64.8 | 62.3 | 77.8 | 74.4 |
| DenseNet-121 | 88.0 | 87.0 | 61.7 | 58.3 | 83.6 | 79.0 |
| MobileNetv2 | 84.5 | 81.2 | 63.2 | 59.7 | 77.5 | 73.5 |
| ShuffleNetv2 | 83.7 | 79.8 | 67.5 | 63.0 | 73.5 | 70.4 |
| ViT-L | 84.3 | 82.2 | 69.1 | 66.6 | 83.5 | 80.6 |
| ViT-B | 91.1 | 89.1 | 68.3 | 66.8 | 85.7 | 82.3 |
| Mean | 87.0 | 84.9 | 65.4 | 62.3 | 80.0 | 76.5 |
| Std | 2.87 | 3.31 | 2.71 | 3.00 | 3.85 | 3.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Bur, A.M.; Kraft, S.; Kavookjian, H.; Renslo, B.; Chen, X.; Luo, B.; Wang, G. Gender, Smoking History, and Age Prediction from Laryngeal Images. J. Imaging 2023, 9, 109. https://doi.org/10.3390/jimaging9060109
Zhang T, Bur AM, Kraft S, Kavookjian H, Renslo B, Chen X, Luo B, Wang G. Gender, Smoking History, and Age Prediction from Laryngeal Images. Journal of Imaging. 2023; 9(6):109. https://doi.org/10.3390/jimaging9060109
Chicago/Turabian StyleZhang, Tianxiao, Andrés M. Bur, Shannon Kraft, Hannah Kavookjian, Bryan Renslo, Xiangyu Chen, Bo Luo, and Guanghui Wang. 2023. "Gender, Smoking History, and Age Prediction from Laryngeal Images" Journal of Imaging 9, no. 6: 109. https://doi.org/10.3390/jimaging9060109
APA StyleZhang, T., Bur, A. M., Kraft, S., Kavookjian, H., Renslo, B., Chen, X., Luo, B., & Wang, G. (2023). Gender, Smoking History, and Age Prediction from Laryngeal Images. Journal of Imaging, 9(6), 109. https://doi.org/10.3390/jimaging9060109







