Time Synchronization of Multimodal Physiological Signals through Alignment of Common Signal Types and Its Technical Considerations in Digital Health
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Data
2.2. Sample Selection
2.3. Time Alignment Algorithm
| Algorithm 1. Algorithm for synchronization delay estimation | ||
| input: Lead I signal , Lead V2 signal | ||
| output: synchronization delay | ||
| 1 | for every time lag that shifts to either positive or negative direction do | |
| 2 | compute correlation coefficient between and shifted signal | |
| 3 | end | |
| 4 | compute maximal and find corresponding time lag | |
| 5 | return | |
2.4. Experiment Setup
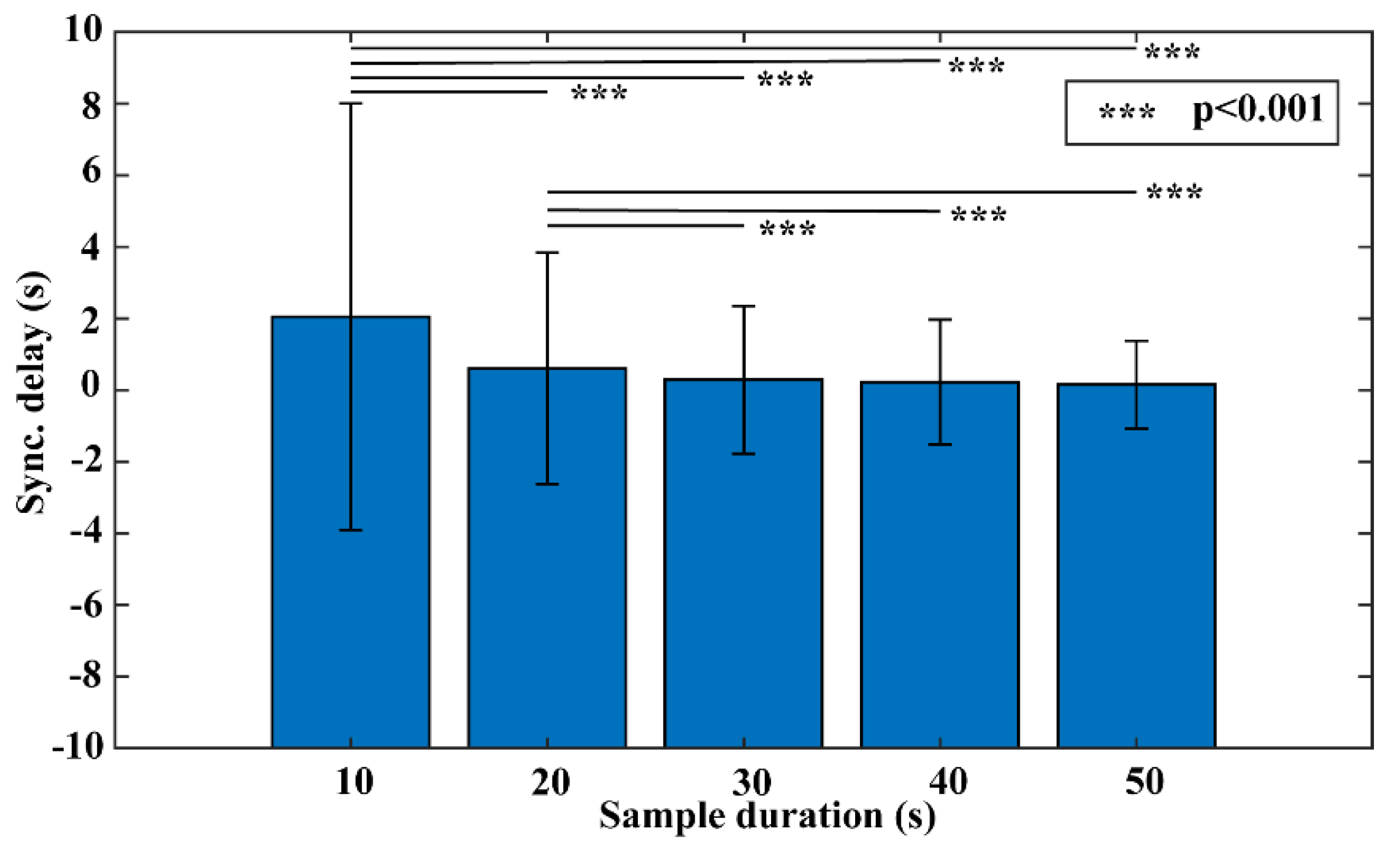
2.4.1. Experiment #1 Sample Duration Test
2.4.2. Experiment #2 Integration of SQI
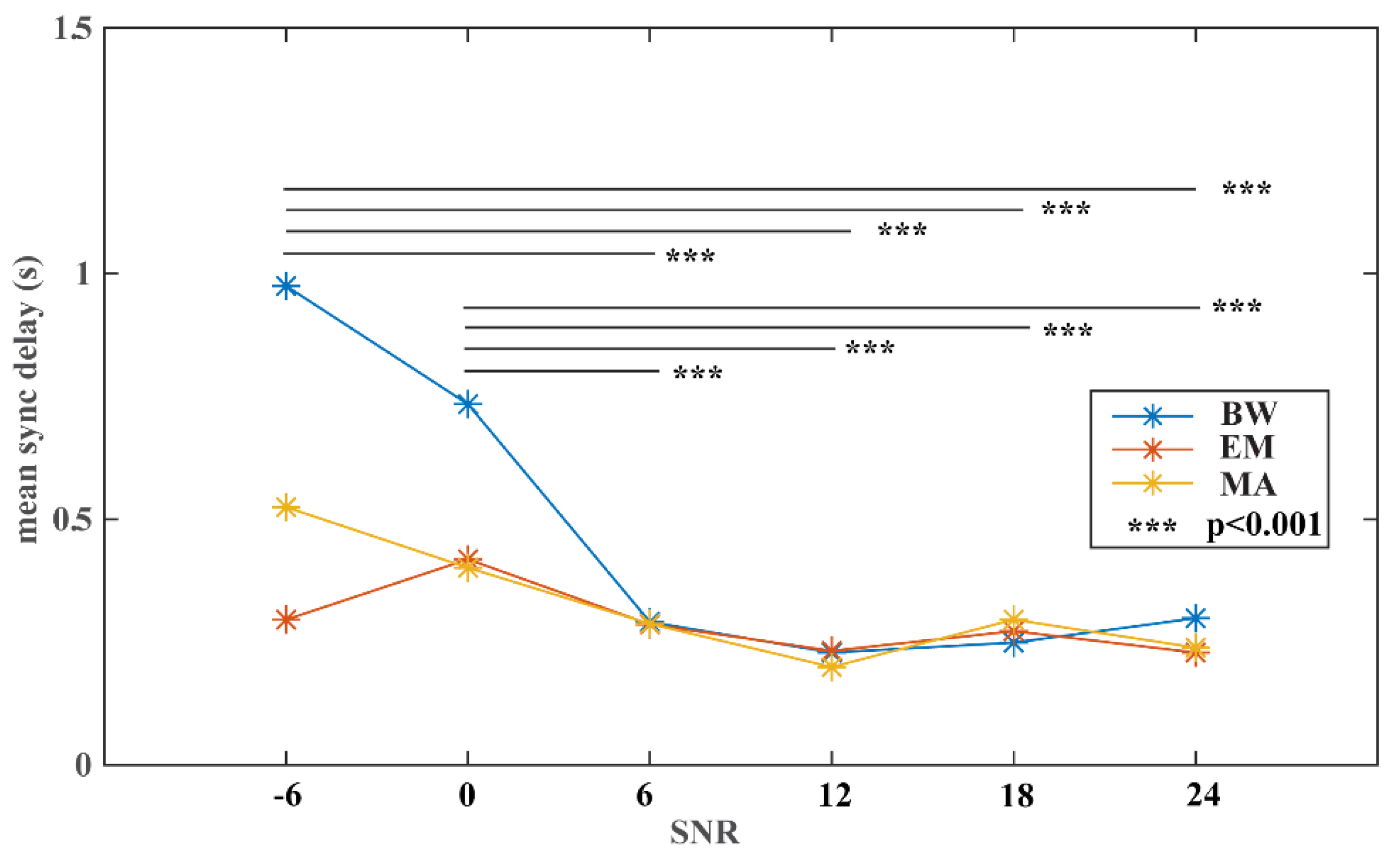
2.4.3. Experiment #3 Noise Stress Test
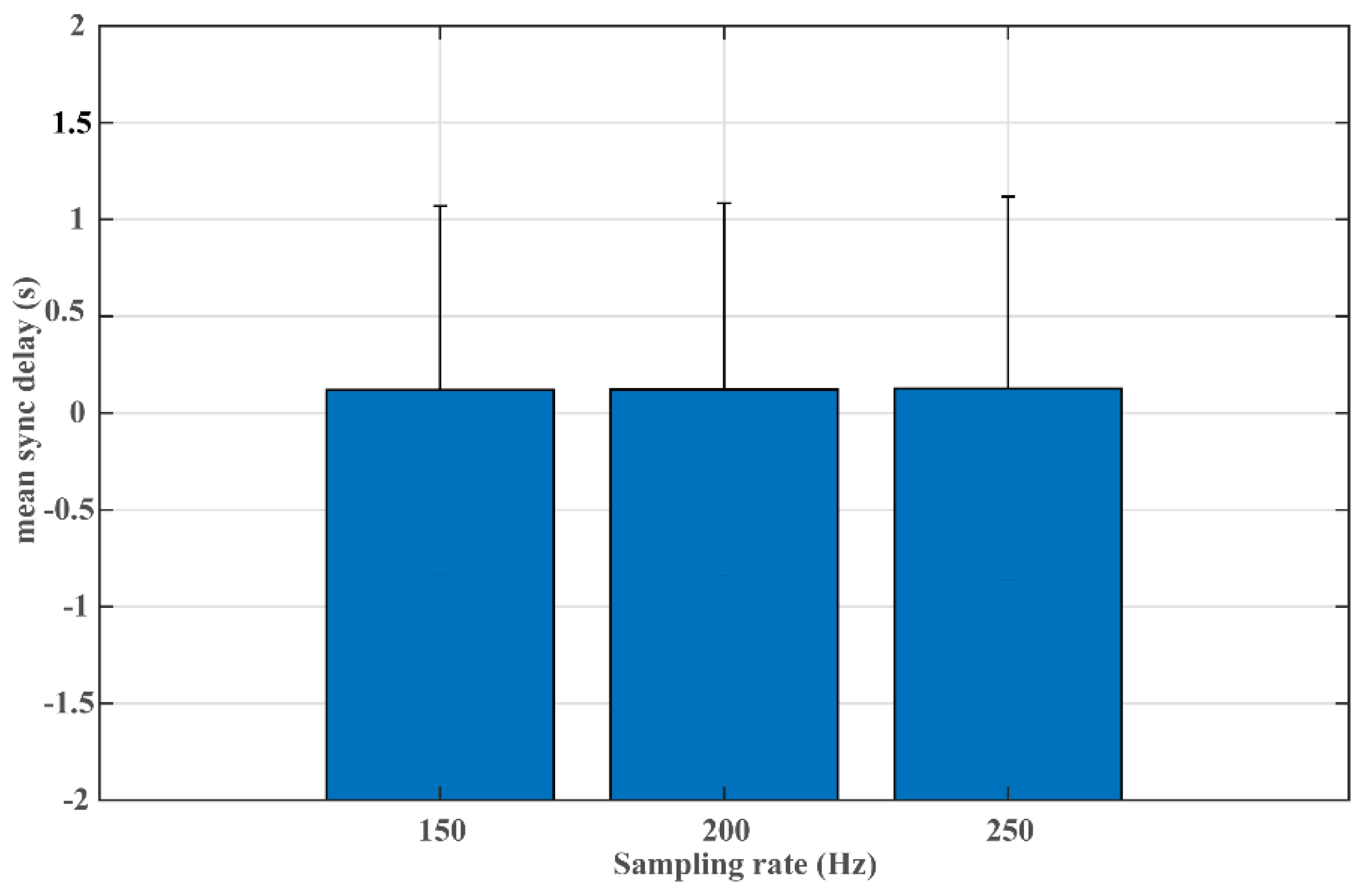
2.4.4. Experiment #4 Sampling Rate Test
2.4.5. Experiment #5 Comparison to DTW-Based Approach
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pu, B.; Li, K.; Li, S.; Zhu, N. Automatic fetal ultrasound standard plane recognition based on deep learning and IIoT. IEEE Trans. Ind. Inform. 2021, 17, 7771–7780. [Google Scholar] [CrossRef]
- Kamruzzaman, M.M.; Alrashdi, I.; Alqazzaz, A. New opportunities, challenges, and applications of edge-AI for connected healthcare in internet of medical things for smart cities. J. Healthc. Eng. 2022, 2022, 2950699. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, R.; Mehrotra, D.; Chandra, S. Potential of Internet of Medical Things (IoMT) applications in building a smart healthcare system: A systematic review. J. Oral Biol. Craniofac. Res. 2022, 12, 302–318. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Torrente-Rodríguez, R.M.; Wang, M.; Gao, W. The era of digital health: A review of portable and wearable affinity biosensors. Adv. Funct. Mater. 2020, 30, 1906713. [Google Scholar] [CrossRef]
- Triantafyllidis, A.K.; Tsanas, A. Applications of machine learning in real-life digital health interventions: Review of the literature. J. Med. Internet Res. 2019, 21, e12286. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yang, Y.; Wang, T.; Sherratt, R.S.; Zhang, J. Big data service architecture: A survey. J. Internet Technol. 2020, 21, 393–405. [Google Scholar]
- Pal, K.; Dack, C.; Ross, J.; Michie, S.; May, C.; Stevenson, F.; Farmer, A.; Yardley, L.; Barnard, M.; Murray, E. Digital health interventions for adults with type 2 diabetes: Qualitative study of patient perspectives on diabetes self-management education and support. J. Med. Internet Res. 2018, 20, e40. [Google Scholar] [CrossRef]
- Shan, R.; Sarkar, S.; Martin, S.S. Digital health technology and mobile devices for the management of diabetes mellitus: State of the art. Diabetologia 2019, 62, 877–887. [Google Scholar] [CrossRef] [Green Version]
- Alwashmi, M.F. The use of digital health in the detection and management of COVID-19. Int. J. Environ. Res. Public Health 2020, 17, 2906. [Google Scholar] [CrossRef]
- Kouroubali, A.; Kondylakis, H.; Katehakis, D.G. Integrated care in the era of COVID-19: Turning vision into reality with digital health. Front. Digit. Health 2021, 3, 647938. [Google Scholar] [CrossRef]
- Hannun, A.Y.; Rajpurkar, P.; Haghpanahi, M.; Tison, G.H.; Bourn, C.; Turakhia, M.P.; Ng, A.Y. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat. Med. 2019, 25, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.V.; Mahaffey, K.W.; Hedlin, H.; Rumsfeld, J.S.; Garcia, A.; Ferris, T.; Balasubramanian, V.; Russo, A.M.; Rajmane, A.; Cheung, L.; et al. Large-scale assessment of a smartwatch to identify atrial fibrillation. N. Engl. J. Med. 2019, 381, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, C.J.; Romano, P.A.; Greisler, L.A.; Brindle, R.A.; Ford, K.R.; Kuennen, M.R. Accuracy and reliability of commercial wrist-worn pulse oximeter during normobaric hypoxia exposure under resting conditions. Res. Q. Exerc. Sport 2021, 92, 549–558. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, C.; Pradhan, N.; Redpath, C.; Adler, A. Validation of the Empatica E4 Wristband. In Proceedings of the 2016 IEEE EMBS International Student Conference (ISC), Ottawa, ON, Canada, 29–31 May 2016; pp. 1–4. [Google Scholar]
- Nelson, B.W.; Allen, N.B. Accuracy of consumer wearable heart rate measurement during an ecologically valid 24-hour period: Intraindividual validation study. JMIR Mhealth Uhealth 2019, 7, e10828. [Google Scholar] [CrossRef]
- Shcherbina, A.; Mattsson, C.M.; Waggott, D.; Salisbury, H.; Christle, J.W.; Hastie, T.; Wheeler, M.T.; Ashley, E.A. Accuracy in wrist-worn, sensor-based measurements of heart rate and energy expenditure in a diverse cohort. J. Pers. Med. 2017, 7, 3. [Google Scholar] [CrossRef]
- Stahl, S.E.; An, H.S.; Dinkel, D.M.; Noble, J.M.; Lee, J.M. How accurate are the wrist-based heart rate monitors during walking and running activities? Are they accurate enough? BMJ Open Sport Exerc. Med. 2016, 2, e000106. [Google Scholar] [CrossRef] [Green Version]
- Bobade, P.; Vani, M. Stress Detection with Machine Learning and Deep Learning using Multimodal Physiological Data. In Proceedings of the 2020 Second International Conference on Inventive Research in Computing Applications (ICIRCA), Coimbatore, India, 15–17 July 2020; pp. 51–57. [Google Scholar]
- Vanneste, P.; Raes, A.; Morton, J.; Bombeke, K.; Van Acker, B.B.; Larmuseau, C.; Depaepe, F.; Van den Noortgate, W. Towards measuring cognitive load through multimodal physiological data. Cogn. Technol. Work 2021, 23, 567–585. [Google Scholar] [CrossRef]
- Yang, C.J.; Fahier, N.; He, C.Y.; Li, W.C.; Fang, W.C. An AI-Edge Platform with Multimodal Wearable Physiological Signals Monitoring Sensors for Affective Computing Applications. In Proceedings of the 2020 IEEE International Symposium on Circuits and Systems (ISCAS), Seville, Spain, 12–14 October 2020; pp. 1–5. [Google Scholar]
- Yin, Z.; Zhao, M.; Wang, Y.; Yang, J.; Zhang, J. Recognition of emotions using multimodal physiological signals and an ensemble deep learning model. Comput. Methods Programs Biomed. 2017, 140, 93–110. [Google Scholar] [CrossRef]
- Liu, Y.; Yao, Y.; Wang, Z.; Plested, J.; Gedeon, T. Generalized Alignment for Multimodal Physiological Signal Learning. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–10. [Google Scholar]
- Jiang, Y.; Qi, Y.; Wang, W.K.; Bent, B.; Avram, R.; Olgin, J.; Dunn, J. EventDTW: An improved dynamic time warping algorithm for aligning biomedical signals of nonuniform sampling frequencies. Sensors 2020, 20, 2700. [Google Scholar] [CrossRef]
- Hong, J.Y.; Park, S.H.; Baek, J. Solving the Singularity Problem of Semiconductor Process Signal Using Improved Dynamic Time Warping. In Proceedings of the 2017 IEEE 11th International Conference on Semantic Computing (ICSC), San Diego, CA, USA, 30 January–1 February 2017; pp. 266–267. [Google Scholar]
- Keogh, E.J.; Pazzani, M.J. Derivative Dynamic Time Warping. In Proceedings of the 2001 SIAM International Conference on Data Mining (SDM), Chicago, IL, USA, 5–7 April 2001; Society for Industrial and Applied Mathematics: Philadelphia, PA, USA, 2001; pp. 1–11. [Google Scholar]
- Zhao, J.; Itti, L. shapeDTW: Shape dynamic time warping. arXiv 2016, arXiv:1606.01601. [Google Scholar] [CrossRef] [Green Version]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, physiotoolkit, and physionet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, E215–E220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gustafsson, F. Determining the initial states in forward-backward filtering. IEEE Trans. Signal. Process. 1996, 44, 988–992. [Google Scholar] [CrossRef] [Green Version]
- Yaghmaie, N.; Maddah-Ali, M.A.; Jelinek, H.F.; Mazrbanrad, F. Dynamic signal quality index for electrocardiograms. Physiol. Meas. 2018, 39, 105008. [Google Scholar] [CrossRef]
- Sun, K.; Jin, T.; Yang, D. An improved time-frequency analysis method in interference detection for GNSS receivers. Sensors 2015, 15, 9404–9426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moody, G.B.; Muldrow, W.E.; Mark, R.G. A noise stress test for arrhythmia detectors. Comput. Cardiol. 1984, 11, 381–384. [Google Scholar]
- Ding, X.; Clifton, D.; Ji, N.; Lovell, N.H.; Bonato, P.; Chen, W.; Yu, X.; Xue, Z.; Xiang, T.; Long, X.; et al. Wearable sensing and telehealth technology with potential applications in the coronavirus pandemic. IEEE Rev. Biomed. Eng. 2021, 14, 48–70. [Google Scholar] [CrossRef]
- Jim, H.S.L.; Hoogland, A.I.; Brownstein, N.C.; Barata, A.; Dicker, A.P.; Knoop, H.; Gonzalez, B.D.; Perkins, R.; Rollison, D.; Gilbert, S.M.; et al. Innovations in research and clinical care using patient-generated health data. CA Cancer J. Clin. 2020, 70, 182–199. [Google Scholar] [CrossRef]
- Wood, W.A.; Bennett, A.V.; Basch, E. Emerging uses of patient generated health data in clinical research. Mol. Oncol. 2015, 9, 1018–1024. [Google Scholar] [CrossRef]
- Jeong, I.C.; Bychkov, D.; Searson, P.C. Wearable devices for precision medicine and health state monitoring. IEEE Trans. Biomed. Eng. 2019, 66, 1242–1258. [Google Scholar] [CrossRef]
- Allen, J. Photoplethysmography and its application in clinical physiological measurement. Physiol. Meas. 2007, 28, R1–R39. [Google Scholar] [CrossRef] [Green Version]
- Pradhan, N.; Rajan, S.; Adler, A. Evaluation of the signal quality of wrist-based photoplethysmography. Physiol. Meas. 2019, 40, 065008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Heart rate variability. Standards of measurement, physiological interpretation, and clinical use. Eur. Heart J. 1996, 17, 354–381. [Google Scholar] [CrossRef] [Green Version]
- Ellis, R.J.; Zhu, B.; Koenig, J.; Thayer, J.F.; Wang, Y. A careful look at ECG sampling frequency and R-peak interpolation on short-term measures of heart rate variability. Physiol. Meas. 2015, 36, 1827–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziemssen, T.; Gasch, J.; Ruediger, H. Influence of ECG sampling frequency on spectral analysis of RR intervals and baroreflex sensitivity using the EUROBAVAR data set. J. Clin. Monit. Comput. 2008, 22, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.; Jeong, J.; Kim, H.B.; Kwon, I.H.; Park, S.Y.; Kim, J.E.; Choi, Y. Electrocardiogram sampling frequency range acceptable for heart rate variability analysis. Healthc. Inform. Res. 2018, 24, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Jager, F.; Taddei, A.; Moody, G.B.; Emdin, M.; Antolic, G.; Dorn, R.; Smrdel, A.; Marchesi, C.; Mark, R.G. Long-term ST database: A reference for the development and evaluation of automated ischaemia detectors and for the study of the dynamics of myocardial ischaemia. Med. Biol. Eng. Comput. 2003, 41, 172–182. [Google Scholar] [CrossRef]
- Sundararaman, B.; Buy, U.; Kshemkalyani, A.D. Clock synchronization for wireless sensor networks: A survey. Ad Hoc Netw. 2005, 3, 281–323. [Google Scholar] [CrossRef]
- Xie, K.; Cai, Q.; Fu, M. A fast clock synchronization algorithm for wireless sensor networks. Automatica 2018, 92, 133–142. [Google Scholar] [CrossRef]
- Hu, B.; Xiang, F.; Wu, F.; Liu, J.; Sun, Z.; Sun, Z. Research on time synchronization method under arbitrary network delay in wireless sensor networks. Comput. Mater. Contin. 2019, 61, 1323–1344. [Google Scholar] [CrossRef]
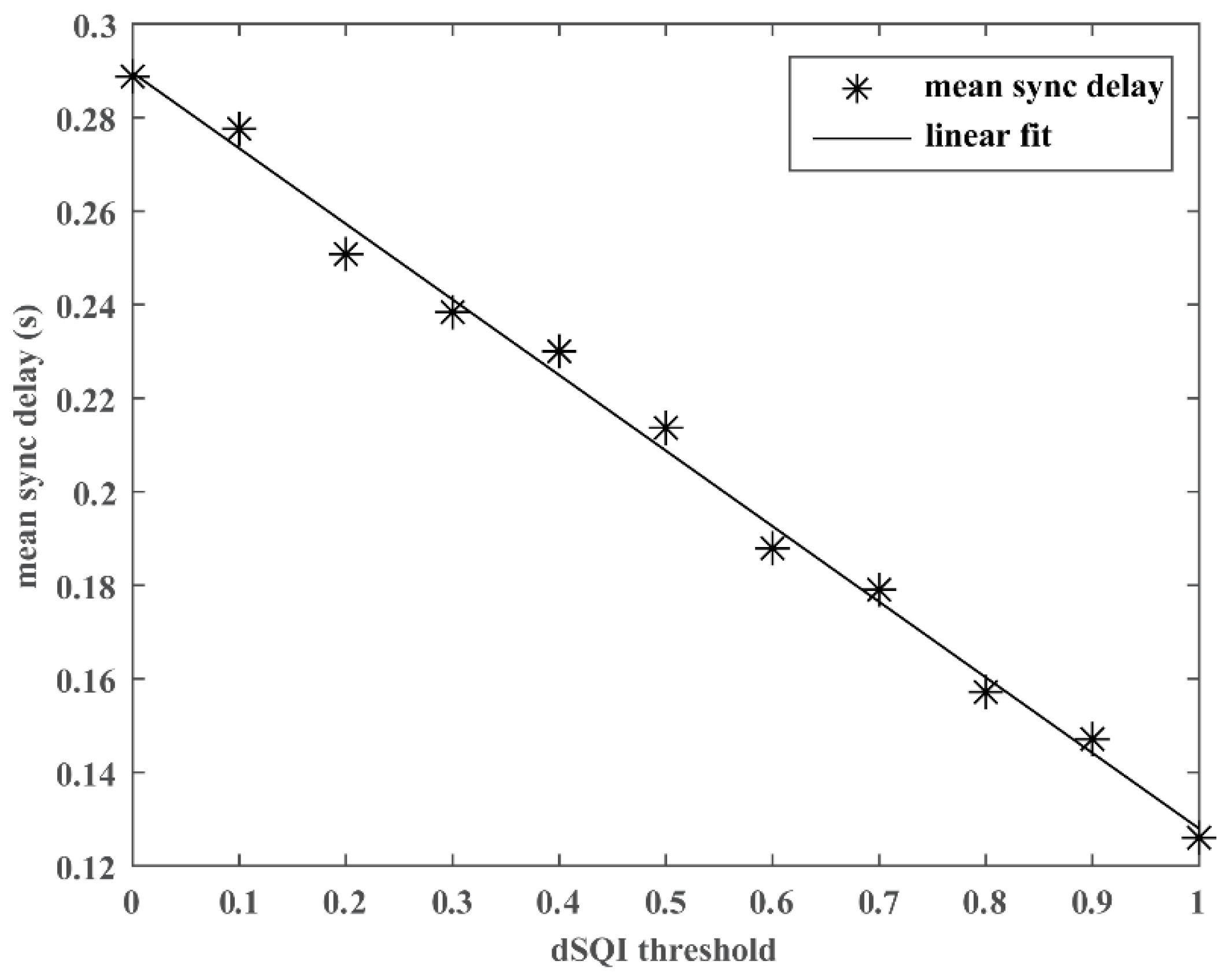
| dSQI Threshold | Sync Delay (Mean/SD, Seconds) | Sample Retaining Ratio (%) |
|---|---|---|
| 0.00 | 0.29/2.06 | 1.00 |
| 0.10 | 0.28/2.00 | 0.94 |
| 0.20 | 0.25/1.82 | 0.92 |
| 0.30 | 0.24/1.75 | 0.89 |
| 0.40 | 0.23/1.70 | 0.86 |
| 0.50 | 0.21/1.61 | 0.82 |
| 0.60 | 0.19/1.40 | 0.78 |
| 0.70 | 0.18/1.34 | 0.73 |
| 0.80 | 0.16/1.13 | 0.68 |
| 0.90 | 0.15/1.08 | 0.61 |
| 1.00 | 0.13/0.99 | 0.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, R.; Ding, C.; Hu, X. Time Synchronization of Multimodal Physiological Signals through Alignment of Common Signal Types and Its Technical Considerations in Digital Health. J. Imaging 2022, 8, 120. https://doi.org/10.3390/jimaging8050120
Xiao R, Ding C, Hu X. Time Synchronization of Multimodal Physiological Signals through Alignment of Common Signal Types and Its Technical Considerations in Digital Health. Journal of Imaging. 2022; 8(5):120. https://doi.org/10.3390/jimaging8050120
Chicago/Turabian StyleXiao, Ran, Cheng Ding, and Xiao Hu. 2022. "Time Synchronization of Multimodal Physiological Signals through Alignment of Common Signal Types and Its Technical Considerations in Digital Health" Journal of Imaging 8, no. 5: 120. https://doi.org/10.3390/jimaging8050120
APA StyleXiao, R., Ding, C., & Hu, X. (2022). Time Synchronization of Multimodal Physiological Signals through Alignment of Common Signal Types and Its Technical Considerations in Digital Health. Journal of Imaging, 8(5), 120. https://doi.org/10.3390/jimaging8050120







