Fully Automated Segmentation Models of Supratentorial Meningiomas Assisted by Inclusion of Normal Brain Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Approval
2.2. Patients
2.3. Pre-Processing of MRI
2.4. Three-Dimensional Neural Network (3D U-Net)
2.5. Loss Function
2.6. Model Training and Selection
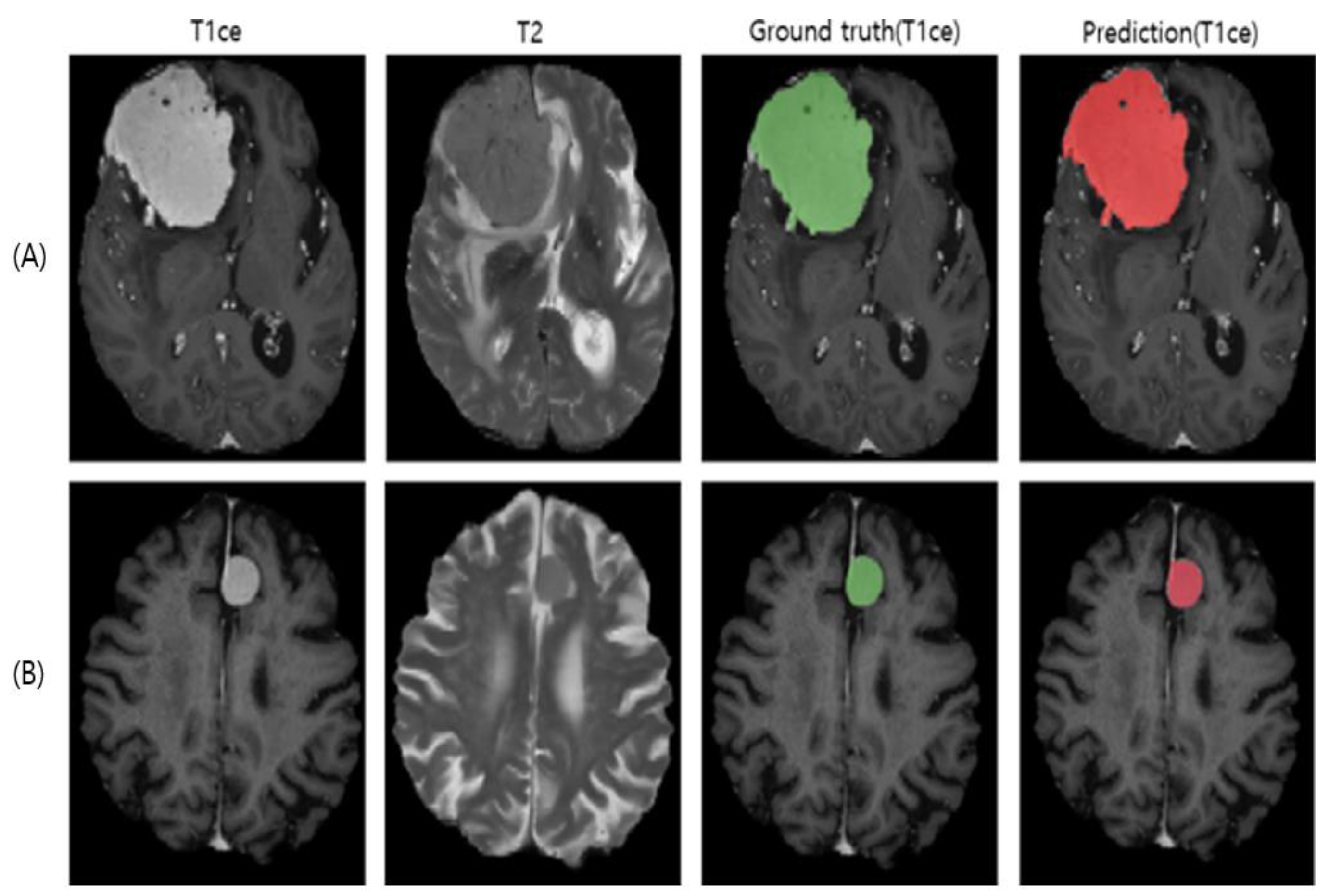
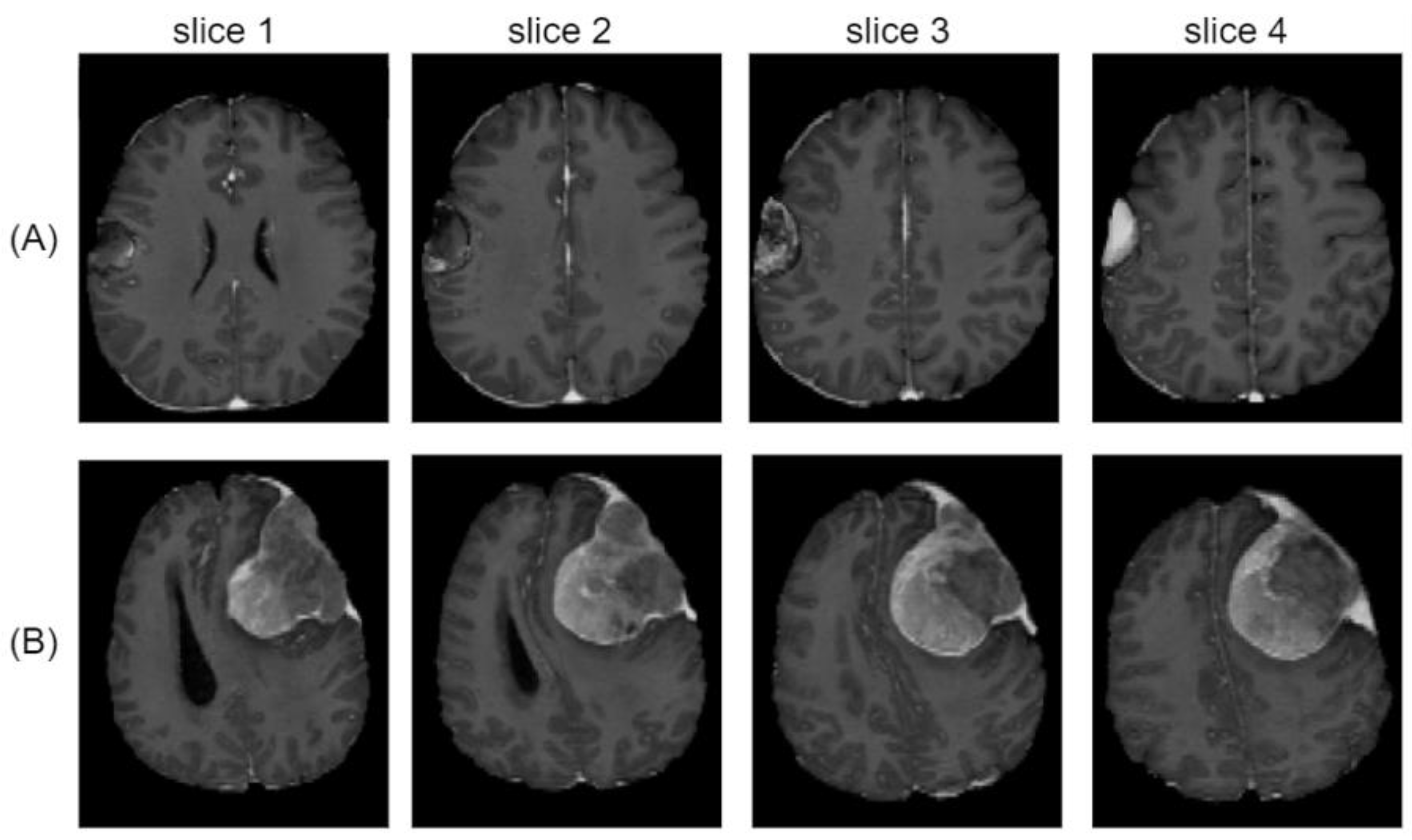
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F., Eds.; Lecture Notes in Computer Science, 9351; Springer International Publishing: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Yeung, M.; Sala, E.; Schönlieb, C.-B.; Rundo, L. Focus U-Net: A Novel Dual Attention-Gated CNN for Polyp Segmentation during Colonoscopy. Comput. Biol. Med. 2021, 137, 104815. [Google Scholar] [CrossRef] [PubMed]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Punn, N.S.; Agarwal, S. Modality Specific U-Net Variants for Biomedical Image Segmentation: A Survey. Artif. Intell. Rev. 2022, 55, 5845–5889. [Google Scholar] [CrossRef]
- Torfi, A.; Shirvani, R.A.; Keneshloo, Y.; Tavaf, N.; Fox, E.A. Natural Language Processing Advancements by Deep Learning: A Survey. arXiv 2021, arXiv:2003.01200. [Google Scholar]
- Galassi, A.; Lippi, M.; Torroni, P. Attention in Natural Language Processing. IEEE Trans. Neural. Netw. Learning Syst. 2021, 32, 4291–4308. [Google Scholar] [CrossRef]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning Where to Look for the Pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention Gated Networks: Learning to Leverage Salient Regions in Medical Images. Med. Image Anal. 2019, 53, 197. [Google Scholar] [CrossRef]
- Ma, J.; Chen, J.; Ng, M.; Huang, R.; Li, Y.; Li, C.; Yang, X.; Martel, A.L. Loss Odyssey in Medical Image Segmentation. Med. Image Anal. 2021, 71, 102035. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BraTS). IEEE Trans Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas Glioma MRI Collections with Expert Segmentation Labels and Radiomic Features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Kamnitsas, K.; Bai, W.; Ferrante, E.; McDonagh, S.; Sinclair, M.; Pawlowski, N.; Rajchl, M.; Lee, M.; Kainz, B.; Rueckert, D.; et al. Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation. arXiv 2017, arXiv:1711.01468. [Google Scholar]
- Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. arXiv 2018, arXiv:1810.11654. [Google Scholar]
- Two-Stage Cascaded U-Net: 1st Place Solution to BraTS Challenge 2019 Segmentation Task. Available online: https://www.springerprofessional.de/en/two-stage-cascaded-u-net-1st-place-solution-to-brats-challenge-2/17993490 (accessed on 22 December 2021).
- Wacker, J.; Ladeira, M.; Nascimento, J.E.V. Transfer Learning for Brain Tumor Segmentation. arXiv 2020, arXiv:1912.12452. [Google Scholar]
- Ouyang, C.; Kamnitsas, K.; Biffi, C.; Duan, J.; Rueckert, D. Data Efficient Unsupervised Domain Adaptation for Cross-Modality Image Segmentation. arXiv 2019, arXiv:1907.02766. [Google Scholar]
- Laukamp, K.R.; Thiele, F.; Shakirin, G.; Zopfs, D.; Faymonville, A.; Timmer, M.; Maintz, D.; Perkuhn, M.; Borggrefe, J. Fully Automated Detection and Segmentation of Meningiomas Using Deep Learning on Routine Multiparametric MRI. Eur. Radiol. 2019, 29, 124–132. [Google Scholar] [CrossRef]
- Laukamp, K.R.; Pennig, L.; Thiele, F.; Reimer, R.; Görtz, L.; Shakirin, G.; Zopfs, D.; Timmer, M.; Perkuhn, M.; Borggrefe, J. Automated Meningioma Segmentation in Multiparametric MRI: Comparable Effectiveness of a Deep Learning Model and Manual Segmentation. Clin. Neuroradiol. 2021, 31, 357–366. [Google Scholar] [CrossRef]
- Bouget, D.; Pedersen, A.; Hosainey, S.A.M.; Vanel, J.; Solheim, O.; Reinertsen, I. Fast Meningioma Segmentation in T1-Weighted MRI Volumes Using a Lightweight 3D Deep Learning Architecture. J. Med. Imag. 2021, 8, 024002. [Google Scholar] [CrossRef]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2020, 18, 203–211. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. arXiv 2016, arXiv:1606.04797. [Google Scholar]
- Jenkinson, M.; Beckmann, C.F.; Behrens, T.E.J.; Woolrich, M.W.; Smith, S.M. FSL. Neuroimage 2012, 62, 782–790. [Google Scholar] [CrossRef] [PubMed]
- Avants, B.B.; Tustison, N.J.; Song, G.; Cook, P.A.; Klein, A.; Gee, J.C. A Reproducible Evaluation of ANTs Similarity Metric Performance in Brain Image Registration. Neuroimage 2011, 54, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Kayalibay, B.; Jensen, G.; van der Smagt, P. CNN-Based Segmentation of Medical Imaging Data. arXiv 2017, arXiv:1701.03056. [Google Scholar]
- Ulyanov, D.; Vedaldi, A.; Lempitsky, V. Instance Normalization: The Missing Ingredient for Fast Stylization. arXiv 2017, arXiv:1607.08022. [Google Scholar]
- Maas, A.L. Rectifier Nonlinearities Improve Neural Network Acoustic Models. In Proceedings of the 30th International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- Keras: Deep Learning for Humans. Available online: https://github.com/keras-team/keras (accessed on 10 January 2021).
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. TensorFlow: A System for Large-Scale Machine Learning. arXiv 2016, arXiv:1605.08695. [Google Scholar]
- Ellis, D.G. 3D U-Net Convolution Neural Network. Available online: https://github.com/ellisdg/3DUnetCNN (accessed on 10 January 2021).
- Pastor-Pellicer, J.; Zamora-Martínez, F.; Boquera, S.E.; Bleda, M.J. F-Measure as the Error Function to Train Neural Networks. In Advances in Computational Intelligence—IWANN 2013; Rojas, I., Joya, G., Gabestany, J., Eds.; Lecture Notes in Computer Science, 7902; Springer: Berlin/Heidelberg, Germany, 2013; pp. 376–384. [Google Scholar]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A Survey of Transfer Learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam. A Method for Stochastic Optimization. arXiv 2017, arXiv:1412.6980. [Google Scholar]
- Huang, R.Y.; Bi, W.L.; Griffith, B.; Kaufmann, T.J.; la Fougère, C.; Schmidt, N.O.; Tonn, J.C.; Vogelbaum, M.A.; Wen, P.Y.; Aldape, K.; et al. International Consortium on Meningiomas. Imaging and Diagnostic Advances for Intracranial Meningiomas. Neuro. Oncol. 2019, 21 (Suppl. S1), i44–i61. [Google Scholar] [CrossRef]
- Rundo, L.; Beer, L.; Ursprung, S.; Martin-Gonzalez, P.; Markowetz, F.; Brenton, J.D.; Crispin-Ortuzar, M.; Sala, E.; Woitek, R. Tissue-Specific and Interpretable Sub-Segmentation of Whole Tumour Burden on CT Images by Unsupervised Fuzzy Clustering. Comput. Biol. Med. 2020, 120, 103751. [Google Scholar] [CrossRef]
- Heo, B.; Chun, S.; Oh, S.J.; Han, D.; Yun, S.; Kim, G.; Uh, Y.; Ha, J.-W. AdamP: Slowing Down the Slowdown for Momentum Optimizers on Scale-Invariant Weights. arXiv 2021, arXiv:2006.08217. [Google Scholar]
- Yu, T.; Zhu, H. Hyper-Parameter Optimization: A Review of Algorithms and Applications. arXiv 2020, arXiv:2003.05689. [Google Scholar]
- Yeung, M.; Sala, E.; Schönlieb, C.B.; Rundo, L. Unified focal loss: Generalising dice and cross entropy-based losses to handle class imbalanced medical image segmentation. Comput. Med. Imaging Graph. 2022, 95, 102026. [Google Scholar] [CrossRef] [PubMed]
| Methods | Training Set | Patients | Total MRIs | Averaged Dice (sd) | Recall (sd) | Precision * (sd) |
|---|---|---|---|---|---|---|
| [A] BraTS | BraTS | 335 | 335 | 0.60 (0.32) | 0.64 (0.35) | 0.71 (0.37) |
| [B] Meningioma | Meningioma | 74 | 154 | 0.72 (0.28) | 0.83 (0.29) | 0.78 (0.27) |
| [C] TL | BraTS (pre-training) | 335 | 335 | 0.76 (0.23) | 0.79 (0.29) | 0.84 (0.19) |
| Meningioma | 74 | 154 | ||||
| [D] TL + Normal | BraTS (pre-training) | 335 | 335 | 0.79 (0.26) | 0.82 (0.28) | 0.81 (0.29) |
| Meningioma | 74 | 154 | ||||
| Normal | 10 | 10 | ||||
| [E] TL + Normal + BDL | BraTS (pre-training) | 335 | 335 | 0.84 (0.15) | 0.89 (0.18) | 0.84 (0.15) |
| Meningioma | 74 | 154 | ||||
| Normal | 10 | 10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, K.; Park, J.; Kwon, Y.-J.; Cho, S.J.; Choi, B.S.; Kim, J.; Kim, E.; Jang, J.; Ahn, K.-S.; Kim, S.; et al. Fully Automated Segmentation Models of Supratentorial Meningiomas Assisted by Inclusion of Normal Brain Images. J. Imaging 2022, 8, 327. https://doi.org/10.3390/jimaging8120327
Hwang K, Park J, Kwon Y-J, Cho SJ, Choi BS, Kim J, Kim E, Jang J, Ahn K-S, Kim S, et al. Fully Automated Segmentation Models of Supratentorial Meningiomas Assisted by Inclusion of Normal Brain Images. Journal of Imaging. 2022; 8(12):327. https://doi.org/10.3390/jimaging8120327
Chicago/Turabian StyleHwang, Kihwan, Juntae Park, Young-Jae Kwon, Se Jin Cho, Byung Se Choi, Jiwon Kim, Eunchong Kim, Jongha Jang, Kwang-Sung Ahn, Sangsoo Kim, and et al. 2022. "Fully Automated Segmentation Models of Supratentorial Meningiomas Assisted by Inclusion of Normal Brain Images" Journal of Imaging 8, no. 12: 327. https://doi.org/10.3390/jimaging8120327
APA StyleHwang, K., Park, J., Kwon, Y.-J., Cho, S. J., Choi, B. S., Kim, J., Kim, E., Jang, J., Ahn, K.-S., Kim, S., & Kim, C.-Y. (2022). Fully Automated Segmentation Models of Supratentorial Meningiomas Assisted by Inclusion of Normal Brain Images. Journal of Imaging, 8(12), 327. https://doi.org/10.3390/jimaging8120327






