SpineDepth: A Multi-Modal Data Collection Approach for Automatic Labelling and Intraoperative Spinal Shape Reconstruction Based on RGB-D Data
Abstract
1. Introduction
2. Materials and Methods
2.1. Choice of Sensor
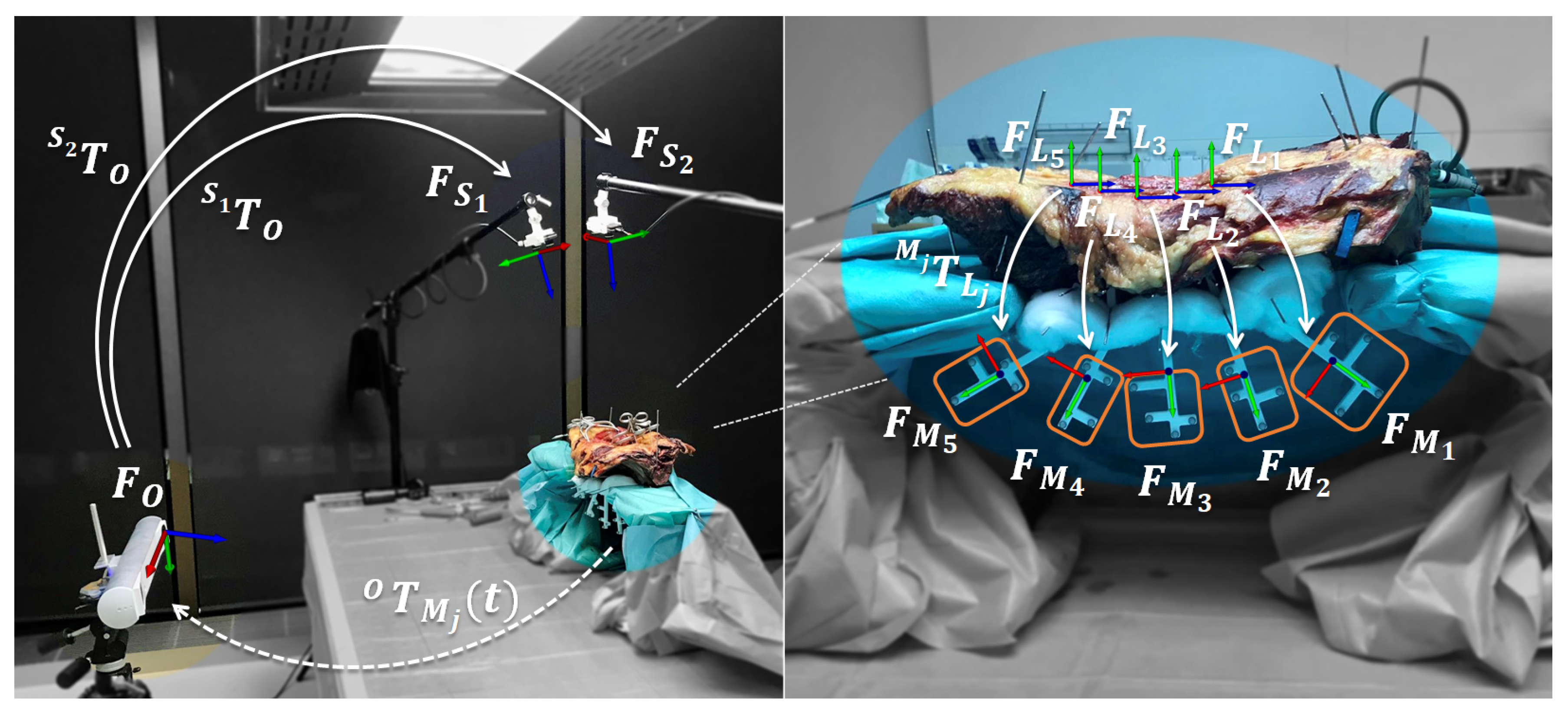
2.2. Data Acquisition Setup
2.3. Extrinsic Calibration
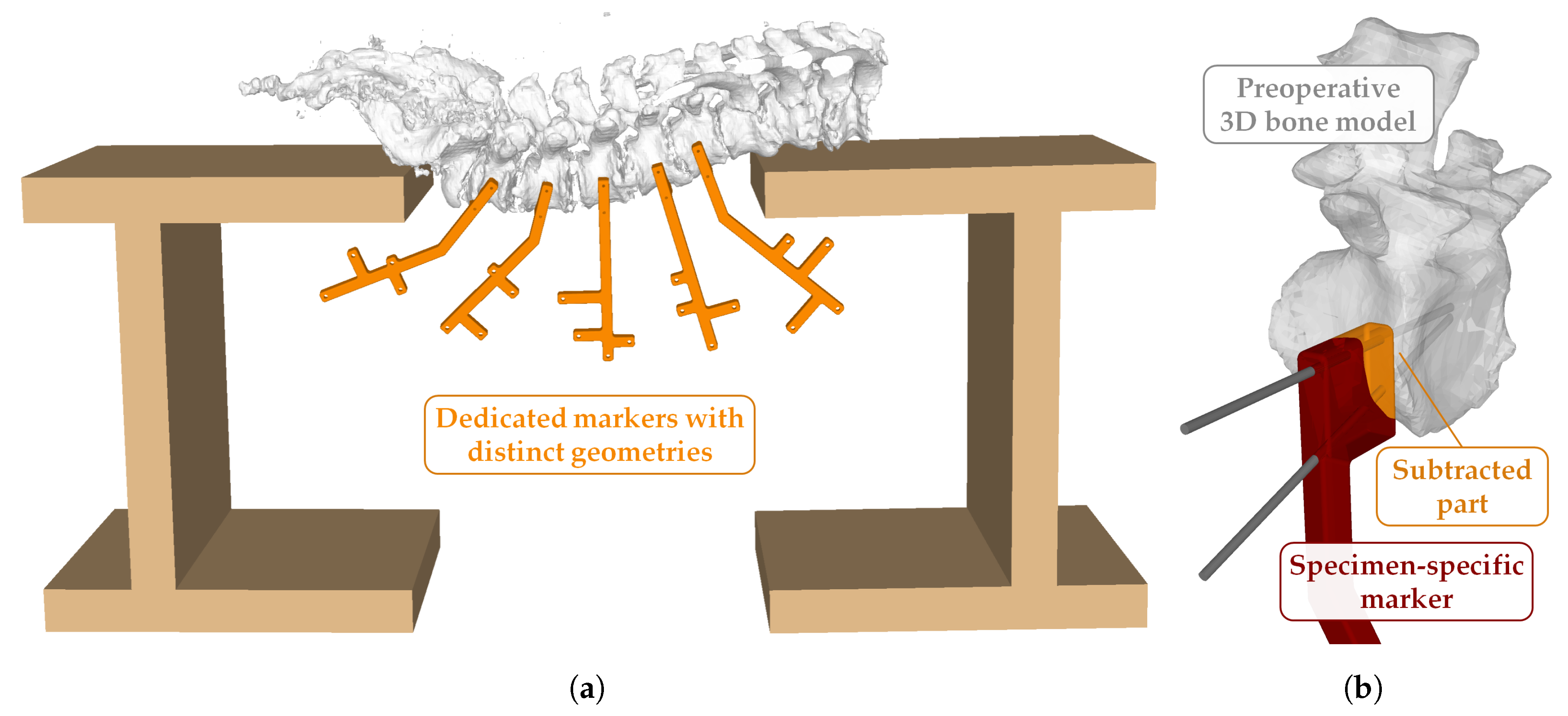
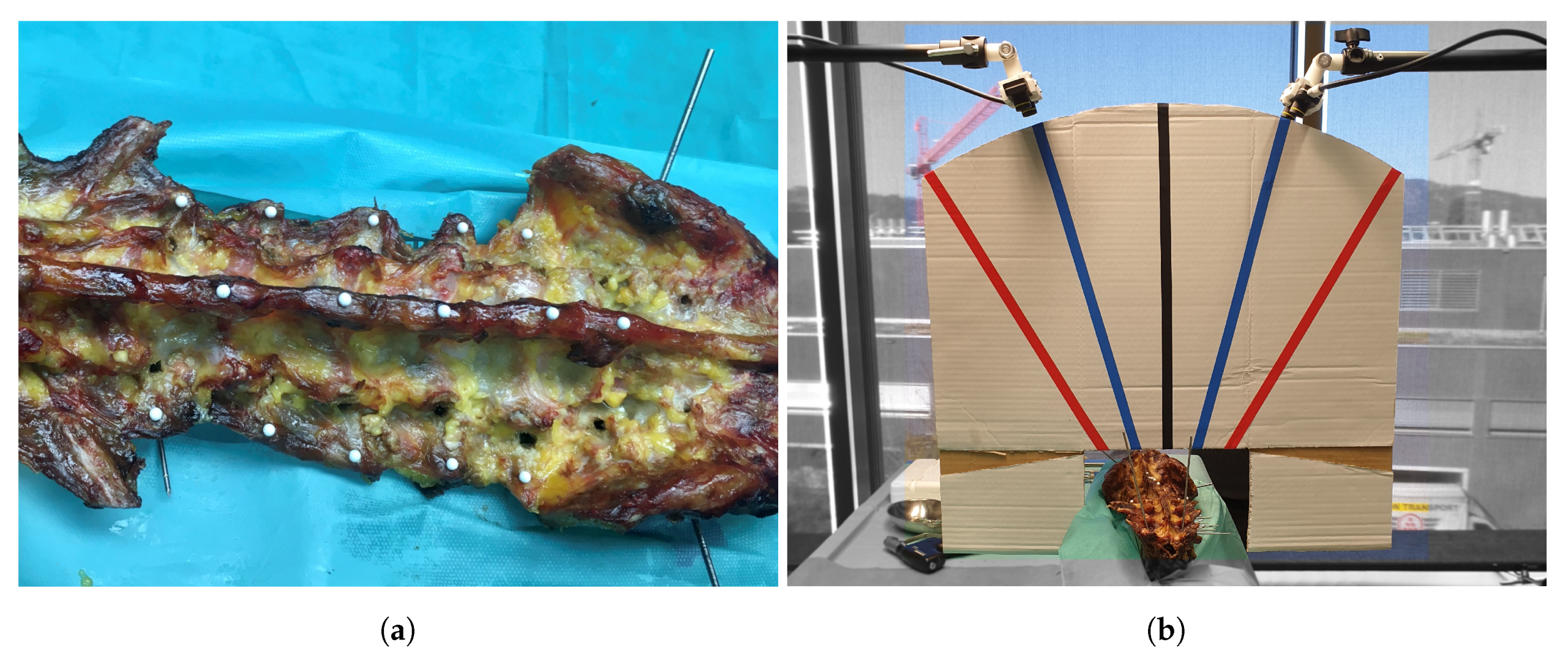
2.4. Cadaver Preparation
2.5. Data Acquisition
2.5.1. Pedicle Screw Placement Recording (Surgical Dataset)
- Reposition RGB-D sensors resembling a realistic surgical viewpoint;
- Perform extrinsic calibration between OTS to RGB-D sensors, store and ;
- For each surgical step:
- (a)
- Start recording;
- (b)
- Perform surgical step;
- (c)
- Stop recording.
2.5.2. TRE Recording (TRE Dataset)
2.5.3. Postoperative CT
2.6. Data Post-Processing
2.7. Outcome Measures
- TRE
- The TRE was defined as the 3D Euclidean distance between the CT-extracted push-pin head centers and their respective counterparts in each RGB-D sensor’s space: and (Section 2.6). As explained in Section 2.6, the 3D fiducial centroid coordinates in the RGB-D space were explicitly triangulated based on the stereo camera setup of the RGB-D sensor (based on intrinsic and extrinsic camera parameters) and we did not rely on the default stereo reconstruction algorithm of the sensor for these measurements. This was done so that the TRE measure only reflected on the errors associated with the data acquisition setup (e.g., extrinsic calibration) and not the inherent point cloud estimation of the sensor. The fiducial coordinates obtained from the CT measurement () were transformed to the space of either RGB-D sensor by (Equation (1)). Note that points were transformed according to the levels they were inserted (i.e., index j) and the pairwise distances were calculated. For each cadaver, 180 datapoints (15 push-pin head centers × 6 viewpoints × 2 sensors) were assessed.
- VBSE
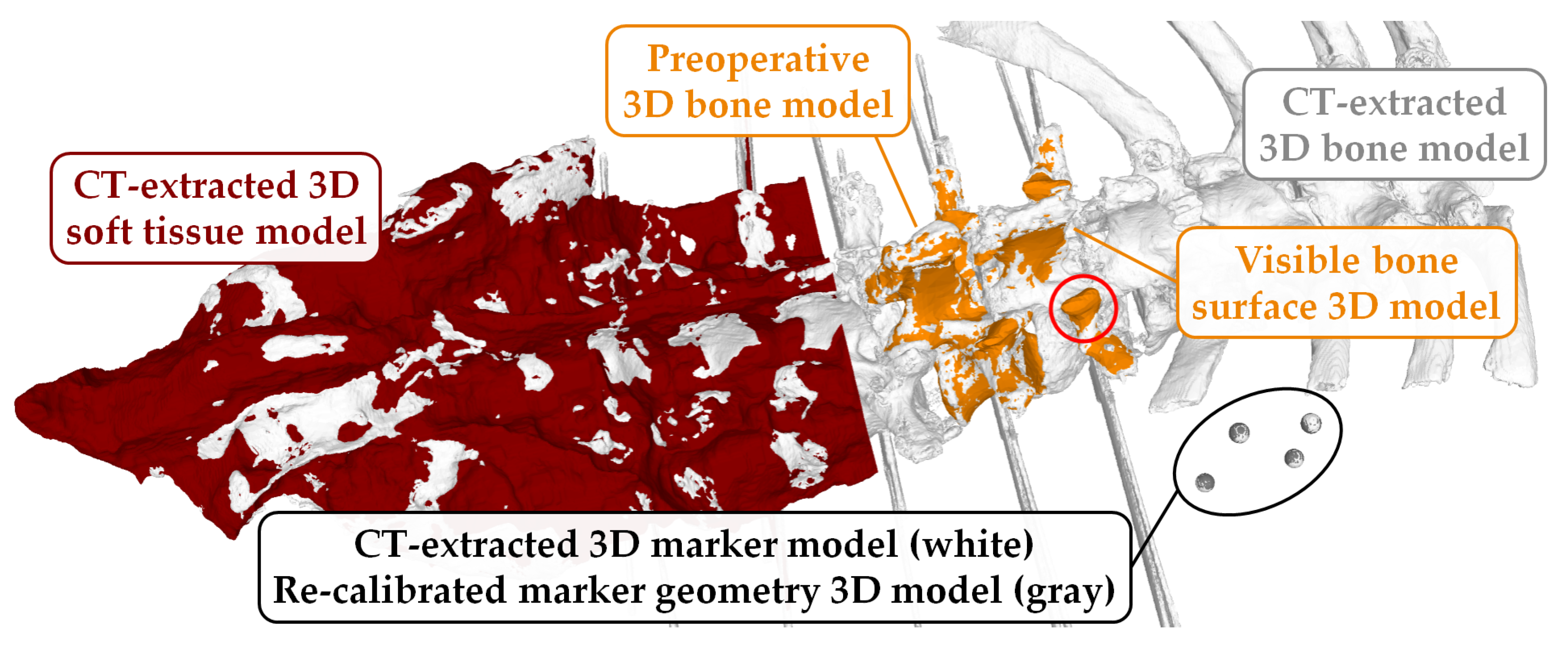
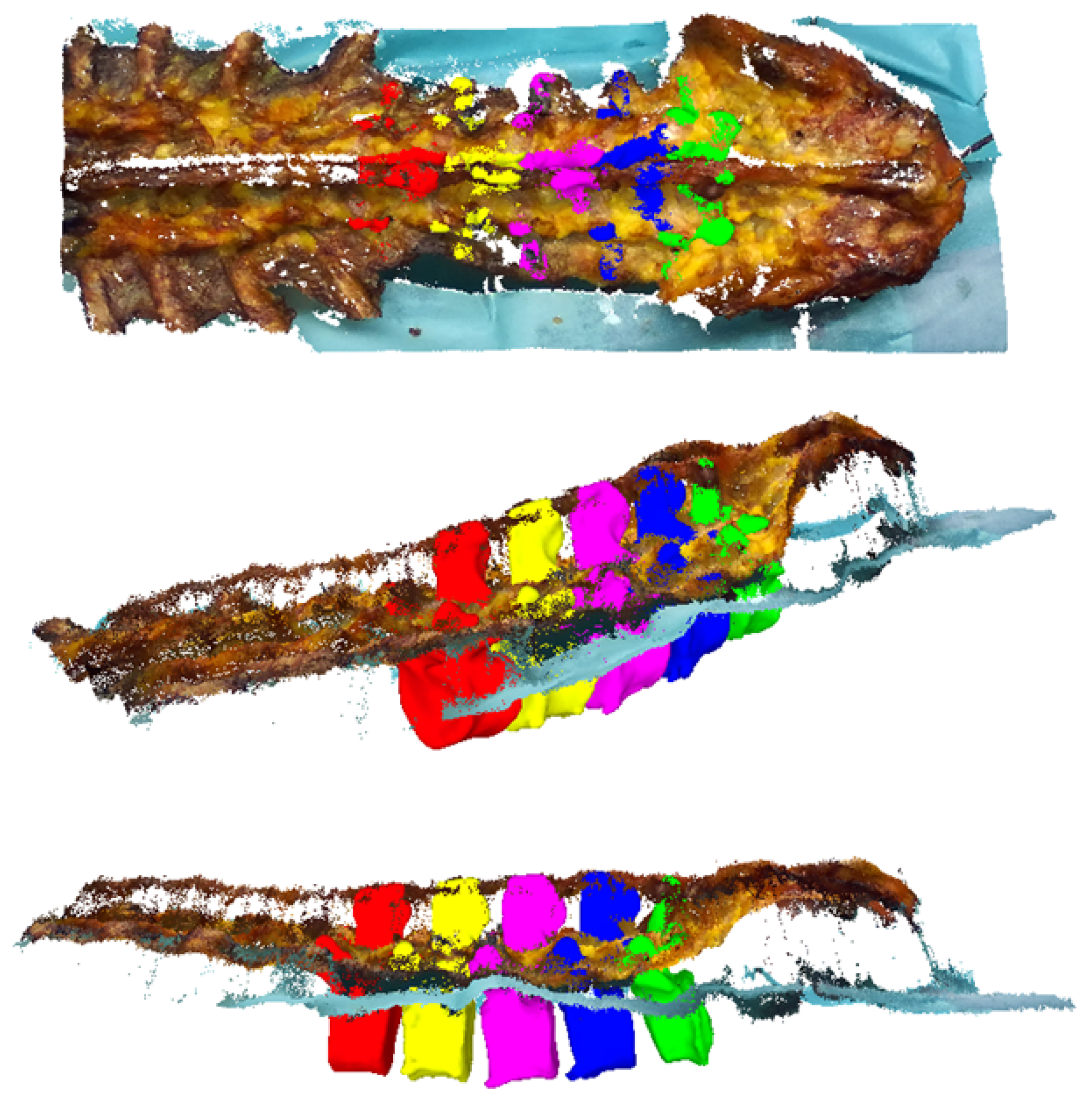
- In contrast to the TRE measure, the VBSE estimates the overall accuracy of our setup including the stereo reconstruction algorithm of the RGB-D sensor in use. The aforementioned visible bone surface 3D models of levels L1–L5 were transformed to the space of either RGB-D sensor using the estimated transformation (Equation (1)), which itself is based on the OTS tracking data, and a depth map was rendered using the code of [33,34]. Ideally, the reconstructed depth map of either RGB-D sensor should be identical to their OTS-based, rendered counterparts. Therefore, any deviations between the two 3D representations can be attributed to inherent reconstruction errors of the RGB-D sensor and the errors within the data acquisition setup (assuming that the OTS measurement errors can be neglected). Furthermore, this error was influenced by the varying presence of screws in the RGB-D recordings and their absence in the postoperative CT as well as the absence of the area around the facet joints and mamillary processes in specimens 2–10 (history of spinal fusion performed within the scope of another cadaveric experiment) in the RGB-D recordings and their presence in the employed preoperative 3D bone models (Figure 5). All such phenomena result in domain mismatch; therefore, the VBSE was defined as the median absolute difference between all non-empty pixels of the rendered depth map and their corresponding pixels in the depth map reconstructed by the RGB-D sensors (except pixels, where no depth was reconstructed). Using the VBSE measure for RGB-D frames where the surgeon’s hand or the surgical tools were occluding the sensors view over the anatomy was not possible. This is due to the fact that the RGB-D stream could not be segmented to parts were exclusively points of the anatomy were present. Therefore, the VBSE measure was calculated for the first 10 frames (roughly s) of each recording, where the surgeon’s hand were not present in the sensors’ field of view.
- SR
- The SR is a qualitative rating by the surgeon. A random sample of four recordings per specimen (10%) was selected for this purpose. For each recording, the first frame was selected (). The aligned preoperative 3D bone models (Figure 5) of levels L1–L5 were transformed to the space of either RGB-D sensor by (Equation (1)). Point clouds were extracted from the respective RGB-D frames. The surgeon then assessed the alignment of the bone models, i.e., the ground truth of our dataset, with the point cloud after the criteria:
- Are the spinous processes in line?
- Are the facet joint in line?
- From a lateral view: are the vertebra at correct height?
- Is the overall alignment correct?
Each criteria received a score between 1 (worst) and 6 (best). The average of the four scores yielded the SR for one recording.
2.8. Depth Correction
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gelalis, I.D.; Paschos, N.K.; Pakos, E.E.; Politis, A.N.; Arnaoutoglou, C.M.; Karageorgos, A.C.; Ploumis, A.; Xenakis, T.A. Accuracy of pedicle screw placement: A systematic review of prospective studies comparing free hand, fluoroscopy guidance and navigation techniques. Eur. Spine J. 2012, 21, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Perdomo-Pantoja, A.; Ishida, W.; Zygourakis, C.; Holmes, C.; Iyer, R.R.; Cottrill, E.; Theodore, N.; Witham, T.F.; Sheng-fu, L.L. Accuracy of current techniques for placement of pedicle screws in the spine: A comprehensive systematic review and meta-analysis of 51,161 screws. World Neurosurg. 2019, 126, 664–678. [Google Scholar] [CrossRef] [PubMed]
- Härtl, R.; Lam, K.S.; Wang, J.; Korge, A.; Kandziora, F.; Audigé, L. Worldwide survey on the use of navigation in spine surgery. World Neurosurg. 2013, 79, 162–172. [Google Scholar] [CrossRef] [PubMed]
- Joskowicz, L. Computer-aided surgery meets predictive, preventive, and personalized medicine. EPMA J. 2017, 8, 1–4. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Joskowicz, L.; Hazan, E.J. Computer Aided Orthopaedic Surgery: Incremental shift or paradigm change? Med. Image Anal. 2016, 100, 84–90. [Google Scholar] [CrossRef]
- Picard, F.; Clarke, J.; Deep, K.; Gregori, A. Computer assisted knee replacement surgery: Is the movement mainstream? Orthop. Muscular Syst. 2014, 3. [Google Scholar] [CrossRef]
- Markelj, P.; Tomaževič, D.; Likar, B.; Pernuš, F. A review of 3D/2D registration methods for image-guided interventions. Med. Image Anal. 2012, 16, 642–661. [Google Scholar] [CrossRef]
- Esfandiari, H.; Anglin, C.; Guy, P.; Street, J.; Weidert, S.; Hodgson, A.J. A comparative analysis of intensity-based 2D–3D registration for intraoperative use in pedicle screw insertion surgeries. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1725–1739. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, P.; Antunes, M.; Raposo, C.; Marques, P.; Fonseca, F.; Barreto, J.P. Deep segmentation leverages geometric pose estimation in computer-aided total knee arthroplasty. Healthc. Technol. Lett. 2019, 6, 226–230. [Google Scholar] [CrossRef]
- Félix, I.; Raposo, C.; Antunes, M.; Rodrigues, P.; Barreto, J.P. Towards markerless computer-aided surgery combining deep segmentation and geometric pose estimation: Application in total knee arthroplasty. Comput. Methods Biomech. Biomed. Eng. Imaging Visualiz. 2020, 9, 271–278. [Google Scholar] [CrossRef]
- Lee, S.C.; Fuerst, B.; Fotouhi, J.; Fischer, M.; Osgood, G.; Navab, N. Calibration of RGBD camera and cone-beam CT for 3D intra-operative mixed reality visualization. Int. J. Comput. Assist. Radiol. Surg. 2016, 11, 967–975. [Google Scholar] [CrossRef]
- Lee, S.C.; Fuerst, B.; Tateno, K.; Johnson, A.; Fotouhi, J.; Osgood, G.; Tombari, F.; Navab, N. Multi-modal imaging, model-based tracking, and mixed reality visualisation for orthopaedic surgery. Healthc. Technol. Lett. 2017, 4, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Shah, K.; Knopf, J.; Navab, N.; Unberath, M. Feasibility of image-based augmented reality guidance of total shoulder arthroplasty using microsoft HoloLens 1. Comput. Methods Biomech. Biomed. Eng. Imaging Visualiz. 2020, 9, 261–270. [Google Scholar] [CrossRef]
- Kalfas, I.H. Machine Vision Navigation in Spine Surgery. Front. Surg. 2021, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, H.; Malacon, K.; Medress, Z.A.; Leung, C.; Sklar, M.; Zygourakis, C.C. First reported use of real-time intraoperative computed tomography angiography image registration using the Machine-vision Image Guided Surgery system: Illustrative case. J. Neurosurg. Case Lessons 2021, 1. [Google Scholar] [CrossRef]
- Cabrera, E.V.; Ortiz, L.E.; da Silva, B.M.; Clua, E.W.; Gonçalves, L.M. A versatile method for depth data error estimation in RGB-D sensors. Sensors 2018, 18, 3122. [Google Scholar] [CrossRef]
- Pratusevich, M.; Chrisos, J.; Aditya, S. Quantitative Depth Quality Assessment of RGBD Cameras At Close Range Using 3D Printed Fixtures. arXiv 2019, arXiv:1903.09169. [Google Scholar]
- Bajzik, J.; Koniar, D.; Hargas, L.; Volak, J.; Janisova, S. Depth Sensor Selection for Specific Application. In Proceedings of the 2020 ELEKTRO, Taormina, Italy, 25–28 May 2020; pp. 1–6. [Google Scholar]
- Cao, M.; Zheng, L.; Liu, X. Single View 3D Reconstruction Based on Improved RGB-D Image. IEEE Sens. J. 2020, 20, 12049–12056. [Google Scholar] [CrossRef]
- Mehta, D.; Sridhar, S.; Sotnychenko, O.; Rhodin, H.; Shafiei, M.; Seidel, H.P.; Xu, W.; Casas, D.; Theobalt, C. Vnect: Real-time 3d human pose estimation with a single rgb camera. ACM Trans. Graph. (TOG) 2017, 36, 1–14. [Google Scholar] [CrossRef]
- Schwarz, M.; Schulz, H.; Behnke, S. RGB-D object recognition and pose estimation based on pre-trained convolutional neural network features. In Proceedings of the 2015 IEEE International Conference on Robotics and Automation (ICRA), Seattle, WA, USA, 26–30 May 2015; pp. 1329–1335. [Google Scholar]
- Hou, J.; Dai, A.; Nießner, M. 3d-sis: 3d semantic instance segmentation of rgb-d scans. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 4421–4430. [Google Scholar]
- Azinović, D.; Martin-Brualla, R.; Goldman, D.B.; Nießner, M.; Thies, J. Neural RGB-D Surface Reconstruction. arXiv 2021, arXiv:2104.04532. [Google Scholar]
- Kaskman, R.; Zakharov, S.; Shugurov, I.; Ilic, S. Homebreweddb: Rgb-d dataset for 6d pose estimation of 3d objects. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) Workshops, Seoul, Korea, 27–28 October 2019. [Google Scholar]
- Hodan, T.; Haluza, P.; Obdržálek, Š.; Matas, J.; Lourakis, M.; Zabulis, X. T-LESS: An RGB-D dataset for 6D pose estimation of texture-less objects. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; pp. 880–888. [Google Scholar]
- Hachenberger, P.; Kettner, L. 3D Boolean Operations on Nef Polyhedra. In CGAL User and Reference Manual, 5.1.1 ed.; CGAL Editorial Board; Utrecht University, Faculty of Mathematics and Computer Science Netherlands: Utrecht, The Netherlands, 2021. [Google Scholar]
- Besl, P.J.; McKay, N.D. Method for registration of 3-D shapes. Sens. Fus. IV Control Paradig. Data Struct. 1992, 1611, 586–606. [Google Scholar] [CrossRef]
- Rusinkiewicz, S.; Levoy, M. Efficient variants of the ICP algorithm. In Proceedings of the Third International Conference on 3-D Digital Imaging and Modeling, Quebec City, QC, Canada, 28 May–1 June 2001; pp. 145–152. [Google Scholar]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000, 120, 122–125. [Google Scholar]
- Horn, B.K. Closed-form solution of absolute orientation using unit quaternions. J. Opt. Soc. Am. A 1987, 4, 629–642. [Google Scholar] [CrossRef]
- Matt, J. Absolute Orientation—Horn’s Method. 2021. Available online: https://www.mathworks.com/matlabcentral/fileexchange/26186-absolute-orientation-horn-s-method (accessed on 5 July 2021).
- Roner, S.; Vlachopoulos, L.; Nagy, L.; Schweizer, A.; Fürnstahl, P. Accuracy and early clinical outcome of 3-dimensional planned and guided single-cut osteotomies of malunited forearm bones. J. Hand Surg. 2017, 42, 1031.e1–1031.e8. [Google Scholar] [CrossRef] [PubMed]
- Guney, F.; Geiger, A. Displets: Resolving stereo ambiguities using object knowledge. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 4165–4175. [Google Scholar]
- Geiger, A.; Wang, C. Joint 3D Object and Layout Inference from a single RGB-D Image. In Proceedings of the 37th German Conference on Pattern Recognition (GCPR), Aachen, Germany, 7–10 October 2015. [Google Scholar]
- Papadopoulos, E.C.; Girardi, F.P.; Sama, A.; Sandhu, H.S.; Cammisa, F.P., Jr. Accuracy of single-time, multilevel registration in image-guided spinal surgery. Spine J. 2005, 5, 263–267. [Google Scholar] [CrossRef]
- Ji, S.; Fan, X.; Paulsen, K.D.; Roberts, D.W.; Mirza, S.K.; Lollis, S.S. Patient registration using intraoperative stereovision in image-guided open spinal surgery. IEEE Trans. Biomed. Eng. 2015, 62, 2177–2186. [Google Scholar] [CrossRef]
- Nottmeier, E.W.; Crosby, T.L. Timing of paired points and surface matching registration in three-dimensional (3D) image-guided spinal surgery. Clin. Spine Surg. 2007, 20, 268–270. [Google Scholar] [CrossRef]

| Name | Quantity | Description | Functionality |
|---|---|---|---|
| 3D marker model | 5 | Infrared-reflecting spheres for each marker attached to levels L1–L5 | Global thresholding, region growing |
| Push-pin head centers | 15 | Push-pin head center coordinates | Manual cursor alignment in axial, coronal and sagittal views |
| 3D bone model | 1 | Bony anatomy | Global thresholding, region growing |
| 3D soft tissue model | 1 | Soft tissue including non-exposed parts of bony anatomy | Global thresholding, smooth mask, smart fill, erosion, manual artifact and push-pin removal, wrapping |
| Specimen | Type | Screw Order | # of Frames |
|---|---|---|---|
| 1 | Midline approach | L1–L5 | 22,553 |
| 2 | Full exposure | L1–L3, L4–L5 | 34,302 |
| 3 | Full exposure | L1–L2, L3–L5 (LLL, RRR) | 33,489 |
| 4 | Full exposure | L1–L5 | 37,497 |
| 5 | Full exposure | L1–L3, L4–L5 | 31,279 |
| 6 | Full exposure | L3–L4, L4–L5, L1–L2 | 30,249 |
| 7 | Full exposure | L4–L5, L1–L3 | 25,746 |
| 8 | Full exposure | L2–L4, L4–L5, L1–L2 | 27,679 |
| 9 | Full exposure | L1–L5 | 24,403 |
| 10 | Full exposure | L4–L5, L1–L3 | 32,359 |
| Specimen | [mm] | [mm] | [1–6] |
|---|---|---|---|
| 1 | N/A | N/A | N/A |
| 2 | |||
| 3 | |||
| 4 | |||
| 5 | |||
| 6 | |||
| 7 | |||
| 8 | |||
| 9 | |||
| 10 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liebmann, F.; Stütz, D.; Suter, D.; Jecklin, S.; Snedeker, J.G.; Farshad, M.; Fürnstahl, P.; Esfandiari, H. SpineDepth: A Multi-Modal Data Collection Approach for Automatic Labelling and Intraoperative Spinal Shape Reconstruction Based on RGB-D Data. J. Imaging 2021, 7, 164. https://doi.org/10.3390/jimaging7090164
Liebmann F, Stütz D, Suter D, Jecklin S, Snedeker JG, Farshad M, Fürnstahl P, Esfandiari H. SpineDepth: A Multi-Modal Data Collection Approach for Automatic Labelling and Intraoperative Spinal Shape Reconstruction Based on RGB-D Data. Journal of Imaging. 2021; 7(9):164. https://doi.org/10.3390/jimaging7090164
Chicago/Turabian StyleLiebmann, Florentin, Dominik Stütz, Daniel Suter, Sascha Jecklin, Jess G. Snedeker, Mazda Farshad, Philipp Fürnstahl, and Hooman Esfandiari. 2021. "SpineDepth: A Multi-Modal Data Collection Approach for Automatic Labelling and Intraoperative Spinal Shape Reconstruction Based on RGB-D Data" Journal of Imaging 7, no. 9: 164. https://doi.org/10.3390/jimaging7090164
APA StyleLiebmann, F., Stütz, D., Suter, D., Jecklin, S., Snedeker, J. G., Farshad, M., Fürnstahl, P., & Esfandiari, H. (2021). SpineDepth: A Multi-Modal Data Collection Approach for Automatic Labelling and Intraoperative Spinal Shape Reconstruction Based on RGB-D Data. Journal of Imaging, 7(9), 164. https://doi.org/10.3390/jimaging7090164







