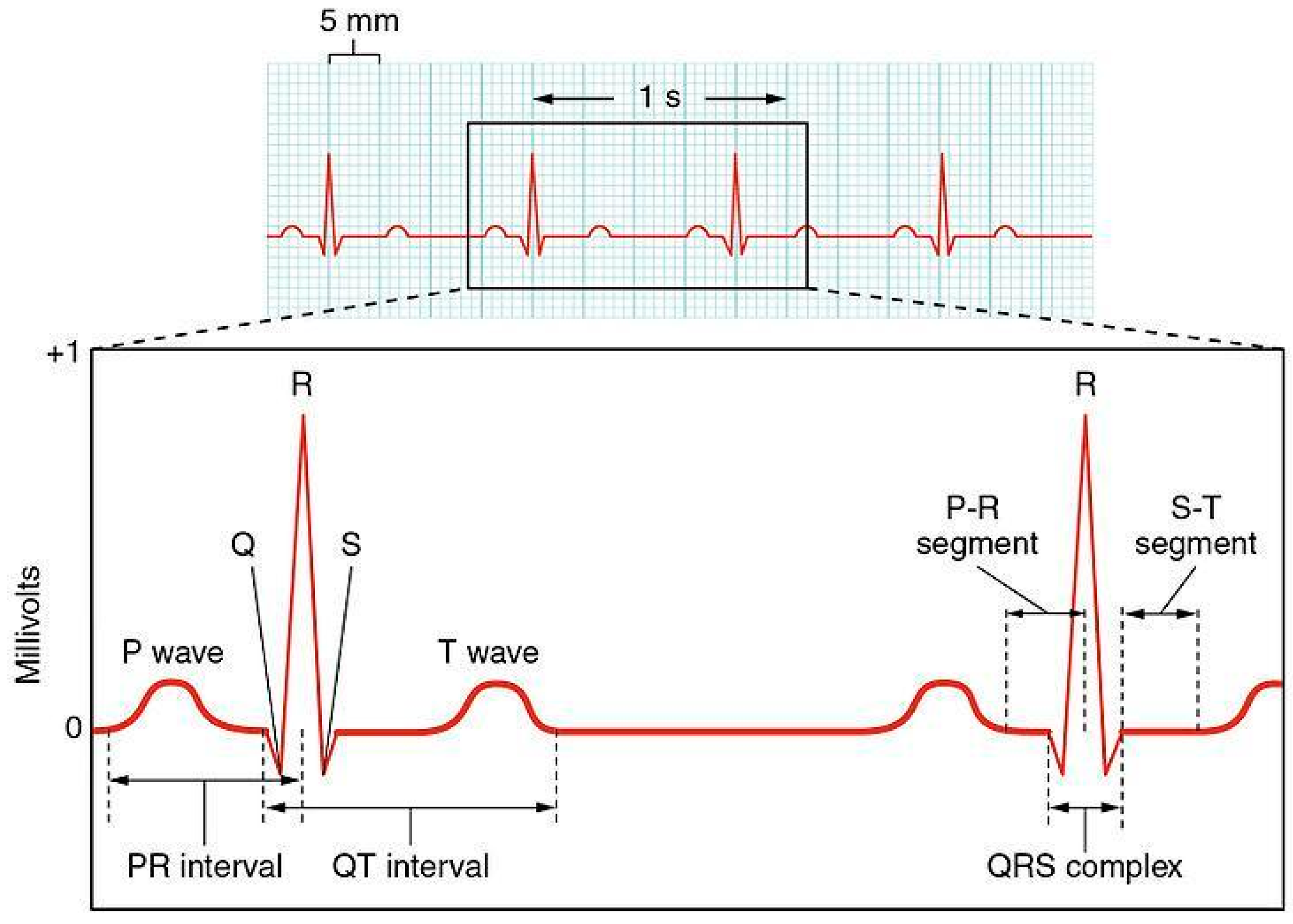
1. Introduction
The aging of the population is leading to an increase in patients suffering from cardiac pathologies, therefore requiring electrocardiographic monitoring. An electrocardiogram (ECG) is an easy, rapid, and non-invasive tool that traces the electrical activity of the heart [
1] revealing the presence of cardiac pathologies such as conduction disease, channelopathies, structural heart disease, and previous ischemic injury [
2]. On the other hand, investigating the altered acoustic characteristics of the cardiac tones, as an example, may allow the early identification of valve malfunction [
3].
Systems able to support the doctors’ work in the diagnosis of pathologies can facilitate health care decision making reducing considerably expenditure of time and money [
4,
5,
6,
7,
8,
9,
10,
11,
12]. The ECG has become the diagnostic procedure most commonly performed in clinical cardiology [
13,
14,
15] and the diffusion of wearable and portable devices has been enabling patients to constantly monitor the cardiac activity, for example of elder people through wireless sensor networks [
16]. Cardiologists cannot examine millions of ECGs daily recorded from portable devices. Thus, systems able to automatically detect suspicious anomalies in ECGs are required, in order to reduce the number of ECGs that need to be manually examined by the cardiologists, identifying those that need a further examination and also the urgency of such examination. For this reason, systems require high detection performance in order to avoid that normal ECGs incorrectly detected as anomalous should be examined by a medical professional, and, even more important, that the presence of an electrocardiographic alteration, which could be the indicator of cardiac pathology, is recognized and does not escape the observation of the cardiologist. To make the anomalous ECGs be examined by the medical specialist and that the proper therapy is administered, the detection system should maximize recall for anomalous ECGs, which is to maximize the number of ECGs correctly classified as anomalous, even losing accuracy.
The future of quick and efficient disease diagnosis lays in the development of reliable non-invasive methods [
17] also through the use of artificial intelligence techniques. Artificial neural networks and deep learning architectures have recently found broad application [
18] achieving striking success in different domains such as image classification [
19,
20,
21,
22,
23,
24], speech recognition [
25], intrusion detection systems [
26,
27], smart city [
28], or biological studies [
29,
30].
Therefore, high expectations are placed in the use of such techniques also for the improvement of health care and clinical practice [
31,
32,
33,
34,
35,
36]. Furthermore, numerous portable devices for personal and frequent monitoring of cardiac activity, such as Kardia [
37], D-hearth [
38], and eKuore [
39], are spreading.
The goal of this paper is to implement a system able to automatically detect the suspect of cardiac pathologies in ECG signals to support personal monitoring devices. We propose a 1D-CNN architecture optimized to detect anomalous ECG recordings, regardless of the kind of cardiac pathology, including in the analysis 21 classes of anomalies.
The system here presented was designed to be implemented on devices for personal use and with the aim to only send to the cardiologist ECGs detected with the suspect of a cardiac alteration for further examination, thus no information about the specific class of anomaly is detected. The proposed system is based on a binary classification model.
In fact, as of now, we want to make it clear that the main goal of our study is not to classify different cardiac pathologies, but to make sure that the suspect of a pathology can be detected and that patients are alarmed: then a correct diagnosis can be carried on with specific tests and the intervention of medical staff. The cardiologist will examine all ECGs detected as anomalous identifying the pathology and prescribing the proper treatment. The system has been implemented with the aim to achieve high levels of recall for anomalous ECGs in order to minimize the possibility that the presence of any kind of cardiac alteration could escape the observation of the cardiologist.
This paper is organized as follows:
Section 2 reports a wide background,
Section 3 describes ECG signals;
Section 4 illustrates material and methods; in
Section 5 are pointed out results and discussions; and
Section 6 sets out conclusions.
2. Background
Many studies have proposed the implementation of artificial neural networks and deep learning architectures for the development of automatic systems able to recognize the suspect of cardiac anomalies [
40,
41,
42]. In the literature the detection of cardiac anomalies has been investigated analyzing both heart sounds acquired by digital stethoscopes and ECG signals from portable devices.
Meintjes et al. [
43] implemented continuous wavelet transform (CWT) scalograms and convolutional neural networks for the correct classification of the fundamental heart sounds in recordings of normal and pathological heart sounds. They implemented a methodology in order to distinguish between the first and second heart sounds using CWT decomposition and convolutional neural network (CNN) features. Results show the high potential in the use of CWT and CNN in the analysis of heart sounds compared to support vector machine (SVM), and k-nearest neighbors (kNN) classifiers. In [
44] authors propose the classification of heart sounds on short, unsegmented recordings and normalized spectral amplitude of 5 s duration phonocardiogram segments was determined by fast Fourier transform and wavelet entropy by wavelet analysis. Spectral amplitude and wavelet entropy features were then combined in a classification tree. They achieved accuracy comparable to other algorithms obtained without the complexity of segmentation. Redlarski et al. [
17] presented a new heart sound classification technique combining linear predictive coding coefficients, used for feature extraction, with a classifier built upon combining support vector machine and the modified cuckoo search algorithm. It showed good performance of the diagnostic system, in terms of accuracy, complexity [
45,
46,
47] and range of distinguishable heart sounds.
With the application of deep learning architectures, also the accuracy of ECG diagnostic analysis has achieved new high levels. The systems implemented using such techniques allow the automated interpretation of ECG signals from portable devices in real time. The common deep learning networks for the analysis of ECG signals are mainly based on recurrent neural networks (RNNs), convolutional neural networks (CNNs), and some other architectures [
1].
Chauhan et al. [
48] investigated the applicability of deep recurrent neural network architectures with long short term memory (LSTM) for detecting cardiac arrhythmias in ECG signals. This approach is quite fast, does not require preprocessing of the data [
49] or hand coded features and do not need prior information about the abnormal signal. The network was tested on the MIT-BIH Arrhythmia Database for the classification of four different types of Arrhythmias showing that LSTMs may be a viable candidate for anomaly detection in ECG signals.
Saadatnejad et al. [
50] proposed an LSTM-based ECG classification algorithm for continuous cardiac monitoring on wearable devices. They preprocessed data extracting RR interval and wavelet features from ECG samples. The ECG signal along with the extracted features were fed into multiple LSTM recurrent neural networks. The MIT-BIH ECG Arrhythmia Database was used for the classification of six different types of anomalies. The proposed algorithm achieved accurate LSTM-based ECG classification to wearable devices with low computational costs.
Thill et al. [
51] presented an unsupervised time series anomaly detection algorithm to detect anomalies in ECG readings. They performed a recurrent LSTM network to predict the normal time series behavior without the usage of the anomaly class labels building a multivariate normal error model for the nominal data. Anomalous events were detected with a high probability through a high Mahalanobis distance. They classified six anomaly classes and obtained good performance achieving high levels of precision and recall.
Although RNN architectures are suitable to process time series data, they present some limitations. The major drawbacks of RNNs are the vanishing gradient and gradient exploding problems that make their training difficult, not allowing the processing of very long sequences. Moreover, due to its recurrent nature, the computation is slow. For this reason, some studies investigated the implementation of 1D-CNNs, with the main aim to design low computational complexity systems to support portable devices. In fact, Kiranyaz et al. [
52] revised the state of the art techniques used in signal processing applications such as patient-specific ECG classification, structural health monitoring, anomaly detection in power electronics circuitry, and motor-fault detection. In particular, they highlighted how the implementation of adaptive and compact 1D-CNN can achieve higher performance than deep conventional 2D with low computational complexity. Adaptive and compact 1D-CNNs can be efficiently trained with a limited data set of 1D signals instead of massive size data sets required by deep 2D CNNs. It can be performed directly to the raw signal without any pre or post processing, such as features extraction, selection, dimensionality reduction, etc., and it is able to extract features from shorter segments of the overall data set. Moreover, due to the low computational requirements, 1D-CNNs are well suited for real-time and low-cost applications, especially on smart mobile devices that can be the proper tools for personal health monitoring [
52,
53]. Yıldırım et al. [
54] proposed a new 1D-convolutional neural network approach for the automatic classification of cardiac arrhythmia on long-duration electrocardiography (ECG) signal analysis. The model was performed on 10-s fragments of ECG signals including 17 different classes of cardiac arrhythmia. The model showed remarkable performance and could be implemented with low computational complexity on mobile devices and cloud computing for tele-medicine, e.g., patient self-monitoring and preventive health. Li et al. [
55] proposed a 1D-CNN based model to classify ECG signals. The model consisted of five layers and realized the classification of five typical kinds of arrhythmia signals. It achieved promising classification accuracy and significantly outperformed several typical ECG classification methods. Zubair et al. [
56] propose an ECG beat classification system using a 1D-CNN model. The proposed classification system efficiently classified ECG beats into five different classes. Results showed that the model achieved a significant classification accuracy and superior computational efficiency than most of the state-of-the-art methods for ECG signal classification. Avanzato et al. [
57] proposed a new neural architecture based on 1D-CNN for the development of automatic heart disease diagnosis systems using ECG signals. The model was performed on 30 s segments and classified three different classes of anomalies. It showed high performance and low complexity implementation. Kamaleswaran et al. [
58] introduced a novel deep learning architecture for detection of normal sinus rhythm, AF, other abnormal rhythms, and noise. They proposed an optimal 13-layer 1D-CNN model with identified normal, AF and other rhythms using single lead short ECG recordings. The architecture was computationally fast and could also be used in real-time cardiac arrhythmia detection applications.
5. Results and Discussion
We performed the proposed 1D convolution neural network on the MIT-BIH Arrhythmia data processed as segments of 15 s consisting of 5400 samples including 21 different classes of anomalies.
In the first experiment, we split the dataset basing on segments as explained in
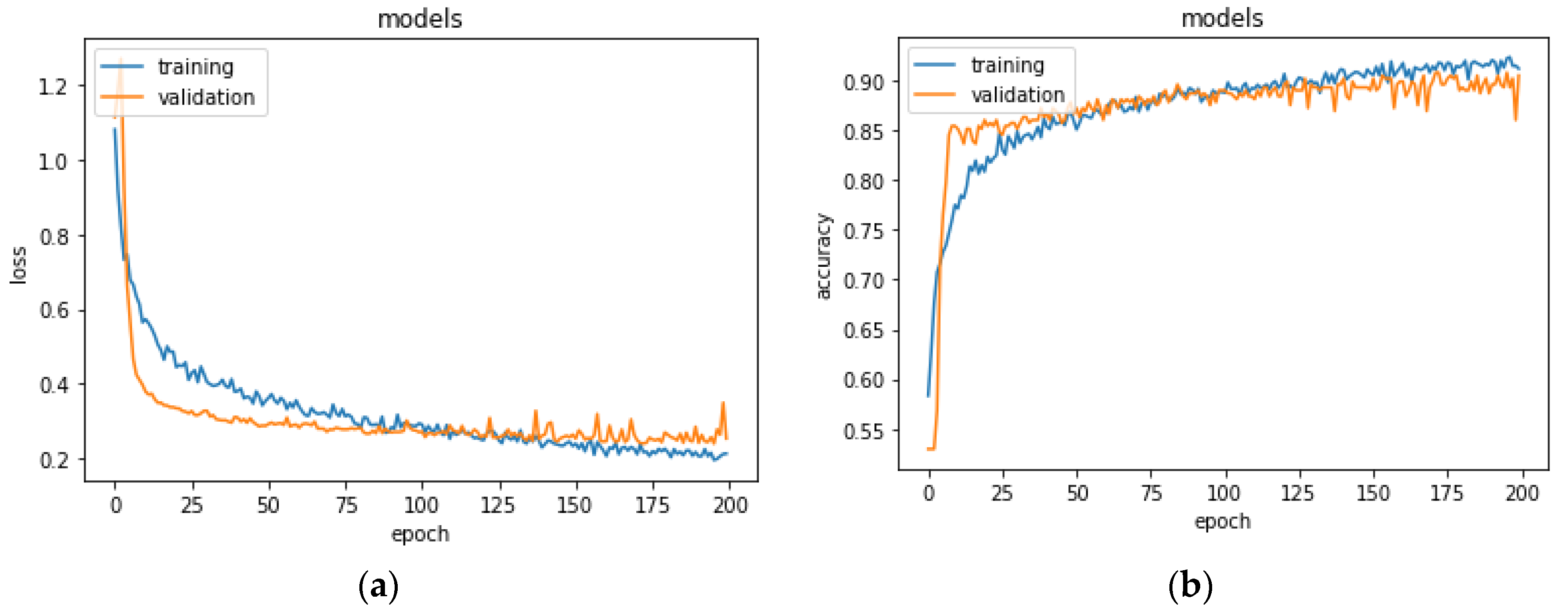
Section 4.2. The network was trained on 70% of the set and was validated on the remaining 30% for 200 epochs. Results for the validation performed on the training set are shown in
Figure 4. The network stabilized in convergence in a training process of 200 epochs. The learning curves of the training and validation loss stabilized below 0.5 and the learning curves of the training and validation accuracy stabilized around 90%, both with a minimal gap between the final values.
In order to validate the stability of the proposed method, we used a k-fold cross validation as explained in
Section 4.4. The average accuracy of the model was 89.3 ± 0.26% of standard deviation and the average loss was of 0.28 ± 0.06% and an average recall 85.6 ± 0.03%.
In order to assess the performance, the network was evaluated on the test set. The confusion matrix and the related metrics were computed (
Table 5). The network showed an accuracy of 89.51%, and a recall of 91.09% for normal and 87.79% for anomalous segments, the precision of 91.81% for normal and 86.78% for anomalous segments, and F1-score 91.45% for normal and 87.28% for anomalous segments.
We compared the proposed network with the studies that, at the state of the art, performed the same 1D convolutional neural network using the MIT-BIH Arrhythmia Dataset with the aim to implement an automatic classification of cardiac pathologies based on ECG signals. The results of this comparison are reported in
Table 6.
Briefly summarizing, Yıldırım et al. [
54] proposed a 1D-convolutional neural network model for cardiac arrhythmia (17 classes) detection based on long-duration electrocardiography (ECG) signal analysis. They designed a complete end-to-end structure with neither hand-crafted feature extraction of the signals nor feature selection at any stage using a 16-layer deep network structure including standard CNN layers. The network was tested on MIT-BIH Arrhythmia database considering 10 s ECG signal segments for one lead (MLII) for a total of 1000 ECG signal segments from 45 persons. The network achieved a detection accuracy of 17 cardiac arrhythmia disorders (classes) at a level of 91.33%. Li et al. [
55] proposed a 1D-convolutional neural network to classify ECG signals. The network consisted of five layers in addition to the input layer and the output layer, in particular, two convolution layers, two down sampling layers and one full connection layer. The model extracted the effective features from data and classified the features automatically. The wavelet threshold method was used to filter the high frequency noise, while the wavelet transform and reconstruction algorithm to correct the baseline drift, which is a low-frequency noise. Subsequently, the segmentation of ECG signals and the reduction of dimensions were performed by using R peaks that were located by the method of the wavelet transform. The network achieved a detection accuracy of five typical kinds of arrhythmia signals at a level of 97.5%. Zubair et al. [
56] proposed an ECG beat classification system based on a 1D-convolutional neural network. The model integrated feature extraction and classification of ECG pattern recognition and consisted of three convolution layers, three pooling layers, a multilayer perceptron and Softmax layer. The classification was performed in three main steps: ECG beat detection, sample extraction, and classification. ECG beat detection stage involves the detection of the individual beat signal from 30 min long ECG recording of each patient, using modified-Lead II signals. Equal numbers of samples (100) on both right and left sides from the Rpeak were extracted and downsampling was performed to represent raw data of each beat by 128 samples. The network classified five different kinds of anomalies with a detection accuracy of 92.7%. Avanzato et al. [
57] proposed an automatic heart disease diagnosis system using ECG signals based on the direct application of a 1D-convolutional neural network. The network consisted of three layers in addition to the input layer and the output layer. Unlike the classic CNN, which use fully connected neurons as their output layer, this network performed a single Average pooling layer and then a Softmax followed by a natural logarithm. The structure of the neural network input consisted of 30-s segments where every second of ECG recording was equivalent to 360 samples, for a total of 10,800 samples. The paper evaluated the performance of the network on three classes of anomalies with an accuracy of 98.33%.
Table 6 shows that the proposed method presents remarkable performance compared to the other studies, considering that it is able to detect anomalies in an ECG recording including 21 different classes. In addition, the proposed method is able to classify anomalies even in presence of more than one kind of cardiac pathology in the same ECG segment. It achieves recall and F1-score, respectively of 87.79% and 86.78%, improving results obtained in [
54], which considered only 17 classes of anomalies. These results were achieved with a detection accuracy of 89.51%, slightly lower than the accuracy obtained using the method proposed in [
54], despite the increase in the number of different kinds of anomalies. The high performance obtained for recall, in spite of a small loss for accuracy, is consistent with the research goal to reduce the number of ECGs sent to the medical specialist for further examination. At the same time, we need to ensure that anomalous ECGs could not escape the medical examination and the prescription of the proper therapy. Although results obtained in [
54,
55,
56] showed higher performance, in those studies the analysis was carried out including a lower number of anomalies. In particular, [
55,
56] included in the analysis five classes and obtained an accuracy, respectively, of 97.5% and 92.7%. In [
57] the analysis was conducted using three classes with an accuracy of 98.33%. Note that the model in [
54] was based on 16 levels architecture (therefore with a large computational complexity to deploy, as an example, on wearable devices) and the models in [
55,
56,
57], although based on four levels, operate only on a very small number of anomalies.
Analyzing the 52 segments containing anomalies and incorrectly classified as normal (false negative, see
Table 5), we observed that, for most of them, at least one of the segments extracted from the same ECG recording was correctly classified as anomalous. Overall, only five ECG recordings from patients affected by cardiac pathologies were not detected at all.
In light of this, we carried out a second experiment, including in the training set only segments extracted from the same ECG recordings, and for the test set, as explained in
Section 4.2. Thus, the proposed 1D-CNN was tested on segments extracted from ECG recordings never presented to the network in the training process.
The training set consisted of 3360 segments, 1594 normal, and 1766 anomalous, extracted from 28 ECG recordings, whereas the remaining recordings were included in the test set. In the test set, there were 1800 segments, 435 normal, and 1365 anomalous. To evaluate the performance of the model, the network was trained on 70% of the set and was validated on the remaining 30% for 200 epochs.
Figure 5 shows the learning process for training and validation loss and accuracy. The model stabilized in convergence in a training process of 200 epochs. Additionally, in this case, the learning curves of the training and validation loss stabilized around zero and the learning curves of the training and validation accuracy stabilized around 90%.
In order to validate the stability of the proposed method, we used a k-fold cross validation as explained in
Section 4.4. The average accuracy of the model was 88.2 ± 0.28% of standard deviation and the average loss was of 0.29 ± 0.07% and an average recall of 87.6 ± 0.03%.
The network was, then, evaluated on the test set. The resulting confusion matrix and the related metrics are shown in
Table 7.
The network showed an accuracy of 84.94%, and a recall of 55.40% for normal and 94.36% for anomalous segments, the precision of 75.79% for normal and 86.91% for anomalous segments, and F1-score 64.01% for normal and 90.48% for anomalous segments.
The model showed a higher recall for anomalous segments compared to the previous test. Analyzing the 77 segments containing anomalies and incorrectly classified as normal, we observed that, for all of them, at least one of the segments extracted from the same ECG recording was correctly classified as anomalous. This result showed a remarkable improvement in the performance of the proposed method since, thinking in terms of patients rather than segments, the system was able to detect 100% of ECG recordings from patients affected by cardiac pathologies.
Considering this performance, we believe that the unavoidable goal to detect anomalous segments and minimize missed alarms was reached. This claim must be considered in the operative context where the model was deployed, in which the cost of a false alarm was considerably less than a missed one.
Moreover, we want to highlight that these results were achieved with the second experiment whose settings represents a closer scenario in terms of model usage. In the second experiment the model was validated on ECG records of different patients never presented to the model during the training. This experiment design is not common since validation is usually carried out with the usual holdout method, that is, segments from the same ECG recordings could be interleaved between training and test sets. With holdout there are chances that some signal patterns have been already presented to the model in the training phase.
However, it should be noted that the system described here still presents a significant false alarm rate. We are currently investigating different strategies to reduce the required workload due to false alarms. One viable solution is to isolate and collect only anomalous detected segments; when they occur in significant quantities can be sent remotely for the expert verification. Looking only at some of the segments, just those classified as anomalous by our model, can be a very quick job for an expert, certainly less demanding than observing an entire ECG, even more so if referred to a Holter exam.
This operational solution is well suited also for continual and active learning techniques, that is to periodically retrain the model based on new annotations from an expert; in this way, as new examples come into the system a probable gradual decrease in false alarms events is expected, with more balanced performances.
6. Conclusions
The diffusion of personal portable monitoring devices could involve the reporting of millions of ECG recordings every day. Systems able to support the cardiologists’ work in the interpretation of ECGs for the diagnosis of cardiac pathologies are required to facilitate health care decision making reducing considerably the expenditure of time and money. In fact, the automated detection of suspicious anomalies in ECG recordings can drastically reduce the number of ECGs that need to be manually examined by the cardiologists, excluding those classified as normal.
In the present paper, we propose a system able to automatically detect the suspect of cardiac pathologies in ECG signals from personal monitoring devices, using a 1D-CNN architecture. The 1D-CNN model overcomes the problems of vanishing gradient and gradient exploding related to recurrent neural networks, making their training difficult. Moreover, the 1D-CNN model allows one to implement real-time and low-cost systems and it is characterized by low computational complexity, feasible implementation on smart devices, and cloud computing. The system was optimized to detect the suspect of anomalies classifying normal and anomalous ECG, regardless of the kind of cardiac pathology. The proposed model was tested on the MIT-BIH ECG Arrhythmia Database, which included 21 different classes of ECG anomalies. Two different experimentations were carried out showing remarkable performance compared to the other studies conducted using the 1D-CNN architecture tested on the MIT-BIH ECG Arrhythmia Database. In particular, the network achieved accuracy and recall, respectively, of 84.94% and 94.36% computed with respect to the ECG signal segments and accuracy and recall of 100% when computed with respect to the patients, therefore considering the detection of anomalies in the entire ECG recordings.
We are now working on a possible personalization of the model, tunable toward a single person. We expect that the performance of this kind of model could be much better than a general one, with an additional cost of model calibration before the actual usage.
In the same study we are also investigating a model trained on “normal” segments of healthy patients and abnormal segments of pathological patients (since it is not possible to obtain abnormal segments from a healthy patient). This will allow us to observe if the model trained on this new dataset presents different characteristics than the model described in this paper. Here, we wanted to compare our study with studies that were as homogeneous as possible, at least in the dataset used, so we did not introduce any further changes regarding the training data.