1. Introduction
In this paper, we compare four different methods, feature engineering, standard convolutional neural networks (networks consisting only of convolutional, pooling and fully connected layers, hereafter referred to as CNN), residual convolutional neural networks (networks consisting only of convolutional, pooling, identity residual connections and fully connected layers, hereafter referred to as ResNet) and deconvolutional neural networks (CNN as an image encoder with a decoder consisting of deconvolutional and max-unpooling layers, hereafter referred to as DCNN), for estimating full spatial resolution muscle fibre orientation in multiple deep muscle layers, directly from B-mode ultrasound images. We provide proof of principal that deep learning methods are a robust, generalized and feasible method for arbitrary full spatial resolution regression analysis, given ground truth labels, within the domain of skeletal muscle ultrasound analysis. Relevant applications of this method include predicting strain maps, motion maps, twitch localisation or muscle/feature segmentation. In recent years, ultrasound has become a valuable and ubiquitous clinical and research tool for understanding the changes which take place within muscle in ageing, disease, atrophy and exercise. Ultrasound has been proposed [
1,
2] as a non-invasive alternative to intramuscular electromyography (iEMG) for measuring twitch frequency, useful for early the diagnosis of motor neuron disease (MND). Ultrasound has also recently demonstrated application to rehabilitative biofeedback [
3]. Other computational techniques have been developed for muscle-ultrasound analysis which would allow estimation of changes in muscle length during contraction [
4] and changes in fibre orientation and length [
5,
6,
7,
8,
9]. Regional muscle fibre orientation and length change when muscle is under active (generated internally through contraction) and/or passive (generated externally through joint movements or external pressure) strain [
10,
11]. Muscle fibre orientation is one of the main identifying features of muscle state [
12].
Previous attempts to automatically estimate fibre orientation directly from B-mode ultrasound images are inadequate, typically requiring manual region identification, a priori feature engineering, and/or typically providing low resolution estimates (i.e., one angle for an entire muscle region). All previous methods depend on some parameterization of features such as Gabor wavelets [
5,
6], and/or edge detectors and vessel enhancement filters [
5,
6,
7,
8,
9,
13,
14,
15]. In all cases, parameters are empirically chosen and/or are based on assumptions about how the descriptive features will present within an image. In the studies we are aware of, parameters are chosen presumptuously or empirically rather than tuned with a multiple-fold cross-validation set. Since the authors unanimously write that results are sensitive to parameter changes, the assumption is that real world performance may not be as optimistic as reported. Further, to our knowledge, nobody has attempted the deep (more challenging) muscles.
Zhou and colleagues [
16] developed a method based on the Revoting Hough Transform (RVHT), which provides an estimate of the overall fibre orientation in a single muscle, the gastrocnemius in the calf. Based upon the incorrect assumption that the fibre paths are straight lines, they use the RVHT to detect an empirically predetermined number of lines within a manually defined region of interest and then take the median orientation of all detected lines as the overall orientation of the fibres. The approach of Zhou and colleagues is fundamentally limited to detecting straight lines, whereas muscle fibres do not always present that way. When observing muscle fibres using ultrasound, there are many other features which can appear as straight lines (blood vessels, noise/dropout, artefacts/reflections, skin/fat layers, connective tissues, etc.). These fundamental facts are a strong limiting factor to the potential of methods such as the RVHT and the Radon Transform [
8].
Rana and colleagues [
6] introduced a method which potentially allowed estimation of local orientation, although after suggesting this, the mean of local orientations is used to produce another method which provides an estimate of the overall fibre orientation. Local orientations were identified by convolving a bank of Gabor Wavelet filters with a processed version of the image. They compare their proposed method with manual annotations from 10 experts and the Radon Transform, which like the Hough Transform (or RVHT) is a method which can be used for detecting straight lines in images. Both the Radon Transform and Wavelet methods require a pre-processing step described as vessel enhancement filtering, which enhances anisotropic features within the image. The vessel enhancement method is parametric, where parameters were chosen empirically with no cross-validation or evaluation of results over varying parameters. Their results showed that the Radon Transform was not significantly different to the expert annotations, whereas the Wavelet method was significantly different. Other than significance values, the only accessible results they report are mean differences (1.41°) between the Wavelet method and the expert annotations of real images and mean differences between both the Radon and Wavelet method applied to synthetic images with known orientations (<0.06°). Although the Wavelet method performed comparatively poorly, the authors rightly suggest that this approach has potential to allow tracking of fibre paths throughout the muscle, whereas Radon and Hough Transform methods do not.
Namburete and colleagues [
7] expanded upon the methods of Rana and colleagues [
6], developing the hypothesis that regional fibre orientation can be tracked using local orientated Gabor Wavelets and vessel enhancement filters, for the first time going beyond linear approximations to the overall orientation. The proposed extension is to convolve the local orientations with a median filter, which effectively smooths the local orientations. The fibre trajectories are then tracked between the muscle boundaries on a continuous coordinate system with linear (following the dominant orientation in a local 15 × 15-pixel region centred on the current fibre track) steps of 15 pixels. Finally, curvature is quantified using the Frenet-Serret curvature formula [
17]. Namburete’s method differs significantly from all preceding methods because it provides an estimation of local fibre orientation, rather than a more global estimate. However, the authors do not provide an estimate of the error for example when comparing to expert annotations; instead they evaluate errors on synthetic images with known orientations, which are trivial. Further, they do not apply this method to the deeper muscles, which are even more challenging.
Several important problems have been identified as unaddressed from a review of previous methods for estimating full regional fibre orientation in multiple layers of muscle; most commonly, the lack of error evaluation on real data, estimating overall fibre orientation and not local orientation, limited application to superficial muscles only and presumptuous justification for chosen parameters. We propose to address these problems by introducing advanced machine learning methods and cross-validation against expert annotation of real data. In recent years the popularity of machine learning (in particular neural networks) has surged since a number of successive algorithmic, methodological and computational hardware developments were introduced [
18,
19,
20,
21,
22]. The application of machine learning to estimating local fibre orientations has thus far not been considered. We extend our preliminary investigations into deep learning [
23] by introducing and comparing deconvolution networks (DCNN) as a way of minimizing parameters in the final regression layer for learning full spatial orientation maps, without the need for segmentation such as [
24]. In recent years, the development of deconvolutional neural networks (DCNN) and variants of DCNN (FCN, U-Net) allows robust regression (heatmaps [
25,
26,
27]) or classification (semantic segmentation [
28,
29]) of pixel level labels in full resolution images, without any feature engineering or pre-processing (i.e., all parameters are learned from data and cross-validated against held-out validation and test sets). It is common knowledge that standard CNN architectures lose precise position information through the max-pooling operation layers. We propose the DCNN architecture as proposed in [
28,
30], which uses max-unpooling layers to recover the spatial information lost during max-pooling.
The next section describes training of three neural network architectures: CNN as a baseline deep learning technique, ResNet to add complexity and allow possibility of position encoding and DCNN to explicitly recover precise position information to reconstruct the fibre orientation maps. For reference to existing literature, we implement a version of the wavelet method described by Namburete and colleagues [
7], since no data are presented on the accuracy of their method on real ultrasound images. All networks were trained with leave one out cross-validation, with and without dropout (50%). For the wavelet method, we tuned some of the parameters to optimize results for each specific muscle segment. We note here that an alternative approach to the methods of Namburete would be to apply wavelets in the frequency domain, but it is not within the scope of this paper to develop that method.
2. Materials and Methods
2.1. Deep Learning Software
All neural network software was developed from scratch by the authors using C/C++ and CUDA-C (Nvidia Corporation, Santa Clara, CA, USA). No libraries other than the standard CUDA libraries (runtime version 8.0 cuda.h, cuda_runtime.h, curand.h, curand_kernel.h, cuda_occupancy.h and device_functions.h) and the C++ 11 standard library were used. All 48 neural networks were trained on an Intel Xeon CPU E5-2697 v3 (2.60 GHz), 64GB (2133 MHz), Nvidia with a single GTX 1080 Ti GPU.
2.2. Data Acquisition
These experiments were approved by the Research Ethics Committee of the Faculty of Science and Engineering, Manchester Metropolitan University (MMU). Participants gave (written) informed consent to these experiments, which conformed to the standards set by the latest revision of the Declaration of Helsinki. Experiments were performed at the Cognitive Motor Function laboratory, in the School of Healthcare Science, Healthcare Science Research Institute, MMU, Manchester, UK.
Ultrasound image data were recorded at 25 Hz (AlokaSSD-5000 PHD, 7.5 MHz) from the calf muscles (medial gastrocnemius and soleus) of 8 healthy volunteers (4 male, ages: 25–36, median 30) during dynamic maximum isometric voluntary contractions. Image settings were constant across participants. Images represented a 6 × 5 cm (horizontal × vertical axes) cross-section of the calf muscles. Volunteers lay prone on a physio bed with their right ankle strapped to an immobilised pedal. Volunteers were asked to push their toes down against the pedal as hard as they could. A dynamometer (Cybex) recorded the torque at 100 Hz at the ankle during the contraction. Matlab (Matlab, R2013a, The MathWorks Inc., Natick, MA, USA) was used to acquire ultrasound frames and a hardware trigger was used to initiate recording at the start of each trial.
2.3. Generating Ground Truth
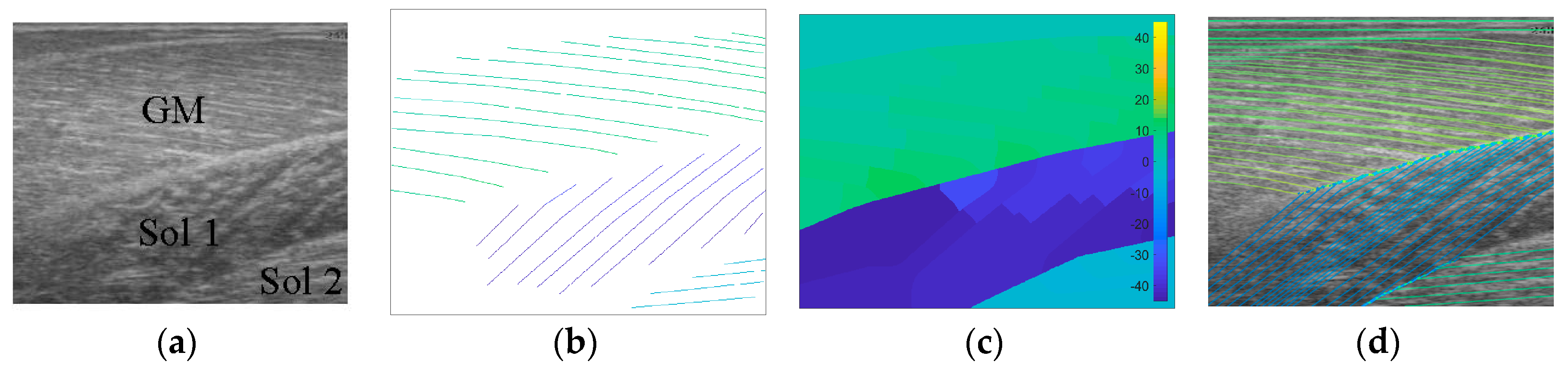
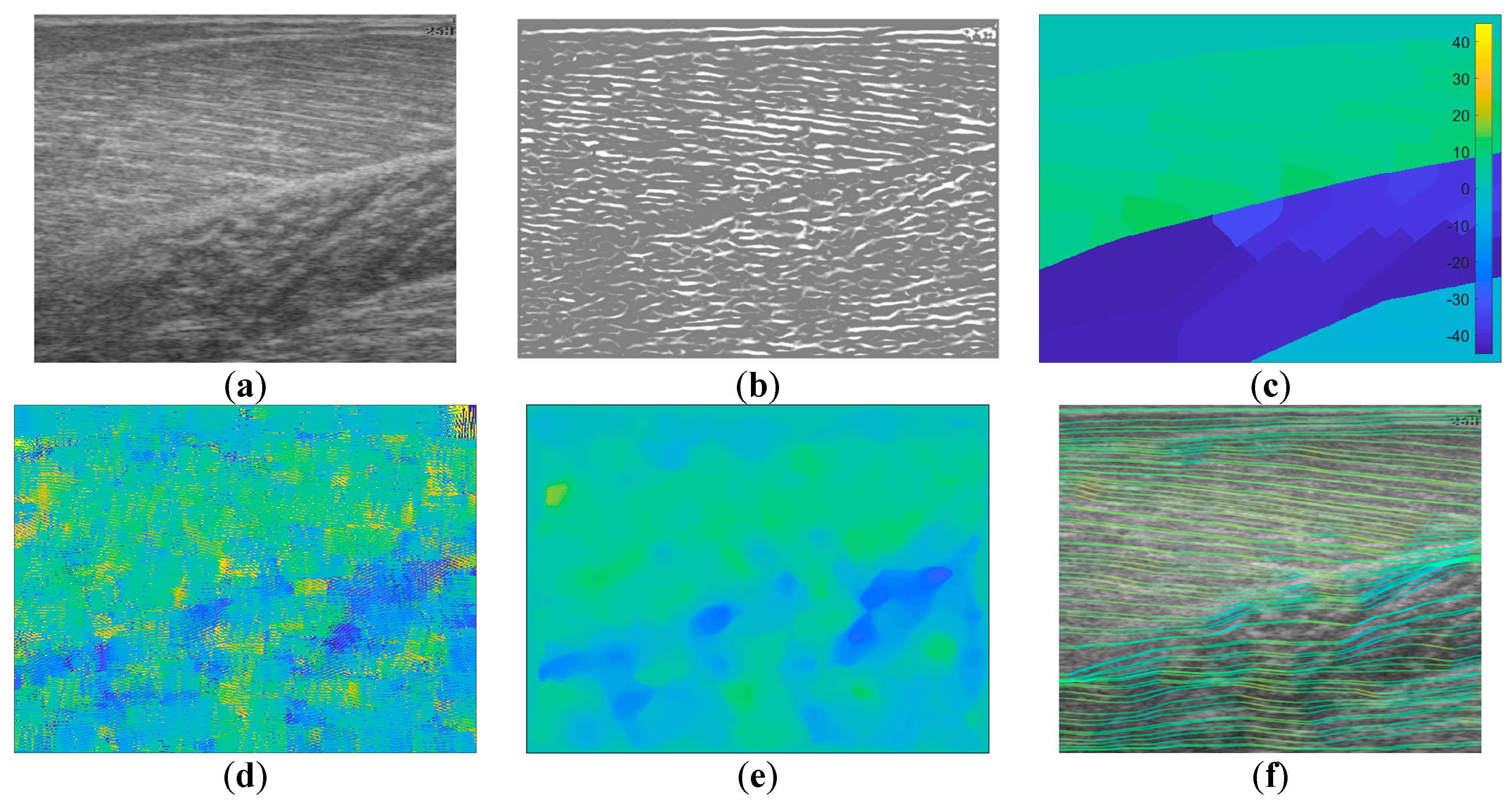
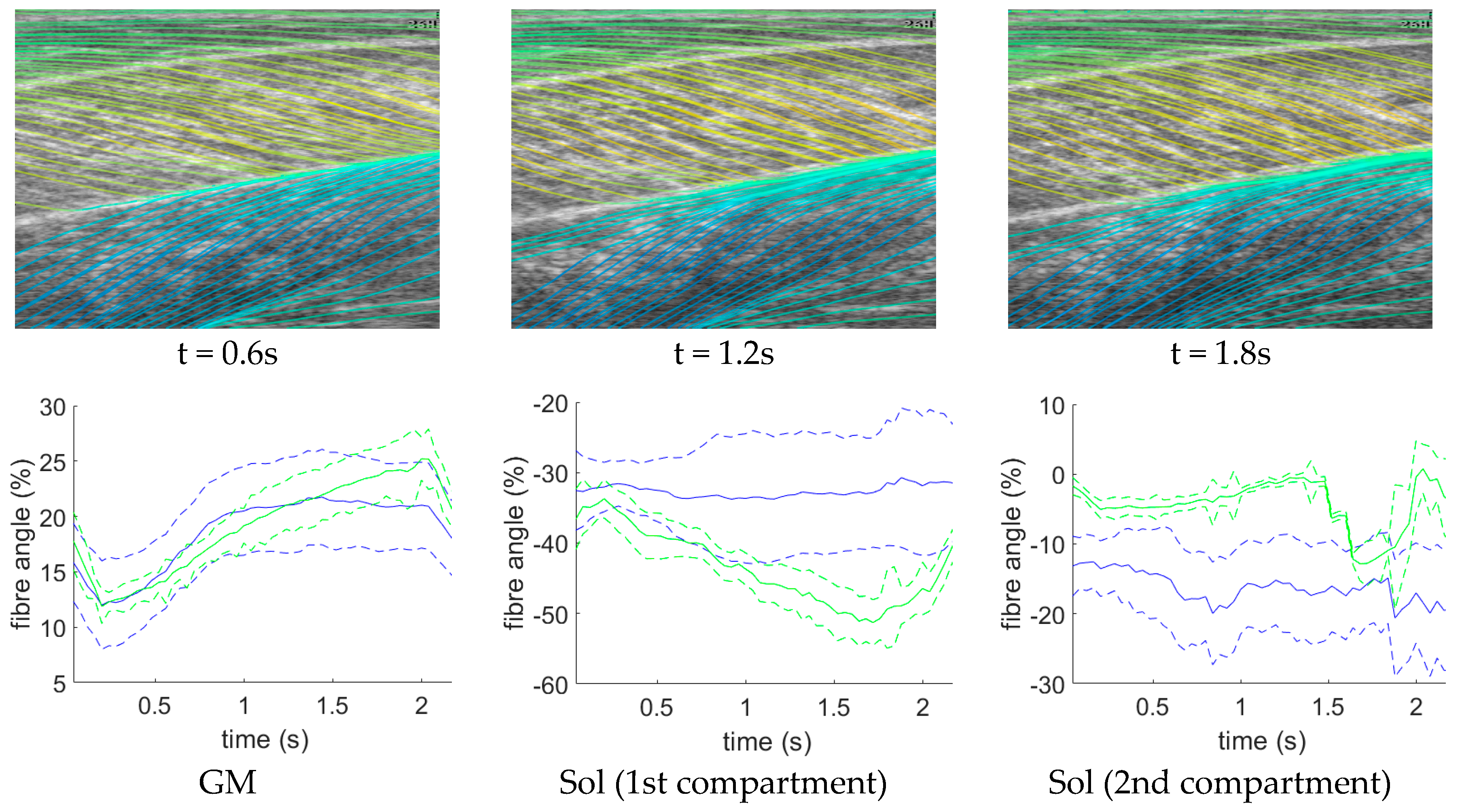
Following data collection, we extracted all frames beginning one frame before contraction started and ending with the frame when contraction peaked, where initial and peak contraction were identified manually. This resulted in a total of 504 images containing spatiotemporal variations in both fibre orientation and muscle thickness. Two experts were asked to manually identify fibre paths in all muscles/compartments in all images by marking polylines (connected straight line segments—typically 20–30 per muscle/compartment). The same experts were asked to identify the boundaries of the medial gastrocnemius (superficial muscle) and the boundaries and internal compartments (where visible) of the Soleus (deep muscle) by marking polylines along the boundaries and visible intramuscular compartments.
To create labels for each pixel, first a blank image (matrix of zeros with equal dimensions to the ultrasound image) was created. Then, the angles along each of the annotated fibre lines were calculated and for each pixel under the lines the calculated angle was stored. The result was an image of mostly zeroes and a few angled lines, where the value of the non-zero pixels represents the local angle of the line intersecting each pixel. Then, within each muscle/compartment (defined by the experts), nearest neighbour interpolation was used to fill the gaps between lines, followed by nearest neighbour extrapolation to fill the muscle/compartment. Between muscles/compartments, nearest neighbour interpolation was also used to fill the gaps. All other pixels (outside muscle, e.g., skin) were set to zero (see
Figure 1). Following, additional data were created to introduce variation in the data/labels and help prevent over-fitting, by randomly (uniform) rotating each image and corresponding labels/angles between −5° and 5°, thus doubling the size dataset to 1008.
2.4. Wavelet Filter Bank Method
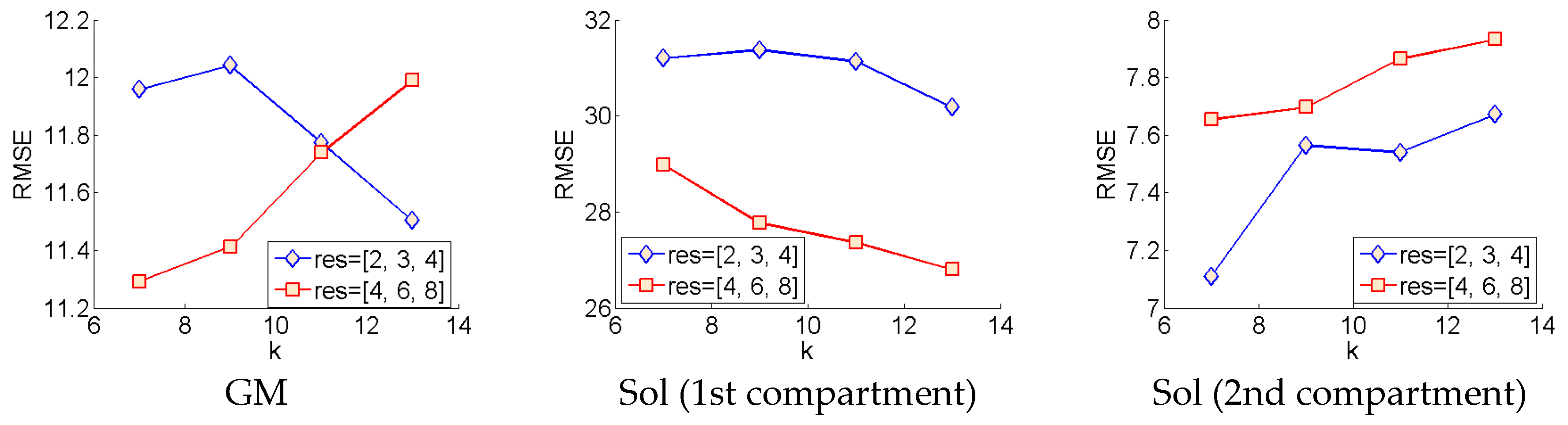
The wavelet method was implemented faithful to the original paper [
7]. A bank of 180 Gabor wavelet filters was generated spanning 0°–179° in 1° increments using,
where
k is the half-width of the kernel size (
k = 20 pixels = 2.2717 mm),
d is a damping term (
d = 51.243),
α is the orientation and
f is the spatial frequency (
f = 7). Initially, all parameters were set as originally described in [
7] and after a first analysis, some parameters (
f and
k) were varied (
f = 9,
f = 11,
f = 13,
k =
f × 3 − 1) to give some scope to the potential of this method on real data. Vessel enhancement was performed using a Matlab version of the Frangi multiscale vessel enhancement filter [
31] which can be found here [
32]. Following [
7], filtering was performed at 3 sigma scales parameter in the code), 2, 3 and 4. Following a first analysis, a second analysis was done at sigma scales, 4, 6 and 8, for all variants of the Gabor wavelet filters described above.
To analyse an image, the image was filtered with the vessel enhancement filter and then convolved with the entire wavelet filter bank. At each spatial location in the image, we take the
α corresponding to the filter with the highest convolution in that location. The result is a map of local orientations (Figure 4). Following [
7], the image was then convolved with a 35 × 35 median filter resulting in the final map of local fibre orientations.
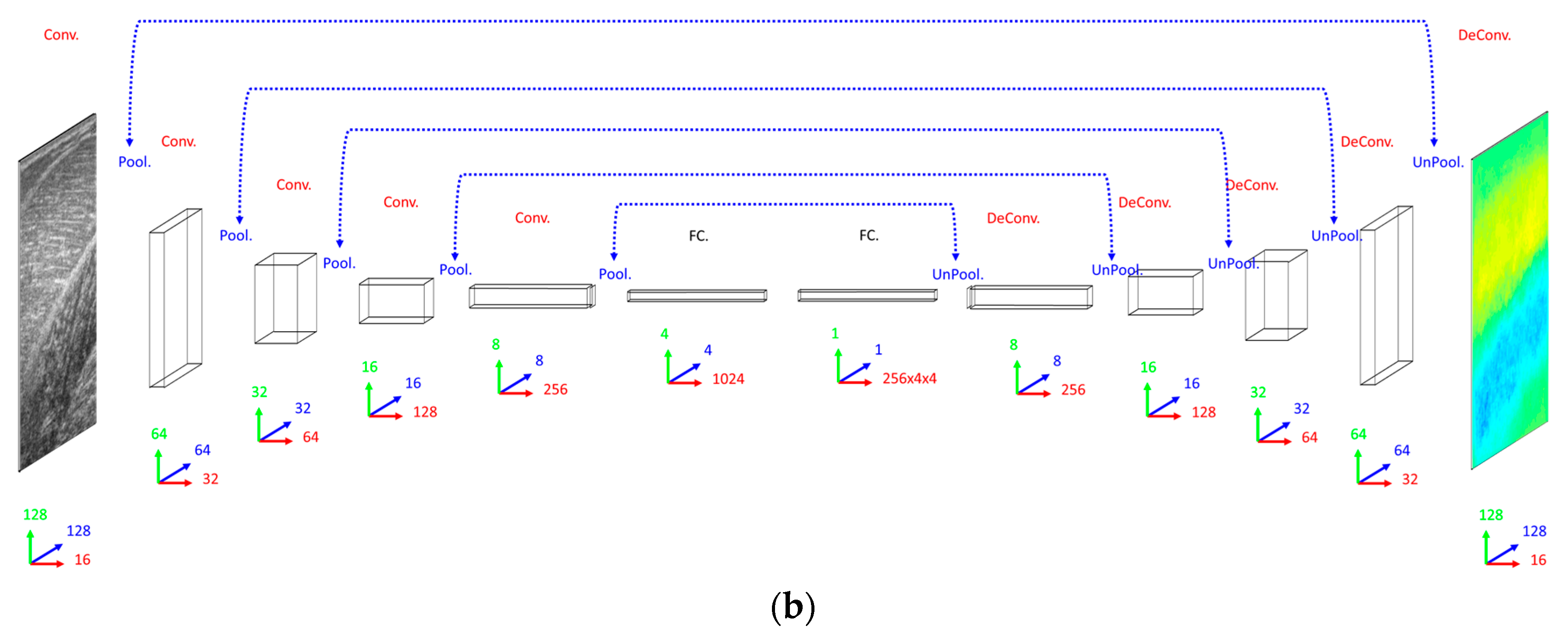
2.5. Deconvolution Neural Network Method
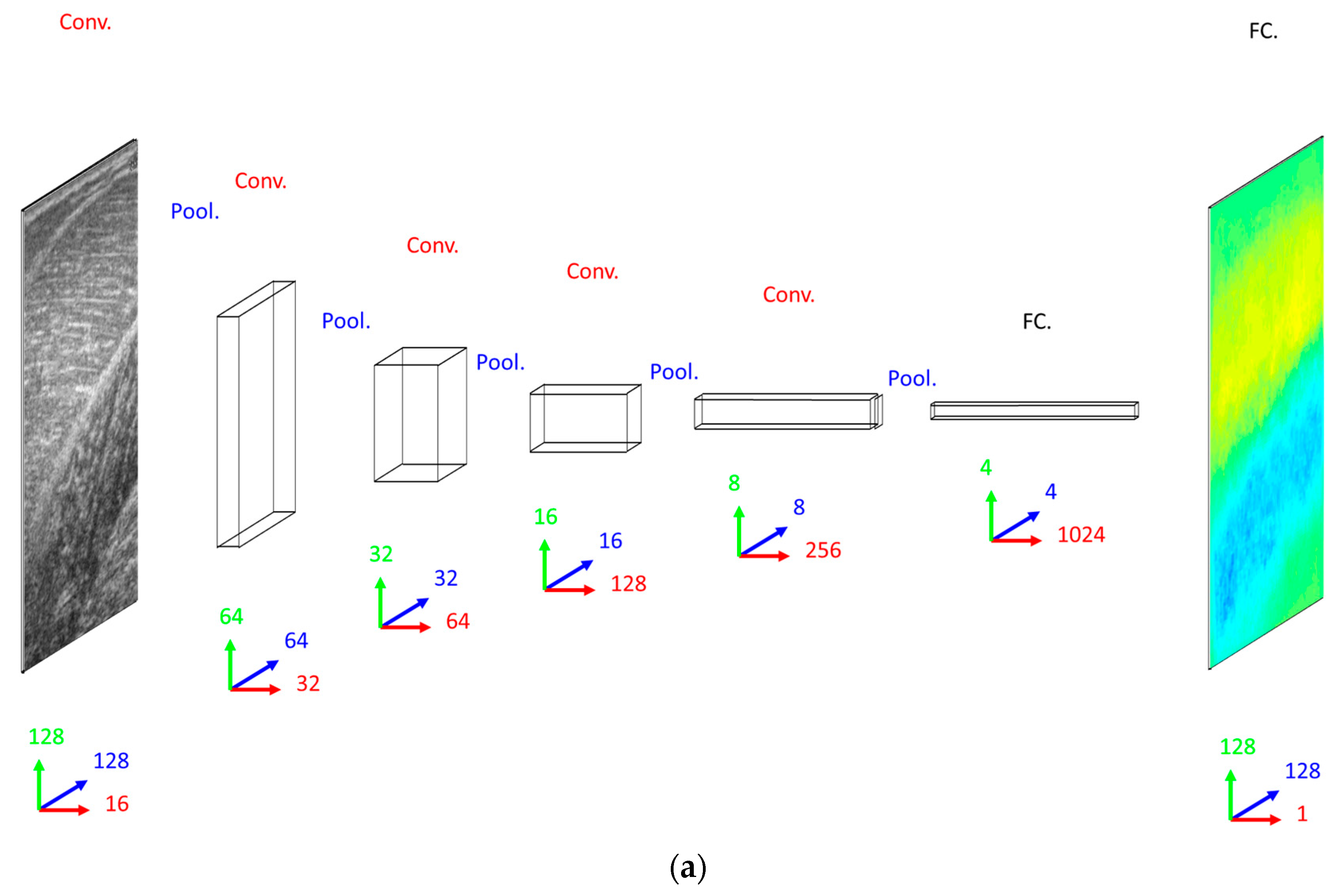
Our primary concern when deciding on a DCNN architecture (number of layer and units per layer) was having a complex enough model to learn the training set. The DCNN implemented 16 convolutional filters in the input layer, with 4 additional convolutional layers, each with double the number of filters in the preceding convolutional layer. Our secondary concern was training time, since we planned to execute an 8-fold cross-validation. Therefore, both input images and labels were down-sampled (bilinear interpolation) to 128 × 128 pixels. Between convolutional layers, the spatial dimensions were down-sampled (max-pooling) by a factor of 2. A dense fully connected layer of 1024 nodes was fully connected to an initial up-sampling (2 × 2 max-un-pooling) layer, followed by a deconvolution layer. An additional 4 up-sampling with deconvolutional layers following, completes the network architecture, with the final layer being a full 128 × 128 resolution regression map. Every layer consisted of rectified linear units (ReLU), with the exception of the final (output) layer, which consisted of linear units (see
Figure 2).
2.6. Convolutional and Residual Neural Network Methods
When deciding on a CNN/ResNet architecture we opted for the same convolutional architecture as the DCNN, with the exception that the ResNet had a factor of 5 additional convolutional layers, ensuring that our primary concern was met by performing empirical tests on small fractions of training data. Following our previous work [
23], for the ResNet, we multiplied the number of convolutional layers in between max-pooling layers by 5, where every other layer we added identity residual connections. Every other aspect of the CNN and ResNet architecture remained the same as the DCNN with the obvious exception of the output layer, which was a dense fully connected layer of linear regression nodes immediately following the 1024 fully connected nodes, rather than deconvolution and up-sampling layers (see
Figure 2).
2.7. Training and Cross-Validation
All networks were trained by minimizing mean square error (MSE) between the labels and the output layer using online stochastic gradient descent (batch size of 1 sample). To optimize and test generalization (estimated real-world performance), leave one person out cross-validation was performed on 2 DCNNs, 2 ResNets and 2 CNNs with different dropout parameter settings (0%, 50%), where dropout was enabled in the fully connected layers only. The weights of the neural networks were initialized using Xavier initialization [
33]. To train the CNN, ResNet and DCNN we used learning rates of 5 × 10
−6, 2.5 × 10
−6 and 2.5 × 10
−2, respectively. Weight gradient momentum of 9.5 × 10
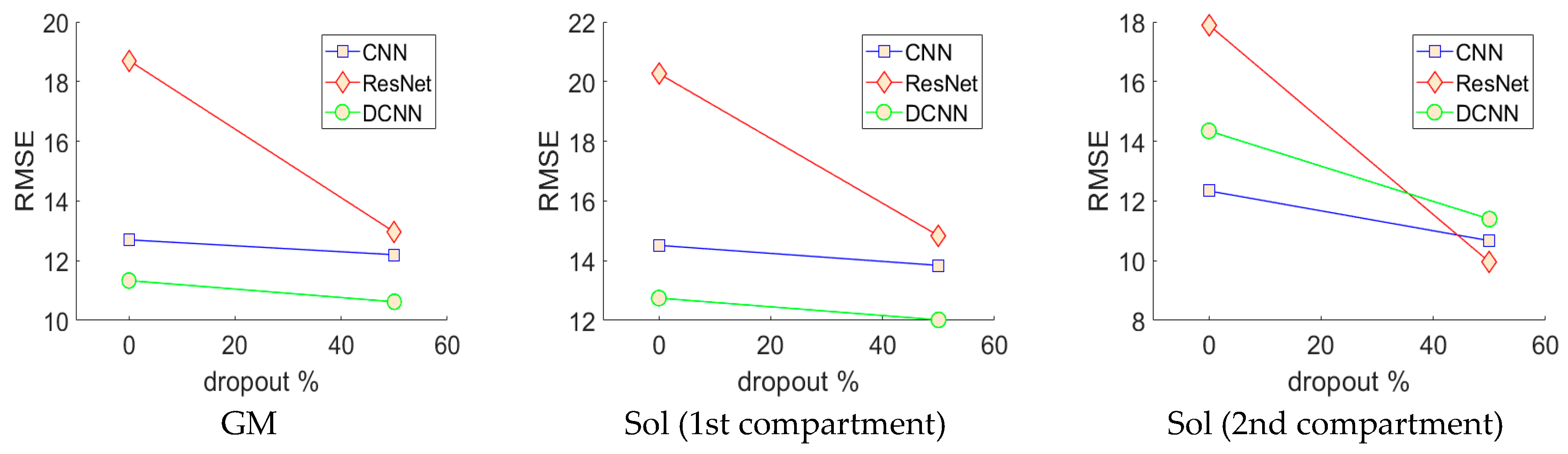
−1 was applied to all networks, which were trained for 100 full batch iterations using stochastic online gradient descent (minibatch = 1). All learning parameters were chosen empirically on a small fraction of the training data. The neural networks were trained 8 distinct times (one for each person), each time reinitializing the networks and leaving a different person out for testing and randomly splitting the remaining data (7 people) into validation and training sets (50% split). Prior to training, the pixels of the input images were normalized over the training (otherwise they were not processed in any way before analysis) set to zero mean and unit variance and pixels of the output images were divided by a constant (45) so that most of the pixels (angles) fell between −1 and +1. Validation and test errors were observed periodically and the test error observed at the lowest validation error (early stopping method) gave results for the validation set and the test set (estimated real-world generalization error). Test results for all held-out participants were combined to reveal the performance of the optimal CNN, ResNet and DCNN.
4. Discussion
In this paper, we presented an investigation into the feasibility of using deep learning methods for developing arbitrary full spatial resolution regression analysis of B-mode ultrasound images of human skeletal muscle. Four different methods were compared, feature engineering, convolutional neural networks (CNN), residual convolutional neural networks (ResNet) and deconvolutional neural networks (DCNN), for estimating full spatial resolution muscle fibre orientation in multiple deep muscle layers, directly from B-mode ultrasound images. The efficacy of this work has shown that the deep learning methods are feasible and particular deep learning methods, DCNNs, work best due to the spatial organization of the output regression map, which intelligently reduces the number of free parameters, preventing over-fitting. The previous established approach, wavelets, lacked accuracy and is limited in application, due to an inherent inability to ignore spurious fibre structures and the requirement for accurate segmentation to apply different wavelet frequencies within different regions.
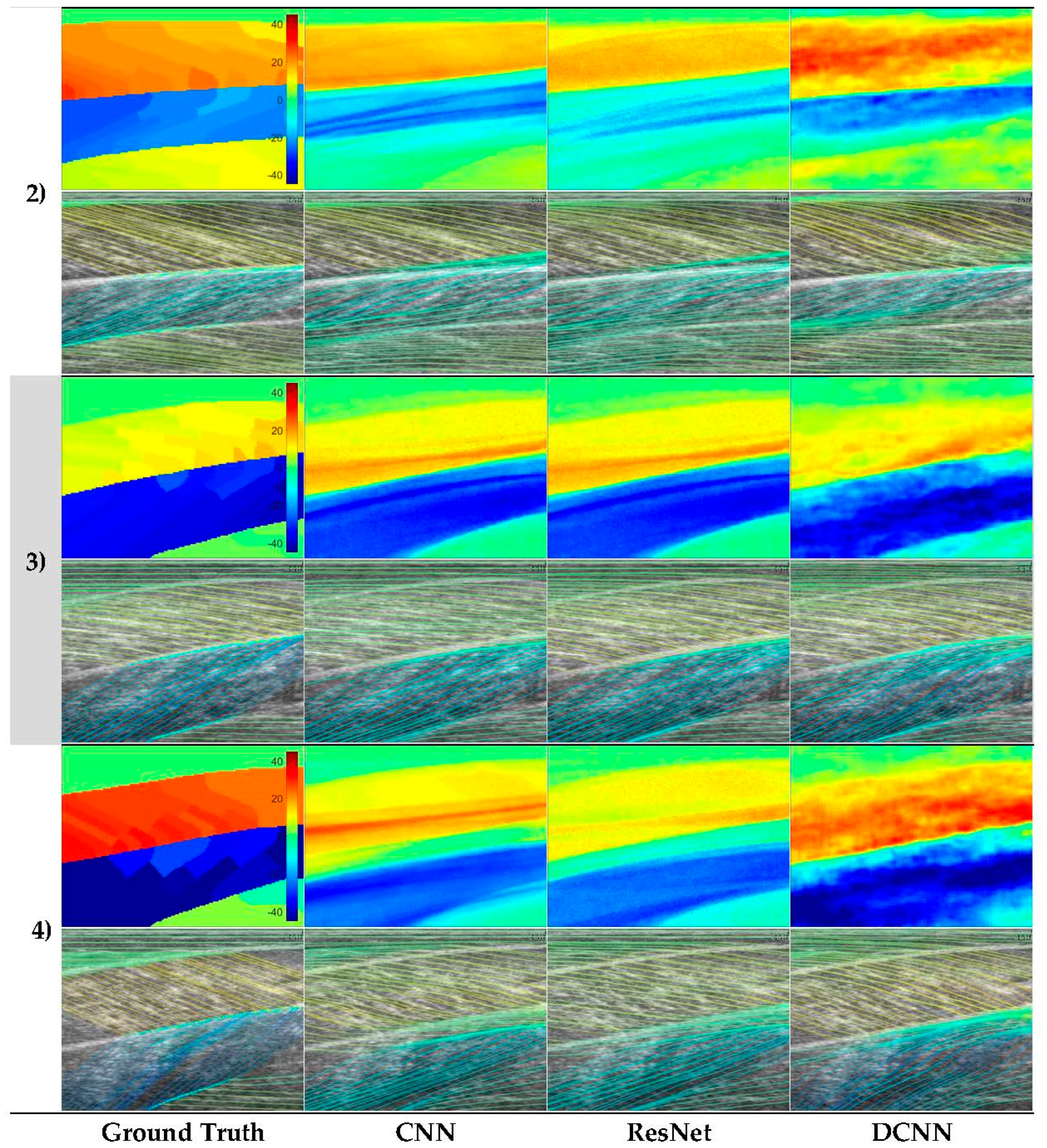
Analysis of each deep learning method revealed the strength of the approach and moreover, the strength of the DCNN approach. Both CNN and ResNets outperformed the wavelet approach (see
Table 1) in GM and Sol 1 muscles. However, the DCNN outperformed the CNN and ResNet markedly. Our interpretation is that the CNN/ResNet methods learned templates of the fibre orientation maps based on muscle dimensions in the training set, resulting in comparatively (to DCNN) poor localization of the muscle regions. Artefacts of this can be seen in
Figure 6, where the heatmaps of CNN/ResNet present with horizontal striping patterns, which are very likely different muscle boundary templates which have been combined to make the new orientation map. The DCNN showed the remarkable ability localize muscle regions and roughly match the colour intensity of the heatmap, which resulted in accurate and visibly pleasing fibre traces (see
Figure 6 and
Figure 7).
Figure 6 shows examples of CNN/ResNet methods performing better than DCNN in specific regions (e.g., rows 3–4 in the Sol 1 region), which can also be explained with the templates argument; i.e., where the test image did not match the templates well, performance was weaker and where the test image matched the templates well, performance was stronger.
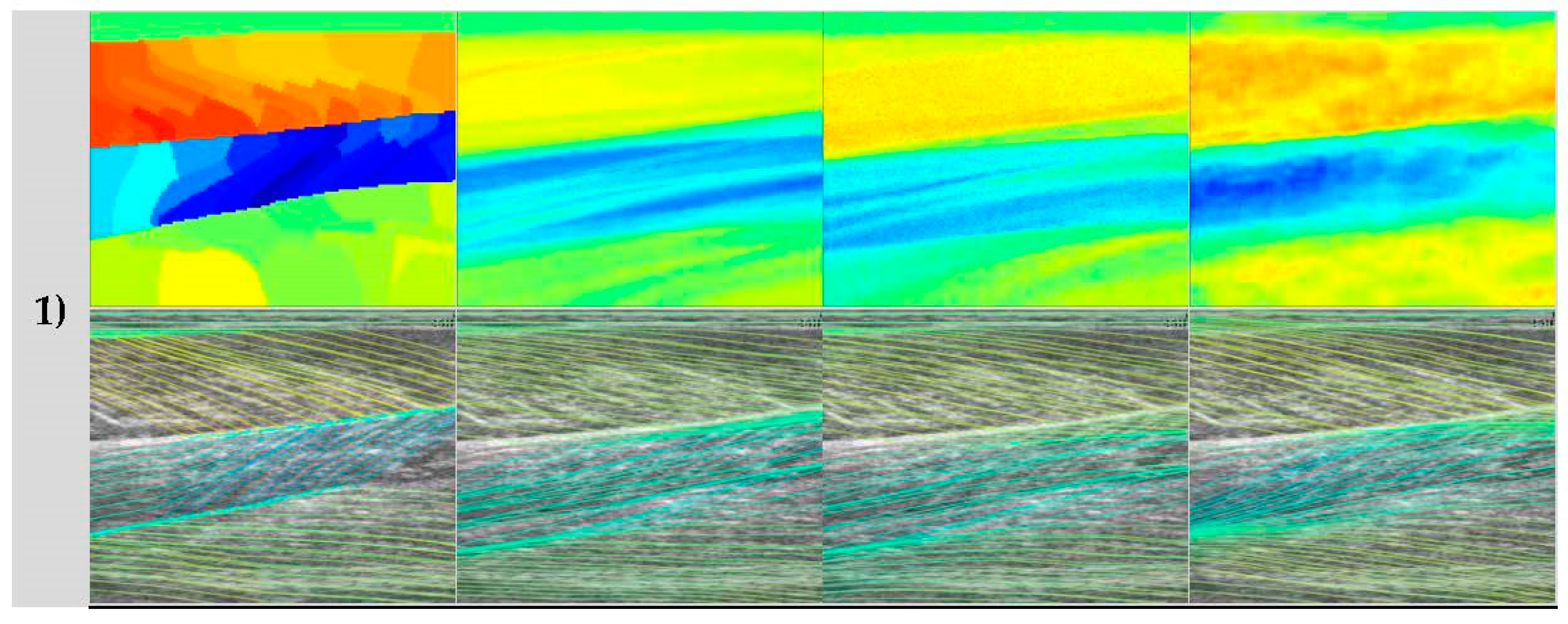
In contrast to the deep learning methods,
Figure 1 shows a representative output from the wavelet method, which appeared to perform well on the GM muscle and not on the other 2 muscles/compartments. In the Sol 1 muscle, where fibres are visible, some the wavelet analysis produces a very localized response which is roughly correct, but unfortunately this type of performance was common over the data set, resulting in very poor results in
Table 1. However, the wavelet method did show comparable performance to the DCNN in the GM muscle, which is certainly because of the texturally well-defined fibre structures in this region. Thee deep muscles in this muscle group often present differently, due to differences in architecture and other arguably more minor reasons like attenuation of the ultrasound beams, reflections, image artefacts and signal dropout. For this reason, results of all four methods are adversely affected in the deeper muscles. In the deepest compartment, defying the logical argument we just presented, the wavelet method produced notably the best performance. It is worth noting that the angles in that muscle region, annotated by the expert, were very commonly 0°, or some positive or negative number close to 0°. The wavelet method used local neighbourhood (35 × 35) median averaging of angles in a range of (–90:90). Therefore, our interpretation is that if we take the median of a set of random positive and negative angles, we would “guess” fibre orientations, close to the ground truth. Although, this could be a genuine result, the other issues with the wavelet approach are more devastating, even when compared to the CNN/ResNet approaches and particularly when compared to the DCNN.
The results presented in
Table 1 for the deep learning methods represent a truly generalized and thoroughly tested expectation of real-world performance. Test results were acquired by leave one participant out testing for all 8 participants (8-fold cross-validation; see
Figure 5), which is the harshest possible test of performance and generalization for this approach, requiring intensive training of 48 neural networks, which took less than a week on the latest GPU compute technology (GTX 1080 Ti). Depending upon the problem, handcrafting features such as Gabor wavelets can be much more time consuming and must be tailored to solve specific problems. What is more, the results we present in
Table 1 on the wavelet method, represent the optimal performance given accurate segmentation, since different wavelet frequencies and image filters were applied to different regions of the image—this would obviously not be possible without accurate and presumably automatic segmentation.
The implications of the efficacy of DCNNs for regression analysis with respect to skeletal muscle ultrasound, is in the application to other, high-impact research problems. The ability to produce a full resolution map of tissue strain, for example, would very likely and very positively impact the way musculoskeletal healthcare was delivered. Currently, elastography is the only non-invasive method for estimating strain within specific skeletal muscles [
35] and it has serious theoretical flaws; elastography currently has a maximum temporal resolution of 1 Hz and due to the anisotropy of skeletal muscle, the theory is only empirically validated for skeletal muscle when the ultrasound probe is parallel to the fibres [
36]. DCNNs can be used to estimate full resolution motion maps [
26]. Such technology would allow localization of muscle twitches, which has applications in the diagnosis and monitoring of motor neurone disease [
1,
2]. Analysis of local strain would also provide a non-invasive technology for diagnosis, targeting and monitoring treatment of conditions such as dystonia (a neurological condition causing painful muscle cramping/spasming). For example, diagnosis of cervical dystonia is currently done by needle electromyography (EMG), which is highly invasive, limiting/preventing treatment and diagnosis of deeper muscles (near the spine in the back and neck). We note that these applications are beyond the scope of the wavelet approach, whereas the DCNN method opens this entire domain for investigation, since we demonstrate feasibility in the application, even when constrained by small data sets. Our DCNN implementation can process an image (copy image to GPU, feedforward through the network and copy predicted fibre orientation map from GPU) at approximately 165 images per second, which is applicable to real-time operation within a clinic, lab or hospital.