1. Introduction
Shape is a distinguishing attribute of an object frequently utilized in image processing and computer vision applications. Measuring the similarity of objects via their shapes is a challenging task due to high within-class and low between-class variations. Within-class variations may be due to transformations such as rotation, scaling, and motion of limbs or independent motion of parts called articulations. Among within-class variations, articulations pose a particular challenge in the sense that articulations can not be modeled as global transformation groups. Hence, one can not define proper global descriptors that are invariant to articulations. Nevertheless, shapes of objects that articulate (referred to as articulated shapes) can be successfully represented by structural representations which are organized in the form of graphs of shape components. These components can be branches or critical points of the shape skeleton, subsets of the shape interior (commonly referred as parts), or fragments of shape boundary (commonly referred as contour fragments). Among many works in the literature that address shape similarity via structural representations, some examples are [
1,
2,
3,
4,
5,
6,
7,
8]. Skeleton based structural representations are the the most common ones; extensive literature reviews can be found in several works, e.g., [
2,
9].
In this work, we develop a novel representation scheme for articulated shapes. The representation scheme is not structural, i.e., it does not explicitly model components and their relationships. Our main argument against structural representations is that it is challenging to extract components and their structural relationships. Moreover, measuring similarity of shapes through their structural representations requires finding a correspondence between a pair of graphs, which is an intricate process necessitating advanced algorithms.
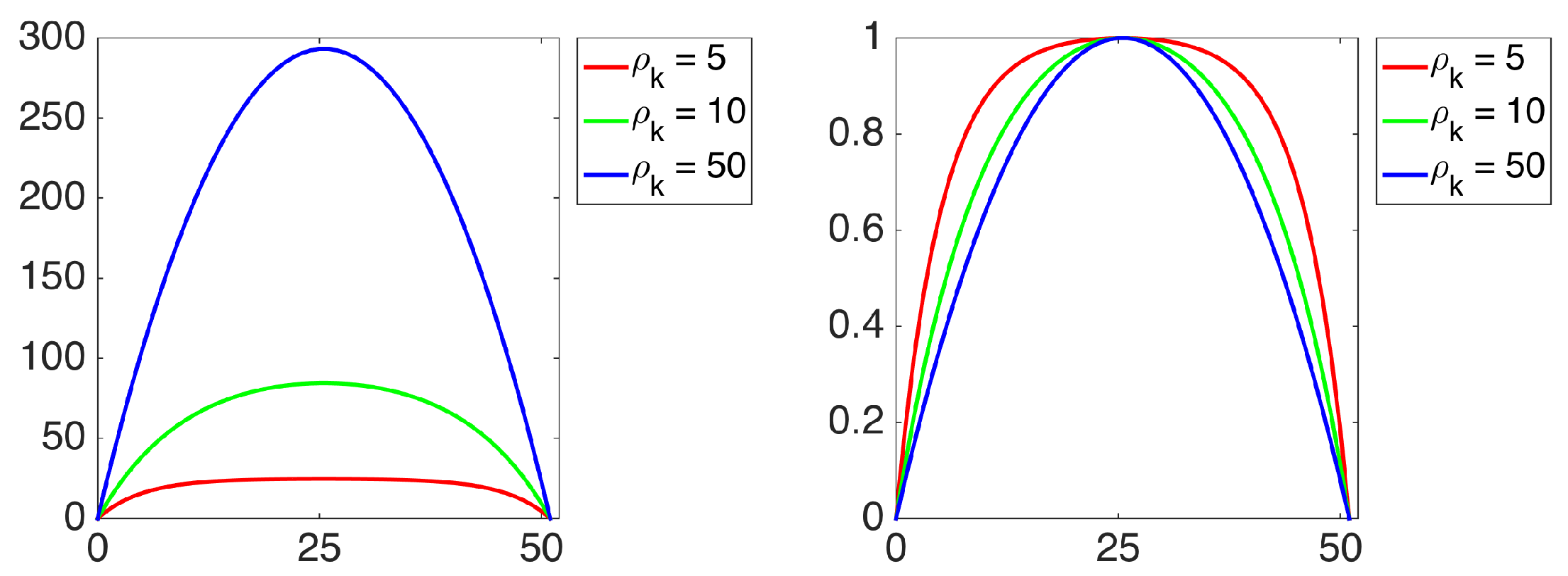
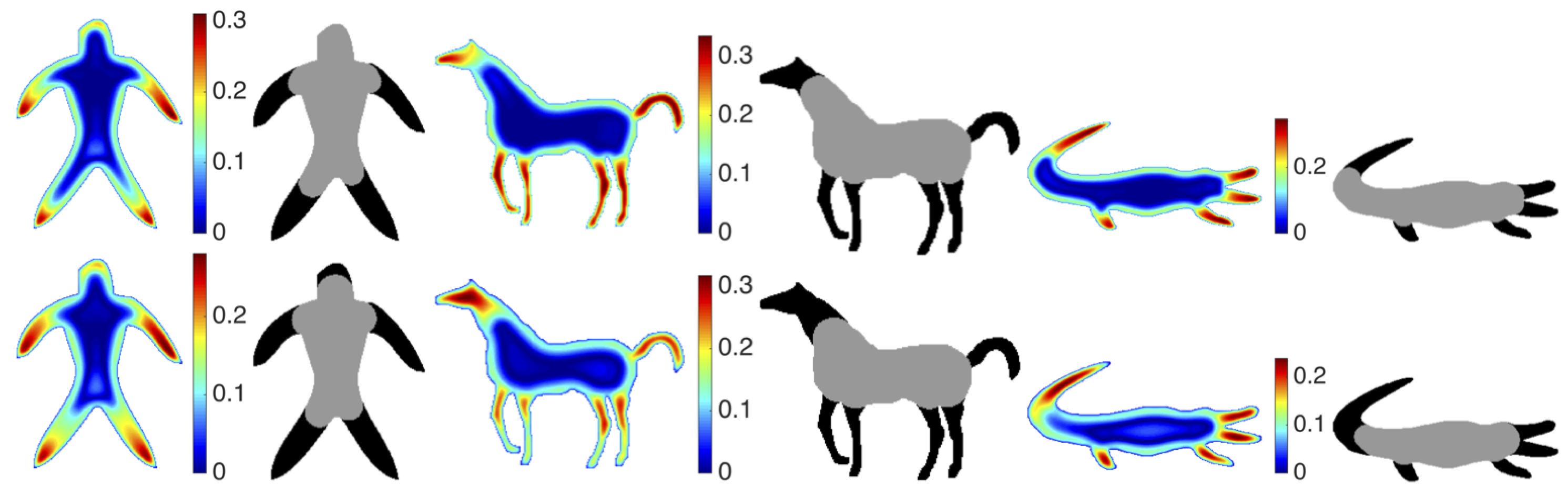
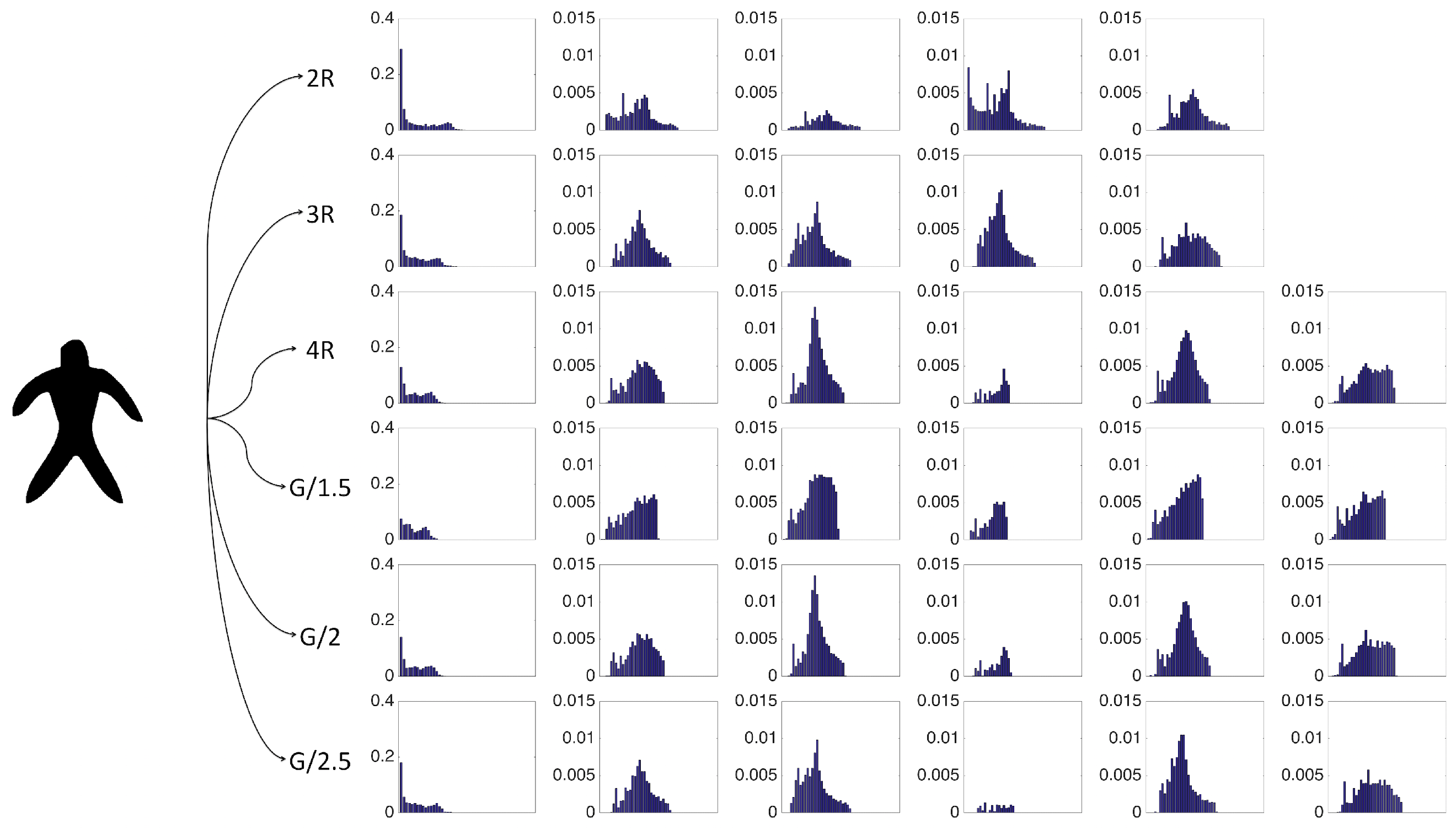
The strength of our representation lies in two factors. First, multiple characterizations of the shape are extracted based on descriptive and complementary shape scales. Second, each point in the shape communicates to the remaining points in the shape, thereby gaining an emergent role with respect to the remaining. We call this emergent role distinctness. The distinctness yields a partitioning of the shape into a set of regions, and the shape regions are described via size normalized probability distribution of the distinctness associated with their constituent points. Ultimately, our final representation is a collection of size normalized probability distribution of the distinctness over regions and over scales.
In order to evaluate the new representation, we use it for shape clustering. The clustering results obtained on three articulated shape datasets show that our method performs comparable to state of the art methods utilizing component graphs or trees, even though we are not explicitly modeling component relations.
Clustering or unsupervised grouping of shapes is a fundamental problem addressed in the literature. In [
10], hierarchical clustering is applied where the closest pair of clusters is merged at each step. A common structure skeleton graph is constructed for each cluster and the distance between the clusters is determined by matching their associated graphs. In [
11], spectral clustering is employed for fusing the similarity measures obtained via skeleton-based and contour-based shape descriptors. In [
12], the distance of shapes is measured using a new shape descriptor obtained by extending a contour-based descriptor to the shape interior and multi-objective optimization is applied to determine both the number of clusters and an optimal clustering result. In [
13], shape is represented via the distances from the centroid of the shape to the boundary points and a nonlinear projection algorithm is applied to group together similar shapes. In [
14], geodesic path constructed between differential geometric representations of the shape boundaries is used for hierarchical clustering of shapes. In [
15], elastic properties of the shape boundaries are considered and clustering is applied based on the elastic geodesic distance with dynamic programming alignment. In [
16], a new distance measure that is defined between a single shape and a group of shapes is utilized in a soft k-means like clustering of shapes.
3. Experimental Evaluation
In order to evaluate clustering results, we used Normalized Mutual Information (NMI). NMI measures the degree of agreement between the ground-truth category partition and the obtained clustering partition by utilizing the entropy measure. Let
denote the number of shapes in cluster
i and category
j,
denote the number of shapes in cluster
i, and
denote the number of shapes in category
j. Then, NMI can be computed as follows:
where
I is the number of clusters,
J is the number of categories, and
N is the total number of shapes. Notice that a high value of NMI indicates that the obtained clustering result matches well with the ground-truth category partition. We experimented with three shape datasets [
2,
3,
5]. These are the most articulated datasets in the literature. The first dataset [
3] consists of 14 shape categories each with 4 shapes, the second dataset [
2] consists of 30 categories each with 6 shapes, and the third dataset [
5] consist of 50 categories each with 20 shapes.
The easiest method of estimating a probability density function is constructing a histogram; an alternative is kernel density estimation (KDE). For comparing probability distributions, a proper distance should be used. Nevertheless, for simplicity we will use city block distance. To make sure that our choices for density estimation method and comparison metric are good, we performed an experiment to compare the chosen combination against the remaining three alternatives. Of course, for experiments, we also need to choose a metric for the final affinity propagation process. We considered four different metrics, hence experimented with
settings for each of the 3 datasets (yielding a total of
NMIs (
Table 1)). Results support the following conclusions. First, for probability distribution estimation, there is no significant difference between the two methods; hence, it is wiser to choose the simpler one, i.e., estimating via histogram (Observe that NMI values only slightly decrease when kernel density estimation is used). Second, for histogram comparison, city block distance is a better alternative. For histogram construction, the bin size is set to 0.01.
A side observation is that all four tested metrics for the affinity propagation seemed to perform comparably. In the next experiment, we enlarged the set of possibilities to 12. NMIs are reported in
Table 2. With the exception of
distance, the metrics seemed to perform comparably.
3.1. Comparison to State of the Art Methods
In
Table 3, we compare NMIs of the state of the art methods of which performances are reported for the considered datasets. The CSD (common structure discovery) [
10] employs hierarchical clustering; a common shape structure is constructed each time two clusters are merged into a single cluster. Building a common shape structure requires matching skeletons. The method (skeleton+spectral) [
11] combines the skeleton path distance [
4] with spectral clustering. The performance of these two skeleton-based methods decreases for the third dataset which contains unarticulated shape categories such as face category. For this data set, the highest performance is obtained via the method (contour+spectral) [
11] which uses the shape context descriptor [
20]. As the shape context descriptor [
20] is not robust to articulations, its performance is lower for the highly articulated first and second datasets. We also report averaged NMIs. In each of the two groups, the first column is the ordinary average whereas the second and third are weighted averages. The weighting is according to total number of categories and total number of shapes, respectively. Our distinctness based method accurately clusters the first set. For the second and the third dataset, respectively, NMIs are at least 99% and 95% of the best score.
Notice that the method in the fifth row (combining Inner Distance Shape Context (IDSC) descriptor IDSC [
22] with normalized cuts) exhibits obviously inferior performance. The NMIs are taken from [
10]. IDSC is not a structural method; the core idea is to build histogram descriptors from pairwise geodesic distance measurements among all boundary points. The expectation is that geodesic distances will be articulation invariant. There are also several other geodesic distance based shape descriptors, one of which is the so called
eccentricity transform [
23]. To compare our distinctness based representation to the geodesic distance based representation in [
23], we performed 36 tests by varying the choice of metric and the dataset. The results are summarized in in
Table 4. Each reported statistics is estimated using 6 NMIs. There seems to be a clear performance difference between the two representations. In order to make sure the observed difference in performance is significant in the sense that it can not be explainable via within group variation as in use of different metric (as in
Table 2), we performed
p-test. Based on the
p-test, the probability of observing the observed difference in mean due to within group variance is calculated to be zero (
Table 5). Dataset specific
values are obtained by pooling the standard deviations for distinctness (
Table 2) and for eccentricity. We used the available implementation of the eccentricity transform given in [
24] and selected the parameters as
. These are the parameter values reported in [
23].
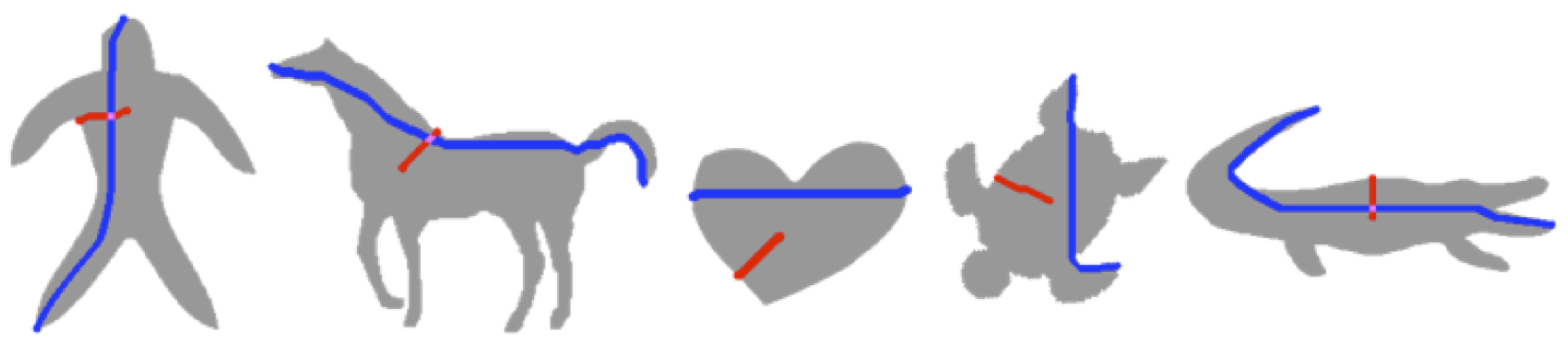
An articulated object is a union of parts and junctions. In the literature, some authors define articulations as transformations that are rigid when restricted to a part. Bending invariant geodesic distance based measures seem to work in a restricted setting when joints are small compared to parts and part transformation are rigid.
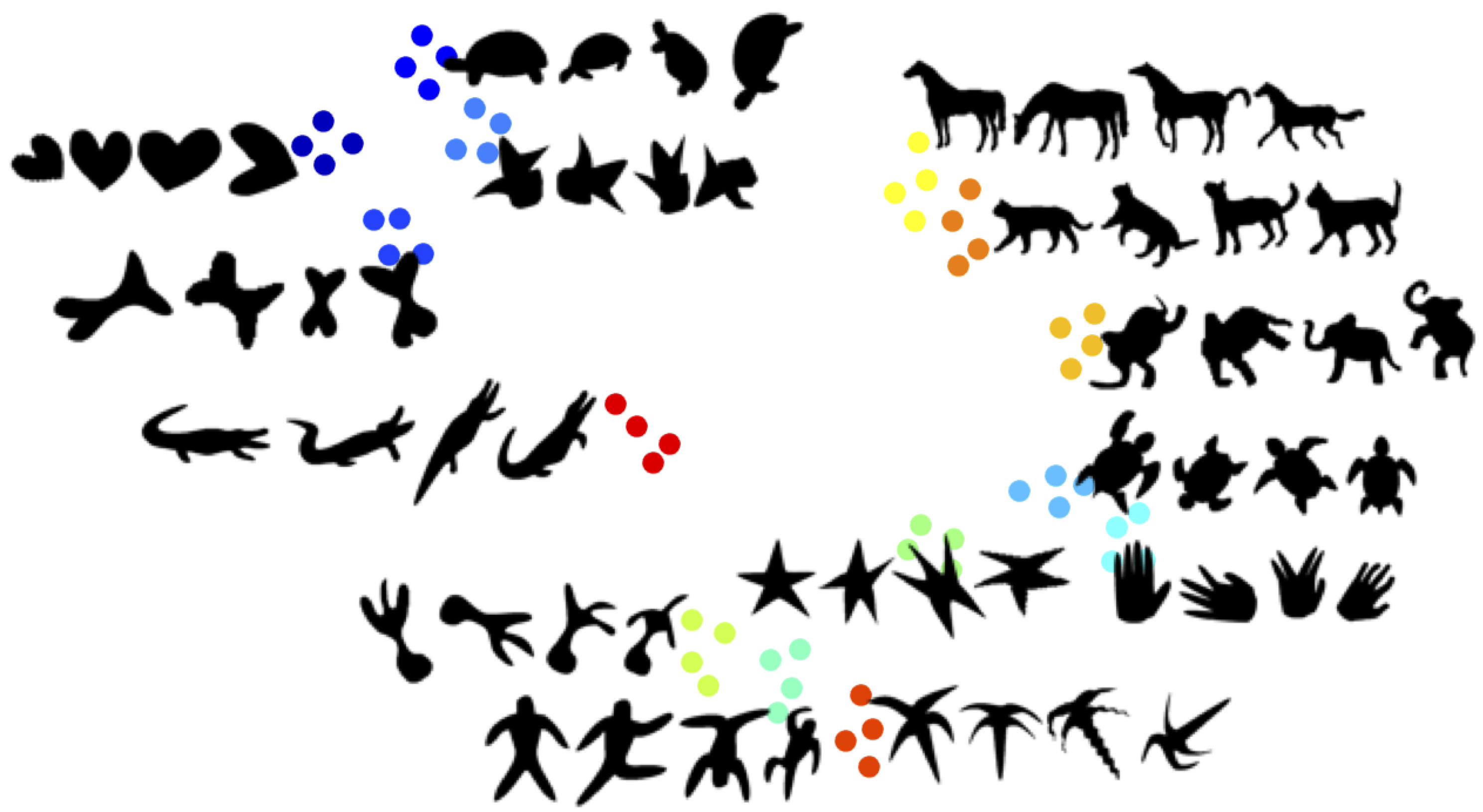
Figure 6 shows few sample shapes on which the geodesic distance based methods (e.g., [
22,
23]) are tested.
3.2. Representing Shapes Using a Single Histogram for Distinctness
In our proposed scheme, each shape is represented by a collection of regional histograms. A simpler alternative is to skip the partitioning step and represent the shape using a single histogram estimated using all of the distinctness values. In
Table 6, we report the clustering performance when shapes are represented by a single global histogram instead of a collection of regional histograms. For each of the three sets, the higher performance is marked as bold. Even when the shape is represented only using a single histogram, the discrepancy outperforms eccentricity.