Detection of Major Depressive Disorder from Functional Magnetic Resonance Imaging Using Regional Homogeneity and Feature/Sample Selective Evolving Voting Ensemble Approaches
Abstract
1. Introduction
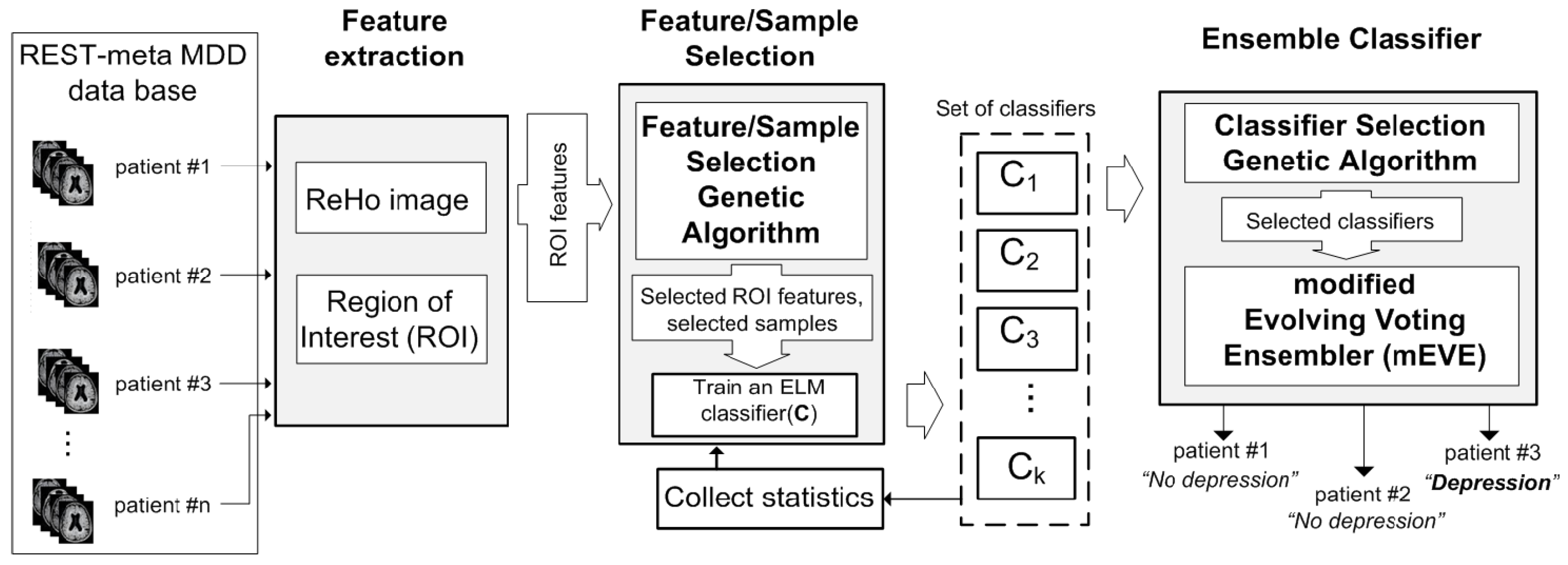
2. Materials and Methods
2.1. Dataset and Preprocessing
2.2. The Feature/Sample Selective Evolving Voting Ensembler
2.2.1. The Feature/Sample Selection Genetic Algorithm
- (1)
- Train set of 100 ELM classifiers using randomly selected features.
- (2)
- Build a decision matrix , where each matrix element is either 0 or 1, 100 is the number of initial ELM classifiers, and is the number of features. refers to misclassified samples, and refers to the correct classification of the j-th sample for the i-th classifier.
- (3)
- Compute opinion score for each sample . Opinion score defines the number of ELM classifiers that correctly classify the j-th sample.
- (4)
- “Sample hardness” is computed as follows:where is the rank of the j-th sample in the sorted row of the opinion scores sorted in ascending order. is the sample hardness of the sample with rank r, D is a scaling factor, is the number of samples, and x is the exponential constant. In this simulation, constants and are selected in the way to maximize the effect of “sample hardness” for further ensemble. The “sample hardness” is higher for samples with lower opinion scores and vice versa.
- (5)
- “Classifier Cost” for p-th classifier is calculated based on the “Sample hardness” as follows:where
| Algorithm 1 Balanced Single-Point Crossover |
|
2.2.2. Modified Evolving Voting Ensembler
| Algorithm 2 Modified Evolving Voting Ensembler |
|
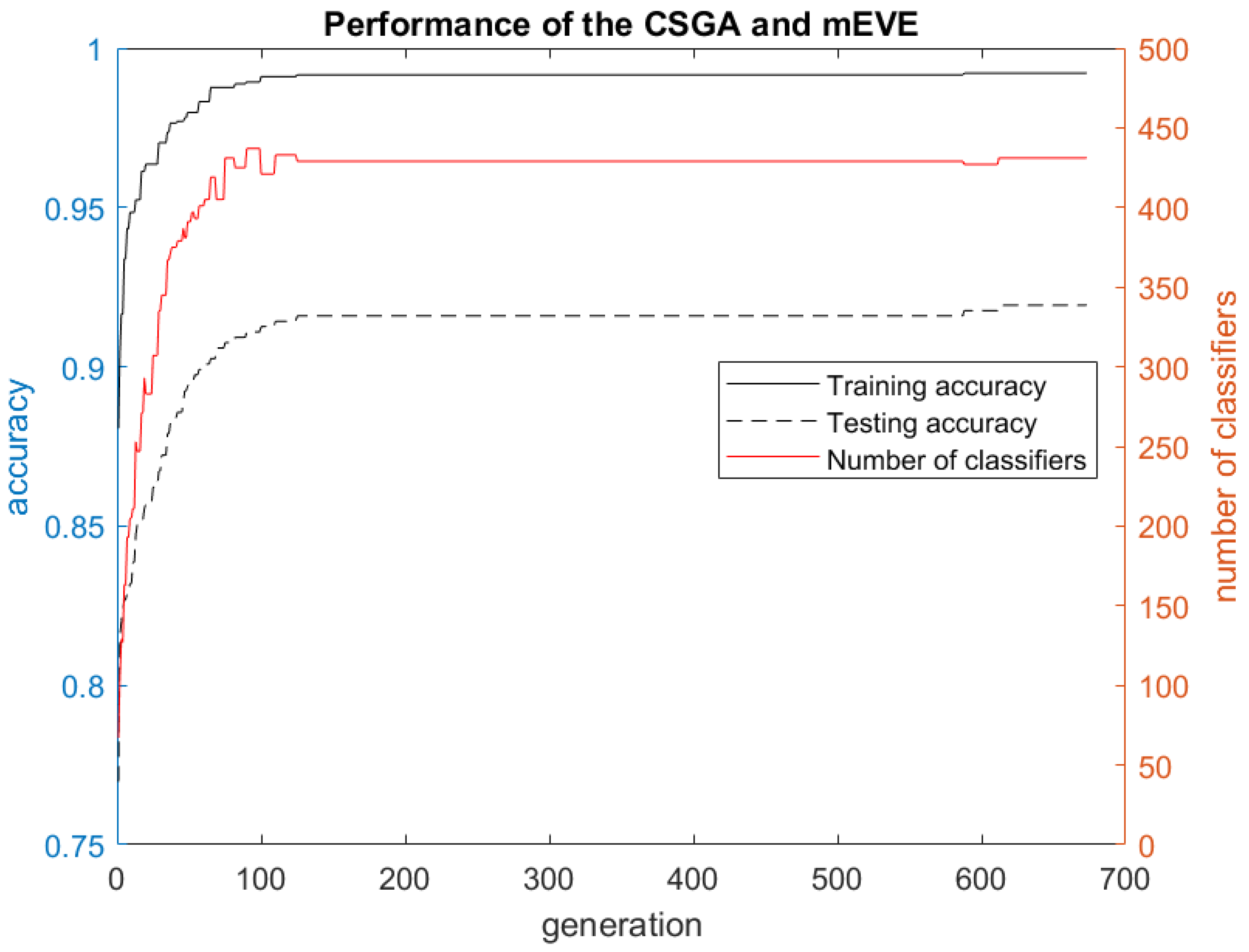
3. Results and Discussion
3.1. Experimental Results
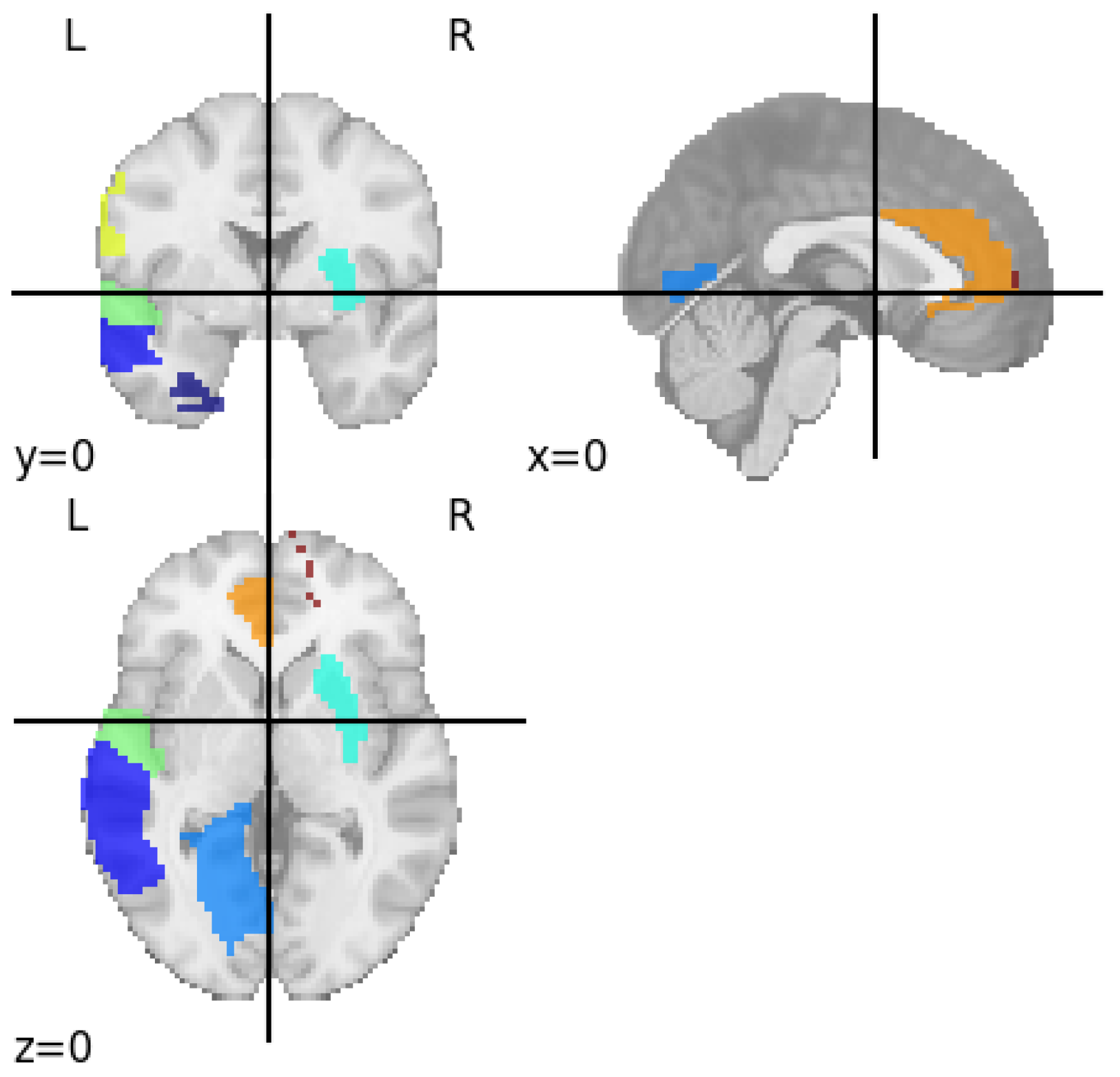
3.2. Identifying Brain Regions Responsible for Major Depressive Disorder
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MDD | Major Depressive Disorder |
| fMRI | Functional Magnetic Resonance Imaging |
| ReHo | Regional Homogeneity |
| HCs | Healthy Controls |
| fsEVE | Feature/Sample Selective Evolving Voting Ensemble |
| mEVE | Modified Evolving Voting Ensemble |
| ELM | Extreme Learning Machine |
| AAL | Automated Anatomical Labeling |
References
- World Health Organization. New Guidance Calls for Urgent Reform of Mental Health Systems. 2025. Available online: https://www.who.int/news/item/25-03-2025-new-who-guidance-calls-for-urgent-transformation-of-mental-health-policies (accessed on 19 April 2025).
- First, M.B.; Gaebel, W.; Maj, M.; Stein, D.J.; Kogan, C.S.; Saunders, J.B.; Poznyak, V.B.; Gureje, O.; Lewis-Fernández, R.; Maercker, A.; et al. An organization-and category-level comparison of diagnostic requirements for mental disorders in ICD-11 and DSM-5. World Psychiatry 2021, 20, 34–51. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, W.; Zhan, L.; Qiu, J.; Jia, T. Multi-feature concatenation and multi-classifier stacking: An interpretable and generalizable machine learning method for MDD discrimination with rsfMRI. Neuroimage 2024, 285, 120497. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M. Overview of fMRI analysis. Br. J. Radiol. 2004, 77, S167–S175. [Google Scholar] [CrossRef]
- Heeger, D.J.; Ress, D. What does fMRI tell us about neuronal activity? Nat. Rev. Neurosci. 2002, 3, 142–151. [Google Scholar] [CrossRef]
- Fan, Z.; Chen, X.; Qi, Z.X.; Li, L.; Lu, B.; Jiang, C.L.; Zhu, R.Q.; Yan, C.G.; Chen, L. Physiological significance of R-fMRI indices: Can functional metrics differentiate structural lesions (brain tumors)? Neuroimage Clin. 2019, 22, 101741. [Google Scholar] [CrossRef]
- Zang, Y.; Jiang, T.; Lu, Y.; He, Y.; Tian, L. Regional homogeneity approach to fMRI data analysis. Neuroimage 2004, 22, 394–400. [Google Scholar] [CrossRef]
- Dai, P.; Zhou, Y.; Shi, Y.; Lu, D.; Chen, Z.; Zou, B.; Liu, K.; Liao, S.; The REST-meta-MDD Consortium. Classification of MDD using a Transformer classifier with large-scale multisite resting-state fMRI data. Hum. Brain Mapp. 2024, 45, e26542. [Google Scholar] [CrossRef]
- Gallo, S.; El-Gazzar, A.; Zhutovsky, P.; Thomas, R.M.; Javaheripour, N.; Li, M.; Bartova, L.; Bathula, D.; Dannlowski, U.; Davey, C.; et al. Functional connectivity signatures of major depressive disorder: Machine learning analysis of two multicenter neuroimaging studies. Mol. Psychiatry 2023, 28, 3013–3022. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, A.; Sakai, Y.; Yamada, T.; Yahata, N.; Kunimatsu, A.; Okada, N.; Itahashi, T.; Hashimoto, R.; Mizuta, H.; Ichikawa, N.; et al. Generalizable brain network markers of major depressive disorder across multiple imaging sites. PLoS Biol. 2020, 18, e3000966. [Google Scholar] [CrossRef]
- Yoshida, K.; Shimizu, Y.; Yoshimoto, J.; Takamura, M.; Okada, G.; Okamoto, Y.; Yamawaki, S.; Doya, K. Prediction of clinical depression scores and detection of changes in whole-brain using resting-state functional MRI data with partial least squares regression. PloS ONE 2017, 12, e0179638. [Google Scholar] [CrossRef]
- Mwangi, B.; Ebmeier, K.P.; Matthews, K.; Douglas Steele, J. Multi-centre diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain 2012, 135, 1508–1521. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Wong, N.M.; Shao, R.; Lee, S.H.; Huang, C.M.; Liu, H.L.; Lin, C.; Lee, T.M. Classification of Major Depressive Disorder using Machine Learning on brain structure and functional connectivity. J. Affect. Disord. Rep. 2022, 10, 100428. [Google Scholar] [CrossRef]
- Yan, C.G.; Chen, X.; Li, L.; Castellanos, F.X.; Bai, T.J.; Bo, Q.J.; Cao, J.; Chen, G.M.; Chen, N.X.; Chen, W.; et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc. Natl. Acad. Sci. USA 2019, 116, 9078–9083. [Google Scholar] [CrossRef]
- Chen, X.; Lu, B.; Li, H.X.; Li, X.Y.; Wang, Y.W.; Castellanos, F.X.; Cao, L.P.; Chen, N.X.; Chen, W.; Cheng, Y.Q.; et al. The DIRECT consortium and the REST-meta-MDD project: Towards neuroimaging biomarkers of major depressive disorder. Psychoradiology 2022, 2, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Tzourio-Mazoyer, N.; Landeau, B.; Papathanassiou, D.; Crivello, F.; Etard, O.; Delcroix, N.; Mazoyer, B.; Joliot, M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002, 15, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Dietterich, T.G. Ensemble methods in machine learning. In Proceedings of the International Workshop on Multiple Classifier Systems, Nanjing, China; Springer: Berlin/Heidelberg, Germany, 2000; pp. 1–15. [Google Scholar]
- Zhang, Y.; Zhang, H.; Cai, J.; Yang, B. A weighted voting classifier based on differential evolution. Abstr. Appl. Anal. 2014, 2014, 376950. [Google Scholar]
- Huang, G.B.; Zhou, H.; Ding, X.; Zhang, R. Extreme learning machine for regression and multiclass classification. IEEE Trans. Syst. Man Cybern. Part (Cybern.) 2011, 42, 513–529. [Google Scholar] [CrossRef]
- Holland, J.H. Adaptation in Natural and Artificial Systems: An Introductory Analysis with Applications to Biology, Control, and Artificial Intelligence; MIT Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Suresh, S.; Omkar, S.; Mani, V.; Prakash, T.G. Lift coefficient prediction at high angle of attack using recurrent neural network. Aerosp. Sci. Technol. 2003, 7, 595–602. [Google Scholar] [CrossRef]
- Sachnev, V.; Sundararajan, N.; Suresh, S. A new approach for JPEG steganalysis with a cognitive evolving ensembler and robust feature selection. Cogn. Comput. 2023, 15, 751–764. [Google Scholar] [CrossRef]
- Guo, Z.P.; Chen, L.; Tang, L.R.; Gao, Y.; Qu, M.; Wang, L.; Liu, C.H. The differential orbitofrontal activity and connectivity between atypical and typical major depressive disorder. NeuroImage Clin. 2025, 45, 103717. [Google Scholar] [CrossRef]
- Ni, S.; Gao, S.; Ling, C.; Jiang, J.; Wu, F.; Peng, T.; Sun, J.; Zhang, N.; Xu, X. Altered brain regional homogeneity is associated with cognitive dysfunction in first-episode drug-naive major depressive disorder: A resting-state fMRI study. J. Affect. Disord. 2023, 343, 102–108. [Google Scholar] [CrossRef]
- Li, M.; Liu, M.; Kang, J.; Zhang, W.; Lu, S. Depression recognition method based on regional homogeneity features from emotional response fMRI using deep convolutional neural network. In Proceedings of the 2021 3rd International Conference on Intelligent Medicine and Image Processing, Tianjin, China, 23–26 April 2021; pp. 45–49. [Google Scholar]
- Noman, F.; Ting, C.M.; Kang, H.; Phan, R.C.W.; Ombao, H. Graph autoencoders for embedding learning in brain networks and major depressive disorder identification. IEEE J. Biomed. Health Inform. 2024, 28, 1644–1655. [Google Scholar] [CrossRef]
- Dai, P.; Shi, Y.; Lu, D.; Zhou, Y.; Luo, J.; He, Z.; Chen, Z.; Zou, B.; Tang, H.; Huang, Z.; et al. Classification of recurrent major depressive disorder using a residual denoising autoencoder framework: Insights from large-scale multisite fMRI data. Comput. Methods Programs Biomed. 2024, 247, 108114. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.S.H.; Wang, J.; Tay, G.W.N.; Ho, R.; Husain, S.F.; Chiang, S.K.; Lin, H.; Cheng, X.; Li, Z.; Chen, N. Interpretable deep learning model for major depressive disorder assessment based on functional near-infrared spectroscopy. Asian J. Psychiatry 2024, 92, 103901. [Google Scholar] [CrossRef]
- Dong, C.; Zheng, H.; Shen, H.; Wan, Y.; Xu, Y.; Li, Y.; Ping, L.; Yu, H.; Liu, C.; Cui, J.; et al. Cortical thickness alternation in obsessive-compulsive disorder patients compared with healthy controls. Brain Imaging Behav. 2025, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; She, S.; Qiu, Y.; Li, Z.; Wu, X.; Hu, H.; Zheng, W.; Huang, R.; Wu, H. Multi-modal MRI measures reveal sensory abnormalities in major depressive disorder patients: A surface-based study. NeuroImage Clin. 2023, 39, 103468. [Google Scholar] [CrossRef]
- Hellewell, S.C.; Welton, T.; Maller, J.J.; Lyon, M.; Korgaonkar, M.S.; Koslow, S.H.; Williams, L.M.; Rush, A.J.; Gordon, E.; Grieve, S.M. Profound and reproducible patterns of reduced regional gray matter characterize major depressive disorder. Transl. Psychiatry 2019, 9, 176. [Google Scholar] [CrossRef]
- Ulmer, S. Neuroanatomy and cortical landmarks. In fMRI: Basics and Clinical Applications; Springer: Berlin/Heidelberg, Germany, 2013; pp. 7–16. [Google Scholar]
- Liu, W.; Jiang, X.; Deng, Z.; Xie, Y.; Guo, Y.; Wu, Y.; Sun, Q.; Kong, L.; Wu, F.; Tang, Y. Functional and structural alterations in different durations of untreated illness in the frontal and parietal lobe in major depressive disorder. Eur. Arch. Psychiatry Clin. Neurosci. 2024, 274, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Peng, W.; Sun, H.; Kuang, W.; Li, W.; Jia, Z.; Gong, Q. High-field magnetic resonance imaging of structural alterations in first-episode, drug-naive patients with major depressive disorder. Transl. Psychiatry 2016, 6, e942. [Google Scholar] [CrossRef]
- Fan, Q.; Zhang, H. Functional connectivity density of postcentral gyrus predicts rumination and major depressive disorders in males. Psychiatry Res. Neuroimaging 2025, 347, 111939. [Google Scholar] [CrossRef]
- Wu, D.; Li, J.; Wang, J. Altered neural activities during emotion regulation in depression: A meta-analysis. J. Psychiatry Neurosci. 2024, 49, E334–E344. [Google Scholar] [CrossRef]
- Bush, G.; Luu, P.; Posner, M.I. Cognitive and emotional influences in anterior cingulate cortex. Trends Cogn. Sci. 2000, 4, 215–222. [Google Scholar] [CrossRef]
- Apps, M.A.; Rushworth, M.F.; Chang, S.W. The anterior cingulate gyrus and social cognition: Tracking the motivation of others. Neuron 2016, 90, 692–707. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.M.; Kulikova, A.; Ly, H.; Rush, A.J.; Brown, E.S. Anterior cingulate cortex in individuals with depressive symptoms: A structural MRI study. Psychiatry Res. Neuroimaging 2022, 319, 111420. [Google Scholar] [CrossRef]
- Radua, J.; Phillips, M.L.; Russell, T.; Lawrence, N.; Marshall, N.; Kalidindi, S.; El-Hage, W.; McDonald, C.; Giampietro, V.; Brammer, M.J.; et al. Neural response to specific components of fearful faces in healthy and schizophrenic adults. Neuroimage 2010, 49, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Liu, H.L.; Huang, C.M.; Chen, Y.L.; Gao, M.; Lee, S.H.; Lin, C.; Lee, T.M. Loneliness and depression dissociated on parietal-centered networks in cognitive and resting states. Psychol. Med. 2020, 50, 2691–2701. [Google Scholar] [CrossRef] [PubMed]
- Lemogne, C.; le Bastard, G.; Mayberg, H.; Volle, E.; Bergouignan, L.; Lehéricy, S.; Allilaire, J.F.; Fossati, P. In search of the depressive self: Extended medial prefrontal network during self-referential processing in major depression. Soc. Cogn. Affect. Neurosci. 2009, 4, 305–312. [Google Scholar] [CrossRef]
- Baker, S.C.; Frith, C.D.; Dolan, R.J. The interaction between mood and cognitive function studied with PET. Psychol. Med. 1997, 27, 565–578. [Google Scholar] [CrossRef]
- Bogousslavsky, J.; Miklossy, J.; Deruaz, J.P.; Assal, G.; Regli, F. Lingual and fusiform gyri in visual processing: A clinico-pathologic study of superior altitudinal hemianopia. J. Neurol. Neurosurg. Psychiatry 1987, 50, 607–614. [Google Scholar] [CrossRef]
- Hong, C.; Ding, C.; Chen, Y.; Cao, S.; Hou, Y.; Hu, W.; Yang, D. Mindfulness-based intervention reduce interference of negative stimuli to working memory in individuals with subclinical depression: A randomized controlled fMRI study. Int. J. Clin. Health Psychol. 2024, 24, 100459. [Google Scholar] [CrossRef]
- Jung, J.; Kang, J.; Won, E.; Nam, K.; Lee, M.S.; Tae, W.S.; Ham, B.J. Impact of lingual gyrus volume on antidepressant response and neurocognitive functions in major depressive disorder: A voxel-based morphometry study. J. Affect. Disord. 2014, 169, 179–187. [Google Scholar] [CrossRef]
- Viñas-Guasch, N.; Wu, Y.J. The role of the putamen in language: A meta-analytic connectivity modeling study. Brain Struct. Funct. 2017, 222, 3991–4004. [Google Scholar] [CrossRef] [PubMed]
- DeLong, M.; Alexander, G.; Georgopoulos, A.; Crutcher, M.; Mitchell, S.; Richardson, R. Role of basal ganglia in limb movements. Hum. Neurobiol. 1984, 2, 235–244. [Google Scholar]
- Husain, M.M.; McDonald, W.M.; Doraiswamy, P.M.; Figiel, G.S.; Na, C.; Escalona, P.R.; Boyko, O.B.; Nemeroff, C.B.; Krishnan, K.R.R. A magnetic resonance imaging study of putamen nuclei in major depression. Psychiatry Res. Neuroimaging 1991, 40, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Liu, Z.; Fan, Z.; Zuo, J.; Xi, C.; Yang, J. Dynamical regional activity in putamen distinguishes bipolar type I depression and unipolar depression. J. Affect. Disord. 2022, 297, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; He, Q.; Zhu, X.; Hu, Y.; Yao, Y.; Hommel, B.; Beste, C.; Liu, J.; Yang, Y.; Zhang, W. Smaller putamen volumes are associated with greater problems in external emotional regulation in depressed adolescents with nonsuicidal self-injury. J. Psychiatr. Res. 2022, 155, 338–346. [Google Scholar] [CrossRef]
- Weiner, K.S.; Zilles, K. The anatomical and functional specialization of the fusiform gyrus. Neuropsychologia 2016, 83, 48–62. [Google Scholar] [CrossRef]
- Lin, S.; Chen, Z.; Zhao, Y.; Gong, Q. Joint and distinct neural structure and function deficits in major depressive disorder with suicidality: A multimodal meta-analysis of MRI studies. J. Psychiatry Neurosci. 2025, 50, E126–E141. [Google Scholar] [CrossRef]
- Yao, C.; Wang, P.; Xiao, Y.; Zheng, Y.; Pu, J.; Miao, Y.; Wang, J.; Xue, S.W. Increased individual variability in functional connectivity of the default mode network and its genetic correlates in major depressive disorder. Sci. Rep. 2025, 15, 8853. [Google Scholar] [CrossRef]
- Onitsuka, T.; Shenton, M.E.; Salisbury, D.F.; Dickey, C.C.; Kasai, K.; Toner, S.K.; Frumin, M.; Kikinis, R.; Jolesz, F.A.; McCarley, R.W. Middle and inferior temporal gyrus gray matter volume abnormalities in chronic schizophrenia: An MRI study. Am. J. Psychiatry 2004, 161, 1603–1611. [Google Scholar] [CrossRef]
- Chen, C.; Liu, Y.; Sun, Y.; Jiang, W.; Yuan, Y.; Qing, Z.; DIRECT Consortium. Abnormal structural covariance network in major depressive disorder: Evidence from the REST-meta-MDD project. Neuroimage Clin. 2025, 46, 103794. [Google Scholar] [CrossRef]
- Zhang, F.F.; Peng, W.; Sweeney, J.A.; Jia, Z.Y.; Gong, Q.Y. Brain structure alterations in depression: Psychoradiological evidence. CNS Neurosci. Ther. 2018, 24, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Pandya, M.; Altinay, M.; Malone, D.A.; Anand, A. Where in the brain is depression? Curr. Psychiatry Rep. 2012, 14, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Helm, K.; Viol, K.; Weiger, T.M.; Tass, P.A.; Grefkes, C.; Del Monte, D.; Schiepek, G. Neuronal connectivity in major depressive disorder: A systematic review. Neuropsychiatr. Dis. Treat. 2018, 14, 2715–2737. [Google Scholar] [CrossRef] [PubMed]
| Healthy Controls | MDD | |
|---|---|---|
| Number of subjects | 1104 | 1276 |
| Female | 642 | 813 |
| Male | 462 | 463 |
| Average age | 36 years Range (12–82) | 36 years Range (14–80) |
| SI No. | Number of ELM Classifiers | Number of Generations | Max Train/ Test Accuracy (Set of ELM) | Min Train/ Test Accuracy (Set of ELM) | mEVE Train/ Test Accuracy | Train/Test F1 Score |
|---|---|---|---|---|---|---|
| 1. | 401 | 193 | 62.41/54.45 | 50.6/49.58 | 98.21/91.09 | 0.98/0.95 |
| 2. | 361 | 799 | 61.70/57.48 | 51.78/49.75 | 97.93/90.76 | 0.98/0.95 |
| 3. | 363 | 943 | 65.04/51.76 | 51.27/50.25 | 97.70/90.92 | 0.98/0.96 |
| 4. | 431 | 408 | 62.95/55.80 | 51.28/51.09 | 99.10/90.59 | 0.99/0.97 |
| 5. | 25 | 673 | 60.34/53.61 | 50.93/50.25 | 99.61/91.93 | 0.99/0.98 |
| SI No. | Authors | Sample Size | Approach | Accuracy |
|---|---|---|---|---|
| 1. | Guo et al. [23] | MDD-101, HC-49 | SVM | 76.42% |
| 2. | Ni et al. [24] | MDD-60, HC-60 | SVM | 90% |
| 3. | Li et al. [25] | MDD-1300, HC-1128 | KELM | 86% |
| 4. | Noman et al. [26] | MDD-250, HC-227 | GAE-FCNN | 65% |
| 5. | Dai et al. [27] | MDD-832, HC-779 | Res-DAE | 70% |
| 6. | Proposed Approach | MDD-1276, HC-1104 | fsEVE | 91.93% |
| SI No | Regions | Functions |
|---|---|---|
| 1. | Left Superior Temporal Gyrus | Social cognition, auditory processing and Language comprehension |
| 2. | Left Postcentral Gyrus | Processing sensory information from the skin, muscles, and joints |
| 3. | Left Anterior Cingulate Gyrus | Motivation and goal directed behaviour, cognition, Visuomotor and auditory |
| 4. | Right Inferior Parietal Lobule | Sensory integration, Language, Socialcognition, Visuomotor and auditory processing |
| 5. | Right Superior Medial Frontal Gyrus | Working memory, impulse control, mood regulation and self-awareness |
| 6. | Left Lingual Gyrus | Linguistic processing |
| 7. | Right Putamen | Addiction, Congnitive function and Learning |
| 8. | Left Fusiform Gyrus | Reading and Emotional perception |
| 9. | Left Middle Temporal Gyrus | Language, Visual perception and Semantic memory |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
A. R., B.; Mahanand, B.S.; Sachnev, V.; DIRECT Consortium. Detection of Major Depressive Disorder from Functional Magnetic Resonance Imaging Using Regional Homogeneity and Feature/Sample Selective Evolving Voting Ensemble Approaches. J. Imaging 2025, 11, 238. https://doi.org/10.3390/jimaging11070238
A. R. B, Mahanand BS, Sachnev V, DIRECT Consortium. Detection of Major Depressive Disorder from Functional Magnetic Resonance Imaging Using Regional Homogeneity and Feature/Sample Selective Evolving Voting Ensemble Approaches. Journal of Imaging. 2025; 11(7):238. https://doi.org/10.3390/jimaging11070238
Chicago/Turabian StyleA. R., Bindiya, B. S. Mahanand, Vasily Sachnev, and DIRECT Consortium. 2025. "Detection of Major Depressive Disorder from Functional Magnetic Resonance Imaging Using Regional Homogeneity and Feature/Sample Selective Evolving Voting Ensemble Approaches" Journal of Imaging 11, no. 7: 238. https://doi.org/10.3390/jimaging11070238
APA StyleA. R., B., Mahanand, B. S., Sachnev, V., & DIRECT Consortium. (2025). Detection of Major Depressive Disorder from Functional Magnetic Resonance Imaging Using Regional Homogeneity and Feature/Sample Selective Evolving Voting Ensemble Approaches. Journal of Imaging, 11(7), 238. https://doi.org/10.3390/jimaging11070238






