Automation of Multi-Class Microscopy Image Classification Based on the Microorganisms Taxonomic Features Extraction
Abstract
1. Introduction
- 1.
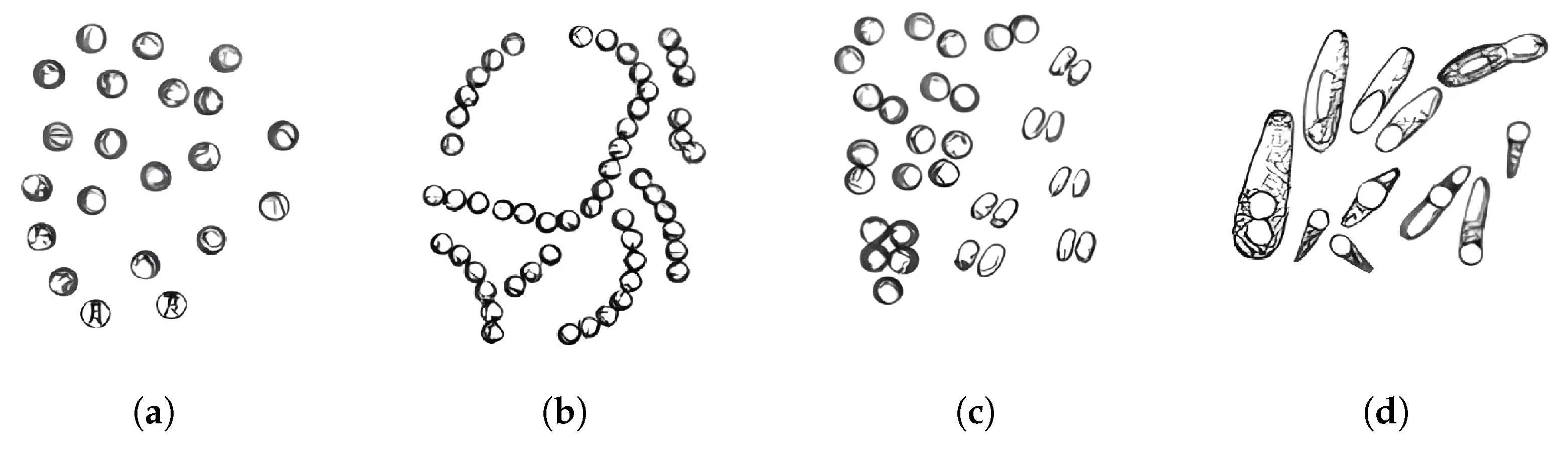
- Micrococci [19]: These are facultatively pathogenic Gram-positive cocci that can cause opportunistic infections, particularly in immunocompromised patients. While generally considered low-virulence organisms, their ability to exploit weakened host defenses makes them a notable concern in healthcare settings [20].
- 2.
- Diplococci [21]: These pathogens are responsible for severe diseases such as pneumonia and meningitis. Notably, Streptococcus pneumoniae alone accounts for approximately 15% of childhood mortality in low-income countries, highlighting its devastating impact on vulnerable populations.
- 3.
- Streptococci [22]: This genus includes pathogens that cause a wide range of diseases, from pharyngitis and scarlet fever to severe post-infectious complications such as rheumatic fever. Globally, streptococcal infections affect over 600 million people annually, underscoring their pervasive public health burden [23].
- 4.
- Bacilli [24]: This group encompasses species such as B. anthracis, the causative agent of anthrax, and B. cereus, a common culprit in foodborne illnesses. While B. anthracis poses significant biosecurity risks due to its potential use as a biological weapon, B. cereus is a frequent cause of gastroenteritis, particularly in improperly stored food. Both species exemplify the dual threat posed by bacilli to both individual health and broader biosecurity [25].
2. Related Work
3. Problem Statement
4. Materials and Methods
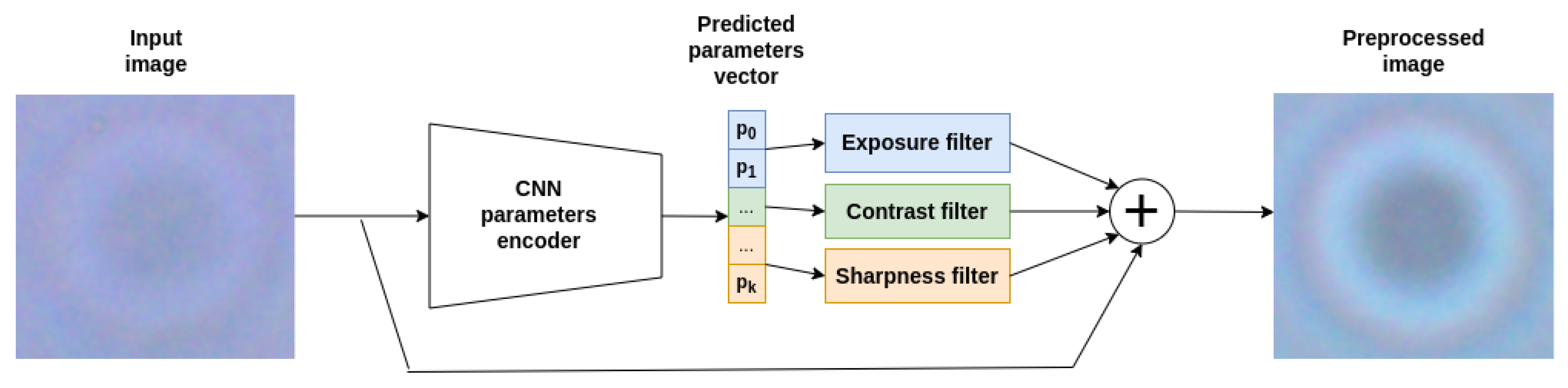
4.1. Image Preprocessing
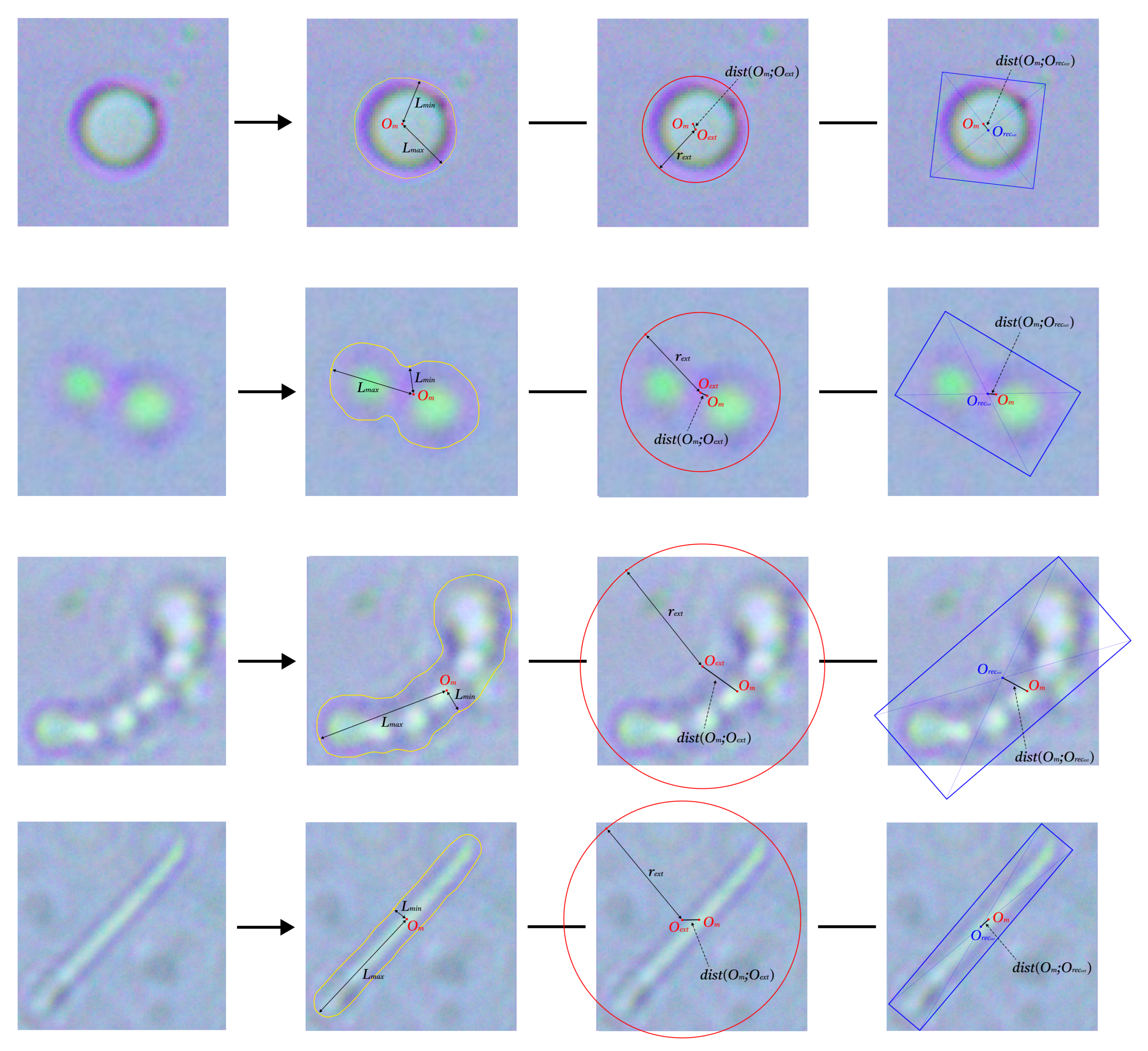
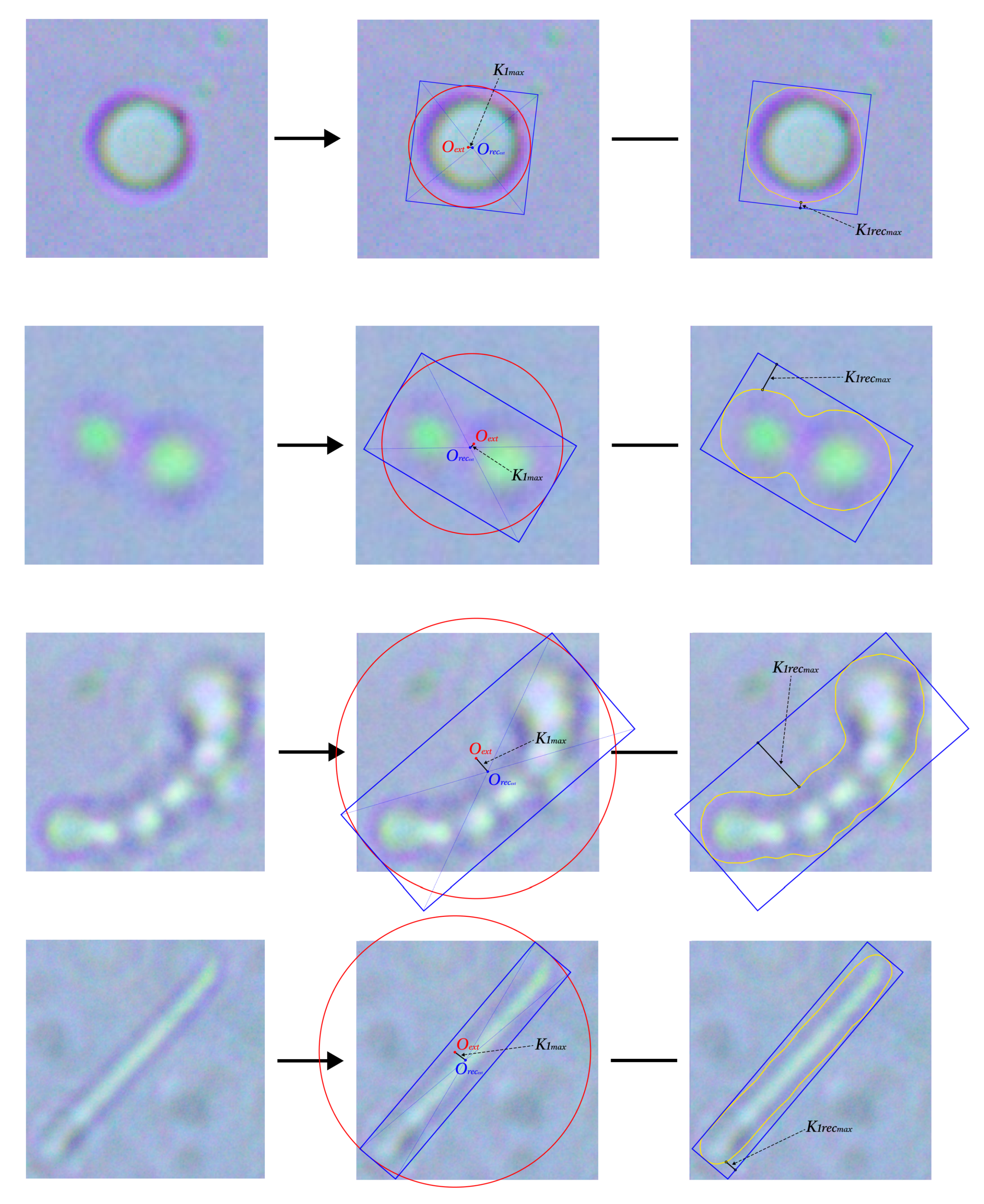
4.2. Contour Primitives Determination
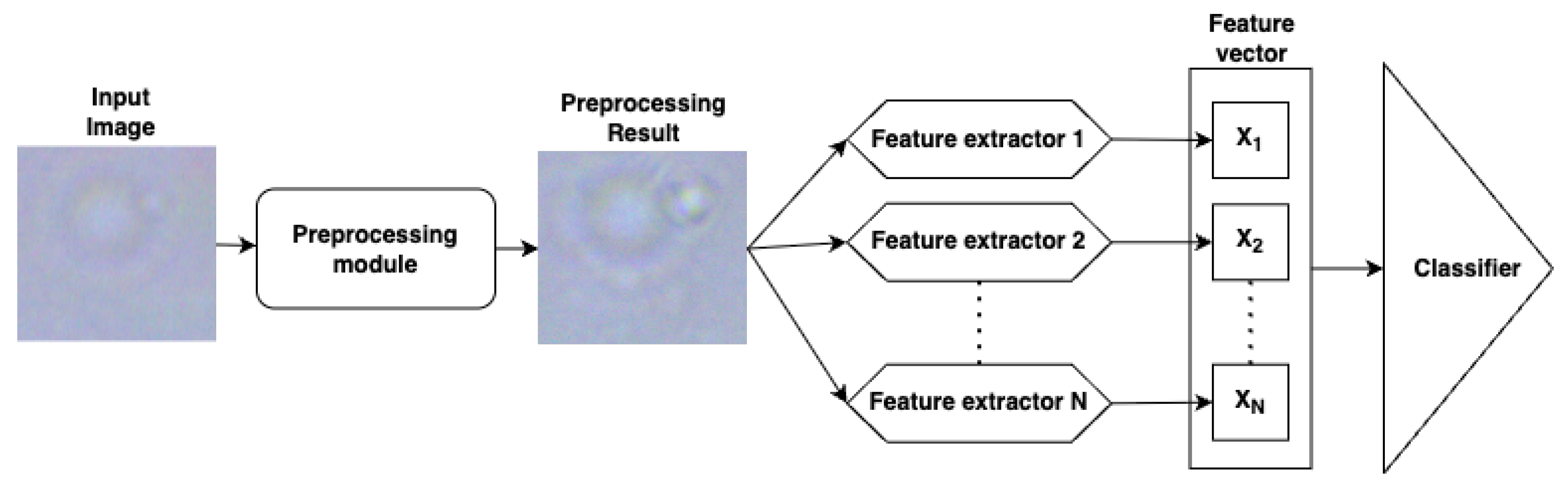
4.3. Automated Feature Generation
4.4. Classifiers
5. Experiments, Results, and Discussion
5.1. Dataset Description
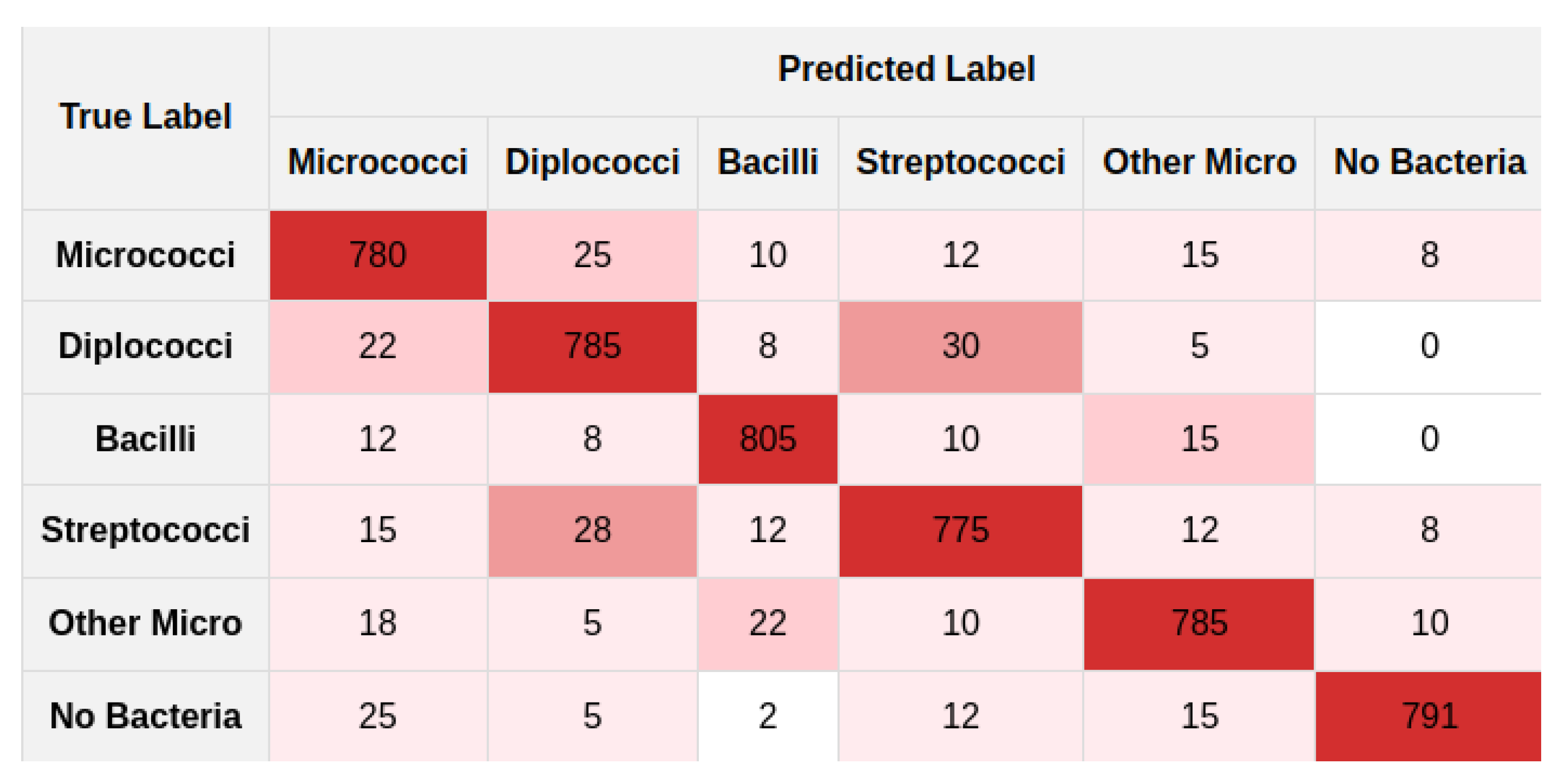
5.2. Experiments
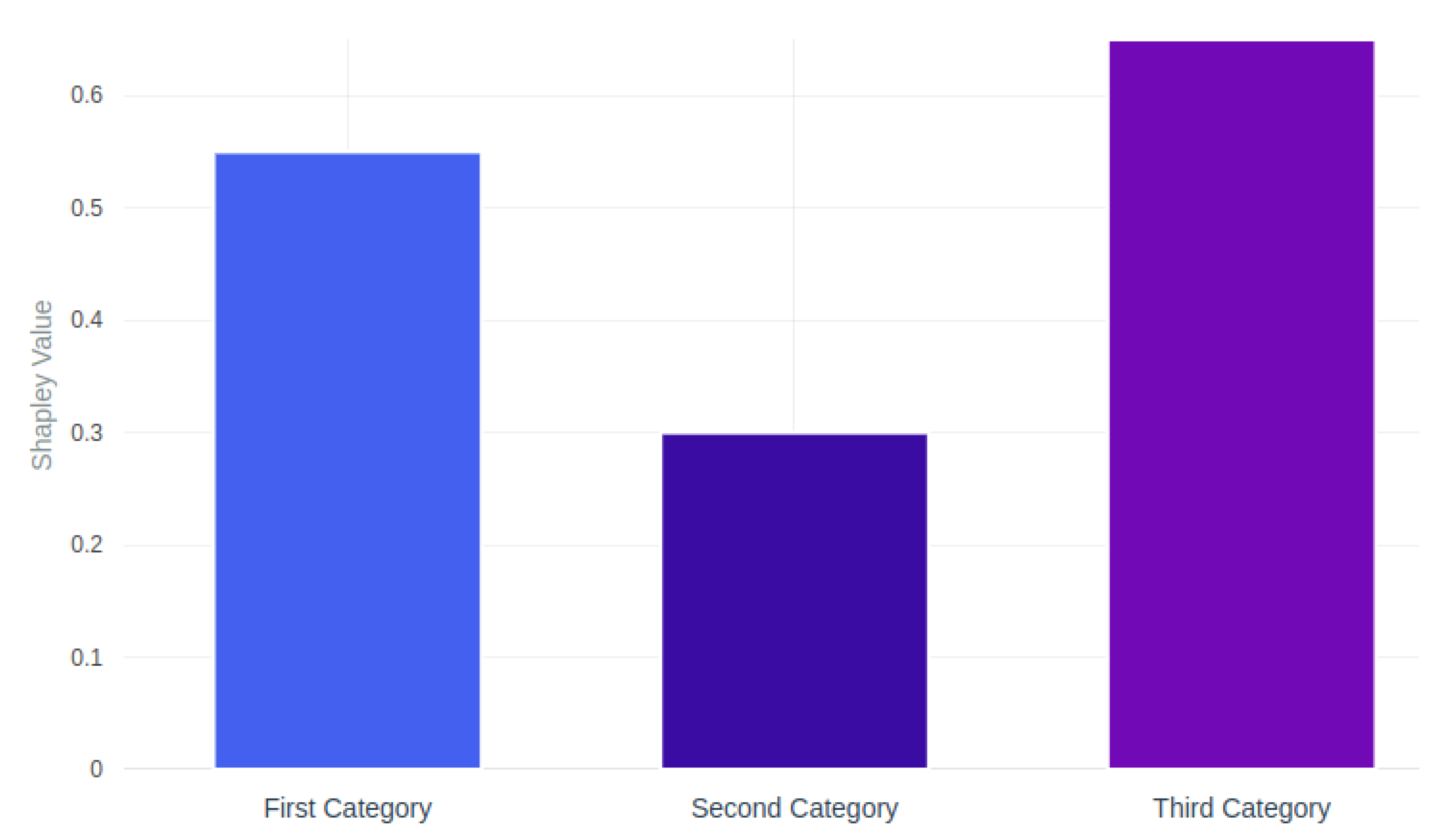
5.3. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moser, F.; Bump, J.B. Assessing the World Health Organization: What does the academic debate reveal and is it democratic? Soc. Sci. Med. 2022, 314, 115456. [Google Scholar] [CrossRef] [PubMed]
- Global Antimicrobial Resistance and Use Surveillance System Report; Technical Report; World Health Organization: Geneva, Switzerland, 2021.
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Technical Report; The Review on Antimicrobial Resistance: London, UK, 2016. [Google Scholar]
- Rodriguez-Morales, A.; Bonilla-Aldana, D.; Tiwari, R.; Sah, R.; Rabaan, A.; Dhama, K. COVID-19, an Emerging Coronavirus Infection: Current Scenario and Recent Developments—An Overview. J. Pure Appl. Microbiol. 2020, 14, 6150. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Alenazy, M.F.; Alshehri, A.A.; Alshahrani, M.A.; Al-Subaie, M.F.; Alrasheed, H.A.; Al Kaabi, N.A.; Thakur, N.; Bouafia, N.A.; Alissa, M.; et al. An updated review on pathogenic coronaviruses (CoVs) amid the emergence of SARS-CoV-2 variants: A look into the repercussions and possible solutions. J. Infect. Public Health 2023, 16, 1870–1883. [Google Scholar] [CrossRef] [PubMed]
- Lansbury, L.; Lim, B.; Baskaran, V.; Lim, W.S. Co-infections in people with COVID-19: A systematic review and meta-analysis. J. Infect. 2020, 81, 266–275. [Google Scholar] [CrossRef]
- Suneja, M.; Beekmann, S.E.; Dhaliwal, G.; Miller, A.C.; Polgreen, P.M. Diagnostic delays in infectious diseases. Diagnosis 2022, 9, 332–339. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the Human Gut Microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Sheikh, J.; Tan, T.S.; Malik, S.; Saidin, S.; Chua, L.S. Bacterial Morphology and Microscopic Advancements: Navigating from Basics to Breakthroughs. Microbiol. Immunol. Commun. 2024, 3, 03–41. [Google Scholar] [CrossRef]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms, 16th ed.; Global Edition; Pearson Education Limited: London, UK, 2021. [Google Scholar]
- Hiremath, P.; Bannigidad, P. Identification and classification of cocci bacterial cells in digital microscopic images. Int. J. Comput. Biol. Drug Des. 2011, 4, 262–273. [Google Scholar] [CrossRef]
- Qian, J.; Wang, Y.; Hu, Z.; Shi, T.; Wang, Y.; Ye, C.; Huang, H. Bacillus sp. as a microbial cell factory: Advancements and future prospects. Biotechnol. Adv. 2023, 69, 108278. [Google Scholar] [CrossRef]
- Podkopaeva, D.; Grabovich, M.; Dubinina, G.; Lysenko, A.; Tourova, T.; Kolganova, T. Two new species of microaerophilic sulfur spirilla, Spirillum winogradskii sp. nov. and Spirillum kriegii sp. nov. Microbiology 2006, 75, 172–179. [Google Scholar] [CrossRef]
- Loughran, A.J.; Orihuela, C.J.; Tuomanen, E.I. Streptococcus pneumoniae: Invasion and Inflammation. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Archambaud, C.; Nunez, N.; da Silva, R.A.G.; Kline, K.A.; Serror, P. Enterococcus faecalis: An overlooked cell invader. Microbiol. Mol. Biol. Rev. 2024, 88, e00069-24. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Medical Microbiology, 9th ed.; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Justice, S.; Hunstad, D.; Cegelski, L.; Hultgren, S. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 2008, 6, 162–168. [Google Scholar] [CrossRef]
- Kocur, M.; Kloos, W.; Schleifer, K. The Genus Micrococcus. In The Prokaryotes; Springer: New York, NY, USA, 2006; Volume 3, pp. 961–971. [Google Scholar] [CrossRef]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Gurvichz, B. Serotherapy for epidemic cerebrospinal meningitis. Kazan Med. J. 2021, 32, 52–53. [Google Scholar] [CrossRef]
- Brouwer, S.; Hernandez, T.; Curren, B.; Harbison-Price, N.; De Oliveira, D.; Jespersen, M.; Davies, M.; Walker, M. Pathogenesis, epidemiology and control of Group A Streptococcus infection. Nat. Rev. Microbiol. 2023, 21, 431–447. [Google Scholar] [CrossRef]
- Chan, H.P.; Samala, R.K.; Hadjiiski, L.M.; Zhou, C. Deep Learning in Medical Image Analysis. In Deep Learning in Medical Image Analysis; Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2020; Volume 1213, pp. 3–21. [Google Scholar] [CrossRef]
- Mohamad, N.A.; Jusoh, N.A.; Htike, Z.Z.; Win, S.L. Bacteria Identification From Microscopic Morphology: A Survey. Int. J. Soft Comput. Artif. Intell. Appl. (IJSCAI) 2014, 3, 12. [Google Scholar] [CrossRef]
- Bottone, E.J. Bacillus cereus, a Volatile Human Pathogen. Clin. Microbiol. Rev. 2010, 23, 382–398. [Google Scholar] [CrossRef]
- Singhal, N.; Kumar, M.; Kanaujia, P.; Virdi, J. MALDI-TOF mass spectrometry: An emerging technology for microbial identification and diagnosis. Front. Microbiol. 2015, 6, 791. [Google Scholar] [CrossRef]
- Rahman, M.; Uddin, M.; Sultana, R.; Moue, A.; Setu, M. Polymerase Chain Reaction (PCR): A Short Review. Anwer Khan Mod. Med. Coll. J. 2013, 4, 30–36. [Google Scholar] [CrossRef]
- Mandlik, J.; Patil, A.; Singh, S. Next-Generation Sequencing (NGS): Platforms and Applications. J. Pharm. Bioallied Sci. 2024, 16, S41–S45. [Google Scholar] [CrossRef] [PubMed]
- Truong, A.; Walters, A.; Goodsitt, J.; Hines, K.; Bruss, C.; Farivar, R. Towards Automated Machine Learning: Evaluation and Comparison of AutoML Approaches and Tools. In Proceedings of the 2019 IEEE 31st International Conference on Tools with Artificial Intelligence (ICTAI), Portland, OR, USA, 4–6 November 2019; pp. 1471–1479. [Google Scholar] [CrossRef]
- He, X.; Zhao, K.; Chu, X. AutoML: A Survey of the State-of-the-Art. arXiv 2019, arXiv:1908.00709. [Google Scholar] [CrossRef]
- Baratchi, M.; Wang, C.; Limmer, S.; van Rijn, J.N.; Hoos, H.H.; Bäck, T.; Olhofer, M. Automated machine learning: Past, present and future. Artif. Intell. Rev. 2024, 57, 122. [Google Scholar] [CrossRef]
- Kumar, S.; Das, P. Duration of bacterial infections and the need for rapid diagnostics. J. Clin. Microbiol. 2006, 44, 2578–2583. [Google Scholar]
- Wang, X.; Shi, Y.; Guo, S.; Qu, X.; Xie, F.; Duan, Z.; Hu, Y.; Fu, H.; Shi, X.; Quan, T.; et al. A Clinical Bacterial Dataset for Deep Learning in Microbiological Rapid On-Site Evaluation. Sci. Data 2024, 11, 608. [Google Scholar] [CrossRef]
- Shaily, T.; Kala, S. Bacterial Image Classification Using Convolutional Neural Networks. In Proceedings of the 2020 IEEE 17th India Council International Conference (INDICON), New Delhi, India, 10–13 December 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Visitsattaponge, S.; Bunkum, M.; Pintavirooj, C.; Paing, M.P. A Deep Learning Model for Bacterial Classification Using Big Transfer (BiT). IEEE Access 2024, 12, 15609–15621. [Google Scholar] [CrossRef]
- Spahn, C.; Laine, R.; Pereira, P.; Gómez de Mariscal, E.; Chamier, L.; Conduit, M.; Pinho, M.; Holden, S.; Jacquemet, G.; Heilemann, M.; et al. DeepBacs: Bacterial image analysis using open-source deep learning approaches. Commun. Biol. 2022, 5, 688. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, 3, 610–621. [Google Scholar] [CrossRef]
- Hu, M.K. Visual Pattern Recognition by Moment Invariants. IRE Trans. Inf. Theory 1962, 8, 179–187. [Google Scholar]
- Trattner, S.; Greenspan, H.; Tepper, G.; Abboud, S. Statistical Imaging for Modeling and Identification of Bacterial Types. In Medical Imaging 2004: Image Processing; Fitzpatrick, J.M., Reinhardt, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2004; Volume 3117, pp. 329–340. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- O’Shea, K.; Nash, R. An Introduction to Convolutional Neural Networks. arXiv 2015, arXiv:1511.08458. [Google Scholar]
- Purwono, P.; Ma’arif, A.; Rahmaniar, W.; Imam, H.; Fathurrahman, H.I.K.; Frisky, A.; Haq, Q.M.U. Understanding of Convolutional Neural Network (CNN): A Review. Int. J. Robot. Control Syst. 2023, 2, 739–748. [Google Scholar] [CrossRef]
- Talo, M. An Automated Deep Learning Approach for Bacterial Image Classification. arXiv 2019, arXiv:1912.08765. [Google Scholar] [CrossRef]
- Sarker, M.I.; Khan, M.M.R.; Prova, S.; Khan, M.; Morshed, M.; Reza, A.W.; Arefin, M. Utilizing Deep Learning for Microscopic Image Based Bacteria Species Identification. In Proceedings of the 2024 International Conference on Computing, Power and Advanced Systems (COMPAS), Cox’s Bazar, Bangladesh, 25–26 September 2024; IEEE: New York, NY, USA, 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Zhang, H.; Qie, Y. Applying Deep Learning to Medical Imaging: A Review. Appl. Sci. 2023, 13, 10521. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Buda, M.; Saha, A.; Bashir, M.R. Deep learning in radiology: An overview of the concepts and a survey of the state of the art with focus on MRI. J. Magn. Reson. Imaging 2019, 49, 939–954. [Google Scholar] [CrossRef]
- Samek, W.; Montavon, G.; Vedaldi, A.; Hansen, L.K.; Müller, K.-R. Explainable AI: Interpreting, Explaining and Visualizing Deep Learning Models. Digit. Signal Process. 2019, 93, 1–15. [Google Scholar]
- Kotwal, D.; Rani, P.; Arif, T.; Manhas, D. Machine Learning and Deep Learning Based Hybrid Feature Extraction and Classification Model Using Digital Microscopic Bacterial Images. SN Comput. Sci. 2023, 4, 21. [Google Scholar] [CrossRef]
- Tammineedi, V.S.V.; Naureen, A.; Ashraf, M.S.; Manna, S.; Mateen Buttar, A.; Muneeshwari, P.; Ahmad, M.W. Biomedical Microscopic Imaging in Computational Intelligence Using Deep Learning Ensemble Convolution Learning-Based Feature Extraction and Classification. Comput. Intell. Neurosci. 2022, 2022, 3531308. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Virtual Event, 3–7 May 2021. [Google Scholar]
- He, K.; Gan, C.; Li, Z.; Rekik, I.; Yin, Z.; Ji, W.; Gao, Y.; Wang, Q.; Zhang, J. Transformers in Medical Image Analysis: A Review. arXiv 2022, arXiv:2202.12165. [Google Scholar] [CrossRef]
- Hutter, F.; Kotthoff, L.; Vanschoren, J. (Eds.) Automated Machine Learning: Methods, Systems, Challenges; The Springer Series on Challenges in Machine Learning; Springer: Cham, Switzerland, 2019; ISBN 978-3-030-05318-5. [Google Scholar] [CrossRef]
- Demir, C.; Yener, B. Automated Cancer Diagnosis Based on Histopathological Images: A Systematic Survey. Pattern Recognit. 2004, 37, 1033–1049. [Google Scholar] [CrossRef]
- Khan, A.; Dwivedi, P.; Mugde, S.; S a, S.; Sharma, G.; Soni, G. Toward Automated Machine Learning for Genomics: Evaluation and Comparison of State-of-the-Art AutoML Approaches. In Automated and AI-Based Approaches for Bioinformatics and Biomedical Research; Academic Press: Cambridge, MA, USA, 2023; pp. 129–152. [Google Scholar] [CrossRef]
- Elangovan, K.; Lim, G.; Ting, D. A Comparative Study of an On Premise AutoML Solution for Medical Image Classification. Sci. Rep. 2024, 14, 10483. [Google Scholar] [CrossRef] [PubMed]
- Laccourreye, P.; Bielza, C.; Larranaga, P. Explainable Machine Learning for Longitudinal Multi-Omic Microbiome. Mathematics 2022, 10, 1994. [Google Scholar] [CrossRef]
- Smith, K.P.; Kirby, J.E. Human Error in Clinical Microbiology. J. Clin. Microbiol. 2020, 58, e00342-20. [Google Scholar]
- Hameurlaine, M.; Moussaoui, A.; Benaissa, S. Deep Learning for Medical Image Analysis. In Proceedings of the 4th International Conference on Recentt Advances iin Ellecttrriicall Systtems; 2019. Available online: https://www.researchgate.net/publication/338490461_Deep_Learning_for_Medical_Image_Analysis (accessed on 13 June 2025).
- Tatanov, O.; Samarin, A. LFIEM: Lightweight Filter-based Image Enhancement Model. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; pp. 873–878. [Google Scholar] [CrossRef]
- Samarin, A.; Nazarenko, A.; Savelev, A.; Toropov, A.; Dzestelova, A.; Mikhailova, E.; Motyko, A.; Malykh, V. A Model Based on Universal Filters for Image Color Correction. Pattern Recognit. Image Anal. 2024, 34, 844–854. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, H.; Chen, I.; Yang, W. Active Contours with Thresholding Value for Image Segmentation. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 2266–2269. [Google Scholar] [CrossRef]
- Li, Y.; Bi, Y.; Zhang, W.; Sun, C. Multi-Scale Anisotropic Gaussian Kernels for Image Edge Detection. IEEE Access 2020, 8, 1803–1812. [Google Scholar] [CrossRef]
- Schamberger, B.; Ziege, R.; Anselme, K.; Ben Amar, M.; Bykowski, M.; Castro, A.; Cipitria, A.; Coles, R.; Dimova, R.; Eder, M.; et al. Curvature in Biological Systems: Its Quantification, Emergence, and Implications across the Scales. Adv. Mater. 2023, 35, 2206110. [Google Scholar] [CrossRef]
- Hearst, M.; Dumais, S.; Osuna, E.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. Their Appl. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Huang, M. Theory and Implementation of linear regression. In Proceedings of the 2020 International Conference on Computer Vision, Image and Deep Learning (CVIDL), Chongqing, China, 10–12 July 2020; pp. 210–217. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Scabini, L.F.; Bruno, O.M. Structure and performance of fully connected neural networks: Emerging complex network properties. Phys. A: Stat. Mech. Its Appl. 2023, 615, 128585. [Google Scholar] [CrossRef]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q.V. Randaugment: Practical automated data augmentation with a reduced search space. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 14–19 June 2020; pp. 3008–3017. [Google Scholar] [CrossRef]
- Psaroudakis, A.; Kollias, D. MixAugment & Mixup: Augmentation Methods for Facial Expression Recognition. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), New Orleans, LA, USA, 19–20 June 2022; pp. 2366–2374. [Google Scholar] [CrossRef]
- Yun, S.; Han, D.; Oh, S.J.; Chun, S.; Choe, J.; Yoo, Y. CutMix: Regularization Strategy to Train Strong Classifiers with Localizable Features. arXiv 2019, arXiv:1905.04899. [Google Scholar]
- Micikevicius, P.; Narang, S.; Alben, J.; Diamos, G.; Elsen, E.; Garcia, D.; Ginsburg, B.; Houston, M.; Kuchaev, O.; Venkatesh, G.; et al. Mixed Precision Training. arXiv 2017, arXiv:1710.03740. [Google Scholar] [CrossRef]
- Guan, L. Weight Prediction Boosts the Convergence of AdamW. In Advances in Knowledge Discovery and Data Mining; Springer: Cham, Switzerland, 2023; pp. 329–340. [Google Scholar] [CrossRef]
- Liu, Z. Super Convergence Cosine Annealing with Warm-Up Learning Rate. In Proceedings of the CAIBDA 2022; 2nd International Conference on Artificial Intelligence, Big Data and Algorithms, Nanjing, China, 17–19 June 2022; pp. 1–7. [Google Scholar]
- Müller, R.; Kornblith, S.; Hinton, G.E. When Does Label Smoothing Help? arXiv 2019, arXiv:1906.02629. [Google Scholar]
- Zhang, J.; He, T.; Sra, S.; Jadbabaie, A. Why gradient clipping accelerates training: A theoretical justification for adaptivity. arXiv 2020, arXiv:1905.11881. [Google Scholar]
- Bai, Y.; Yang, E.; Han, B.; Yang, Y.; Li, J.; Mao, Y.; Niu, G.; Liu, T. Understanding and Improving Early Stopping for Learning with Noisy Labels. arXiv 2021, arXiv:2106.15853. [Google Scholar]
- Lundberg, S.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. arXiv 2017, arXiv:1705.07874. [Google Scholar] [CrossRef]





| Preprocessing Method Combination | Precision | Recall | F1-Score |
|---|---|---|---|
| No preprocessing | 0.812 | 0.803 | 0.807 |
| Exposure only | 0.845 | 0.832 | 0.838 |
| Sharpness only | 0.827 | 0.818 | 0.822 |
| Contrast only | 0.851 | 0.842 | 0.846 |
| Linear transformation only | 0.836 | 0.824 | 0.830 |
| Trainable kernel only | 0.838 | 0.829 | 0.833 |
| Exposure + Sharpness | 0.867 | 0.854 | 0.860 |
| Exposure + Contrast | 0.882 | 0.871 | 0.876 |
| Sharpness + Contrast | 0.875 | 0.863 | 0.869 |
| Blur + Contrast | 0.841 | 0.833 | 0.837 |
| Linear transformation + Sharpness | 0.853 | 0.841 | 0.847 |
| Trainable kernel + Exposure | 0.864 | 0.853 | 0.858 |
| Exposure + Contrast + Blur | 0.878 | 0.866 | 0.872 |
| Exposure + Linear transformation + Sharpness | 0.881 | 0.870 | 0.875 |
| Exposure + Contrast + Sharpness | 0.910 | 0.901 | 0.905 |
| Exposure + Contrast + Sharpness + Blur | 0.892 | 0.881 | 0.886 |
| All filters combined | 0.885 | 0.874 | 0.879 |
| Filters Configuration | Classifier Model | Params (M) | FLOPs (G) | Precision | Recall | F1 |
|---|---|---|---|---|---|---|
| Exposure + Contrast + Sharpness | MobileNetV3 | 5.4 | 0.22 | 0.723 | 0.698 | 0.710 |
| Exposure + Contrast + Sharpness | InceptionResNetV1 | 27.9 | 5.71 | 0.735 | 0.712 | 0.723 |
| Exposure + Contrast | ResNet152 | 60.2 | 11.31 | 0.748 | 0.725 | 0.736 |
| Exposure + Contrast | EfficientNetB0 | 5.3 | 0.39 | 0.752 | 0.731 | 0.741 |
| Exposure + Contrast | Features Gen + SVM | 0.8 | 0.05 | 0.761 | 0.739 | 0.750 |
| Exposure + Contrast | InceptionResNetV2 | 55.9 | 12.98 | 0.768 | 0.745 | 0.756 |
| Exposure + Contrast + Sharpness | Features Gen + SVM | 0.8 | 0.05 | 0.774 | 0.752 | 0.763 |
| Exposure + Contrast + Sharpness | EfficientNetB0 | 5.3 | 0.39 | 0.781 | 0.760 | 0.770 |
| Exposure + Contrast | ResNet101 | 44.6 | 7.85 | 0.785 | 0.764 | 0.774 |
| Exposure + Contrast + Sharpness | EfficientNetB1 | 7.8 | 0.70 | 0.792 | 0.771 | 0.781 |
| Exposure + Contrast | EfficientNetB2 | 9.2 | 1.01 | 0.798 | 0.778 | 0.788 |
| Exposure + Contrast + Sharpness | ResNet101 | 44.6 | 7.85 | 0.803 | 0.784 | 0.793 |
| Exposure + Contrast + Sharpness | EfficientNetB3 | 12.2 | 1.86 | 0.809 | 0.790 | 0.799 |
| Exposure + Contrast | EfficientNetB4 | 19.3 | 3.39 | 0.815 | 0.796 | 0.805 |
| Exposure + Contrast | CoAtNet | 42.1 | 6.52 | 0.821 | 0.803 | 0.812 |
| Exposure + Contrast | EfficientNetB6 | 43.0 | 10.34 | 0.827 | 0.810 | 0.818 |
| Exposure + Contrast | Features Gen + RF | 1.2 | 0.08 | 0.832 | 0.815 | 0.823 |
| Exposure + Contrast | SE-ResNext50 | 27.6 | 4.25 | 0.838 | 0.821 | 0.829 |
| Exposure + Contrast + Sharpness | ResNet152 | 60.2 | 11.31 | 0.843 | 0.827 | 0.835 |
| Exposure + Contrast + Sharpness | Features Gen + RF | 1.2 | 0.08 | 0.849 | 0.833 | 0.841 |
| Exposure + Contrast | Features Gen + GBM | 1.5 | 0.12 | 0.854 | 0.839 | 0.846 |
| Exposure + Contrast + Sharpness | CoAtNet | 42.1 | 6.52 | 0.860 | 0.845 | 0.852 |
| Exposure + Contrast | ViT-L/16 | 304.3 | 190.7 | 0.866 | 0.852 | 0.859 |
| Exposure + Contrast | EfficientNetB3 | 12.2 | 1.86 | 0.872 | 0.858 | 0.865 |
| Exposure + Contrast + Sharpness | EfficientNetB4 | 19.3 | 3.39 | 0.878 | 0.865 | 0.871 |
| Exposure + Contrast + Sharpness | InceptionResNetV2 | 55.9 | 12.98 | 0.884 | 0.871 | 0.877 |
| Exposure + Contrast + Sharpness | SE-ResNext50 | 27.6 | 4.25 | 0.890 | 0.878 | 0.884 |
| Exposure + Contrast + Sharpness | ViT-L/16 | 304.3 | 190.7 | 0.896 | 0.885 | 0.890 |
| Exposure + Contrast + Sharpness | EfficientNetB6 | 43.0 | 10.34 | 0.902 | 0.892 | 0.897 |
| Exposure + Contrast + Sharpness | Features Gen + AutoML | 1.8 | 0.15 | 0.910 | 0.901 | 0.905 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Samarin, A.; Savelev, A.; Toropov, A.; Dozortseva, A.; Kotenko, E.; Nazarenko, A.; Motyko, A.; Narova, G.; Mikhailova, E.; Malykh, V. Automation of Multi-Class Microscopy Image Classification Based on the Microorganisms Taxonomic Features Extraction. J. Imaging 2025, 11, 201. https://doi.org/10.3390/jimaging11060201
Samarin A, Savelev A, Toropov A, Dozortseva A, Kotenko E, Nazarenko A, Motyko A, Narova G, Mikhailova E, Malykh V. Automation of Multi-Class Microscopy Image Classification Based on the Microorganisms Taxonomic Features Extraction. Journal of Imaging. 2025; 11(6):201. https://doi.org/10.3390/jimaging11060201
Chicago/Turabian StyleSamarin, Aleksei, Alexander Savelev, Aleksei Toropov, Aleksandra Dozortseva, Egor Kotenko, Artem Nazarenko, Alexander Motyko, Galiya Narova, Elena Mikhailova, and Valentin Malykh. 2025. "Automation of Multi-Class Microscopy Image Classification Based on the Microorganisms Taxonomic Features Extraction" Journal of Imaging 11, no. 6: 201. https://doi.org/10.3390/jimaging11060201
APA StyleSamarin, A., Savelev, A., Toropov, A., Dozortseva, A., Kotenko, E., Nazarenko, A., Motyko, A., Narova, G., Mikhailova, E., & Malykh, V. (2025). Automation of Multi-Class Microscopy Image Classification Based on the Microorganisms Taxonomic Features Extraction. Journal of Imaging, 11(6), 201. https://doi.org/10.3390/jimaging11060201






