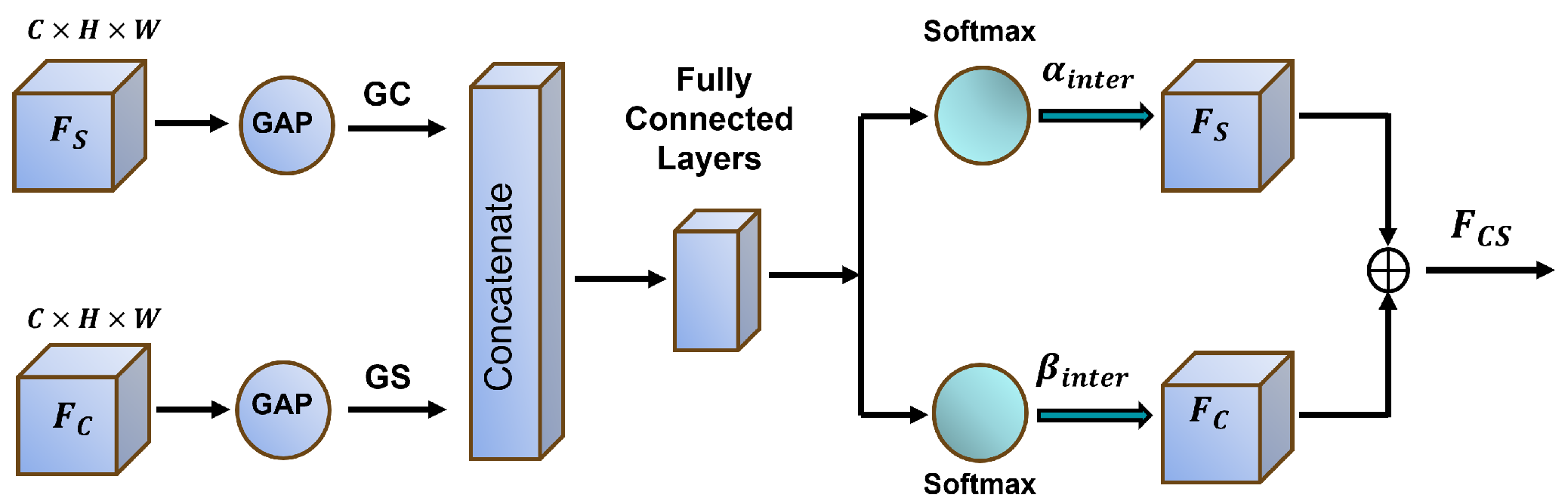
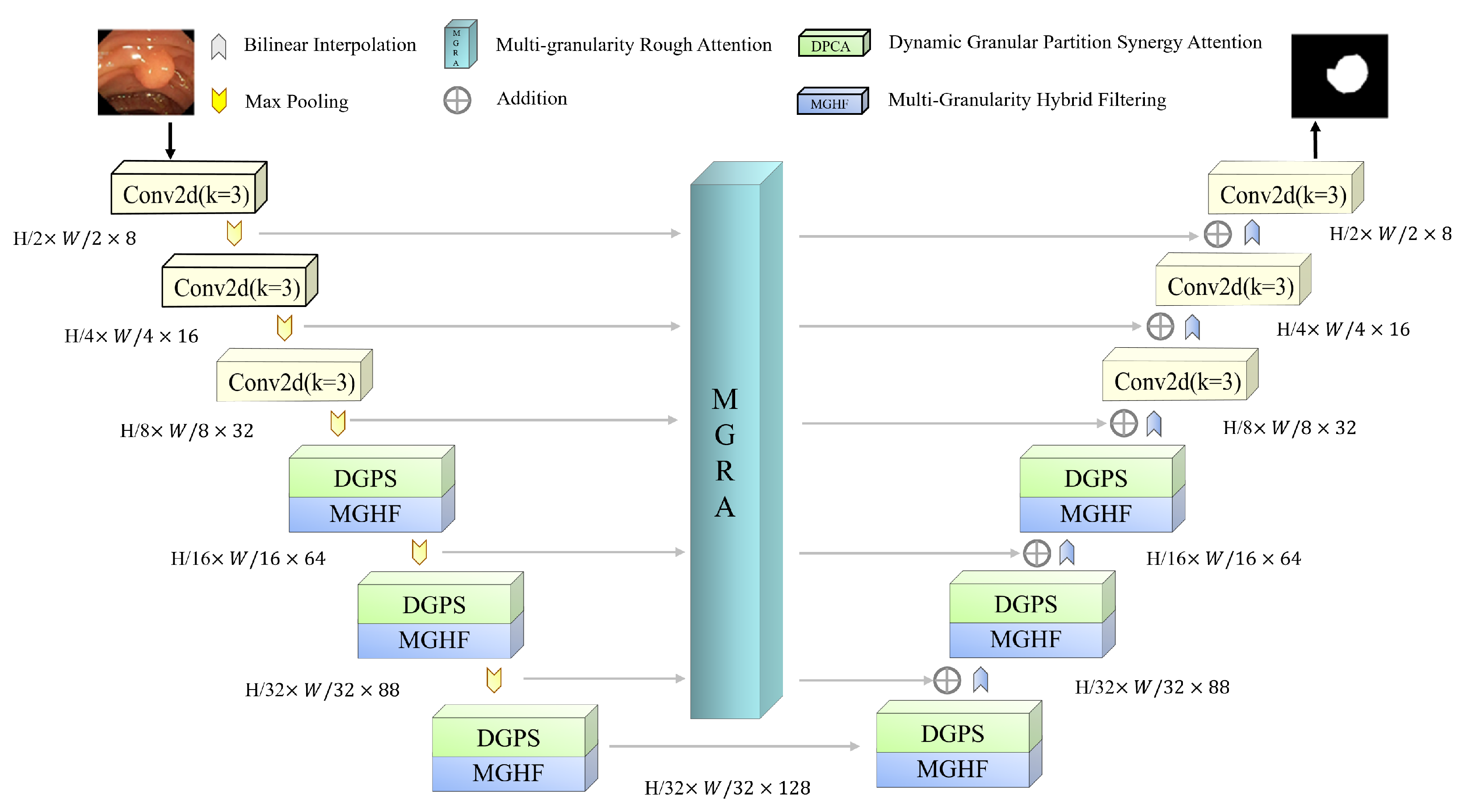
1. Introduction
Polyps are abnormal growths in gastrointestinal tissues and are considered the major precursors of colorectal cancer (CRC), posing a serious threat to patient health [
1]. CRC is one of the most common malignant tumours worldwide, ranking third in incidence and second in mortality [
2]. Although CRC generally develops slowly, early detection and removal of polyps can significantly reduce the risk of malignant transformation [
3]. As the current standard screening method, colonoscopy effectively identifies and removes polyps. However, its detection accuracy is influenced by factors such as bowel preparation quality and physician experience.
In polyp segmentation tasks, complex background interference and the diverse characteristics of polyps pose significant technical challenges [
4]. Polyps exhibit substantial variations in shape and size, with smaller polyps often misclassified as background or noise due to their indistinct features [
5,
6]. Moreover, the color and texture of polyps frequently resemble those of surrounding normal tissues, making boundary identification particularly difficult [
7,
8]. The distribution of polyps is also complex, especially in intestinal folds or curved regions, further complicating the segmentation task [
9]. Therefore, developing efficient and accurate polyp segmentation techniques is essential for improving clinical diagnostic efficiency and preventing colorectal cancer. The accuracy of segmentation directly impacts clinical decision-making and treatment outcomes, making it an indispensable component of modern healthcare systems [
10].
In recent years, deep learning has made significant progress in polyp segmentation. With their powerful feature extraction capabilities, convolutional neural networks (CNNs) have provided practical solutions for medical image segmentation. For example, fully convolutional network (FCNs) [
11] were the first to achieve end-to-end pixel-level prediction, while UNet [
12] introduced an encoder–decoder structure with skip connections, significantly improving segmentation accuracy. Subsequently, U-Net++ [
13] enhanced multi-level feature fusion by incorporating dense skip connections and nested decoders.
Despite the effectiveness of CNNs, traditional methods primarily rely on local convolution operations, which are constrained by a limited receptive field and struggle to capture global contextual information effectively. This limitation is particularly problematic when dealing with polyps with complex morphologies or small sizes, as they are susceptible to background interference, reducing segmentation accuracy. To address these challenges, researchers have proposed various improvements. For instance, PraNet [
14] employs a Recurrent Reverse Attention (RRA) mechanism to enhance boundary regions iteratively, improving segmentation consistency, reducing fragmentation, and mitigating boundary blurring. FANet [
15] design a feedback mechanism that allows the network to adjust feature extraction at different levels adaptively, optimizing feature fusion across layers and enhancing segmentation accuracy. However, despite leveraging contextual information to some extent, PraNet and FANet struggle to network long-range dependencies effectively and lack efficient multi-scale feature modelling, limiting their performance when handling lesions with significant scale variations.
ICGNet [
16] design Contour Contra-information to enhance target region perception in low-contrast scenarios. Its adversarial learning strategy improves the discrimination between target regions and backgrounds, integrating global and local features to refine polyp boundary details. However, this method primarily relies on color features for target recognition, which limits its generalizability in cases with significant color variations or complex backgrounds. To reduce dependency on color information, UM-Net [
17] proposes a morphology-based feature extraction method, incorporating a specialized morphological feature extraction module to enhance polyp edge and structural modeling. However, its contextual modeling remains constrained to a single scale, limiting its ability to handle large-scale or morphologically complex polyps, which affects segmentation accuracy and boundary delineation. DLGRAFE-Net [
18] integrates a graph attention mechanism to improve adaptability to polyps of varying sizes and shapes through global context modeling and multi-level feature fusion. Its graph-based feature interaction mechanism effectively enhances spatial relationship modeling within polyp regions while mitigating the limitations of single-scale information. However, as this method heavily relies on global feature modeling, it may struggle to accurately capture fine-grained details in noisy backgrounds or low-contrast conditions, leading to imprecise boundary localization.
In recent years, Transformer-based networks and self-attention mechanisms have demonstrated powerful capabilities in multi-scale feature extraction and long-range dependency modeling. FCB-SwinV2 [
19] combines the strengths of CNNs and Transformers, leveraging self-attention mechanisms for global feature interaction to improve the accuracy of polyp segmentation. MSNet [
20] further integrates CNN and Transformer architectures by introducing a Multi-Scale Perception Module and a Boundary Enhancement Module to optimize lesion boundary segmentation. However, since self-attention mechanisms tend to emphasize global information, local boundary details may be diluted during feature propagation, reducing the ability to delineate small-scale lesions. Additionally, the high computational complexity of self-attention mechanisms makes it challenging for networks to efficiently adapt to lesion variations across different scales. Therefore, a key challenge remains how to enhance local boundary details while preserving global contextual modeling capabilities to improve the segmentation of ambiguous boundaries.
In computer vision, extracting multi-level and multi-scale features from images is a core task. Deep neural networks inherently achieve multi-granularity feature extraction through hierarchical learning and multi-level transformations, where low-level features are combined to form abstract representations, thereby obtaining optimal feature representations [
21,
22]. Building on this insight, we propose a Synergistic Multi-Granularity Rough Attention U-Net (S-MGRAUNet) to address the aforementioned challenges. This method is based on the classical UNet framework and integrates a Multi-Granularity Hybrid Filtering (MGHF) module, a Dynamic Granular Partition Synergistic Attention (DGPS) mechanism, and a Multi-Granularity Rough Attention (MGRA) mechanism. These components are designed to optimize multi-scale feature extraction for morphologically complex polyps, enhance target recognition in low-contrast and noisy backgrounds, and improve global contextual modeling while preserving local boundary details. The main innovations are as follows:
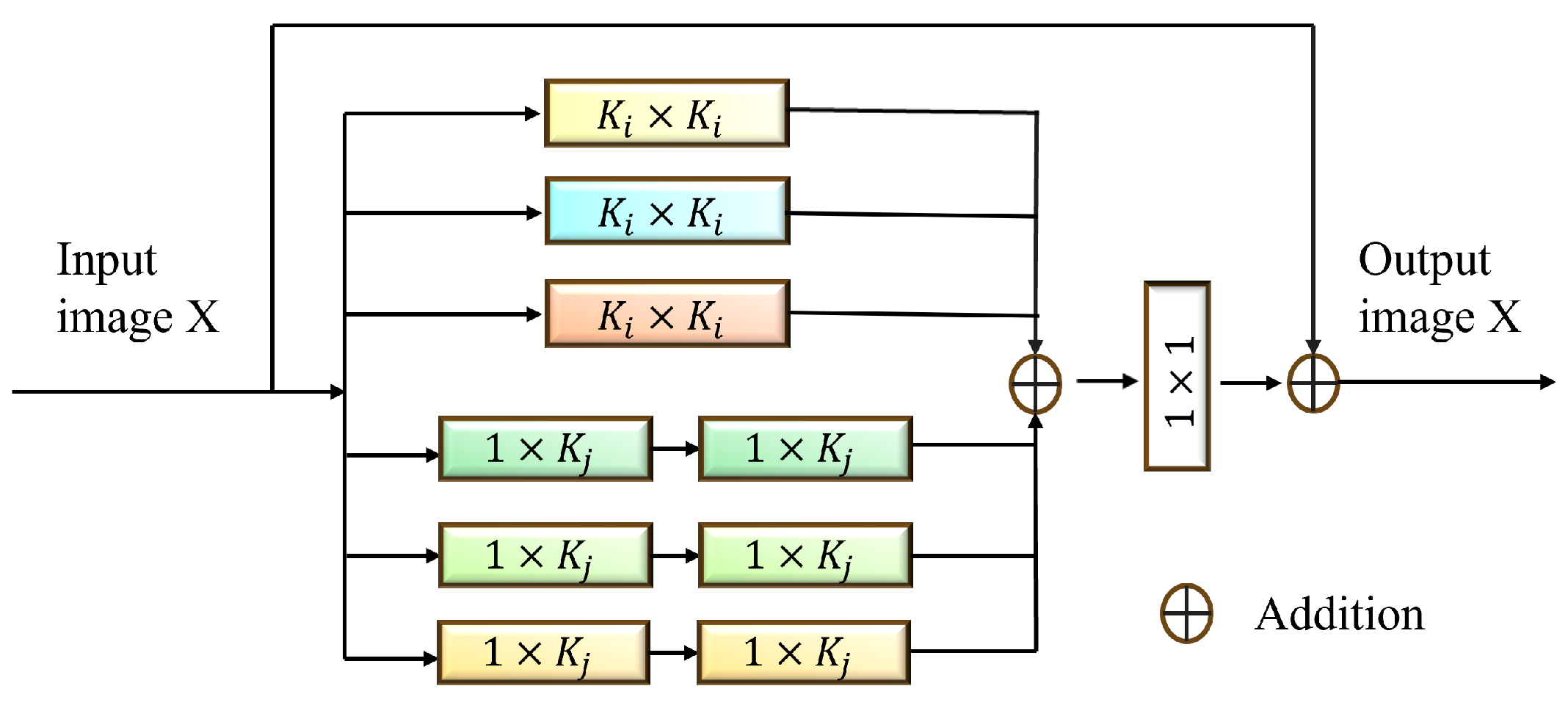
Existing multi-scale fusion methods often suffer from small-scale information loss or large-scale redundancy. To overcome these issues, we designed the MGHF module, which combines multi-scale convolutional kernels and strip convolutions to facilitate cross-granularity information interaction and feature reorganization. MGHF ensures that the network captures the overall structure of polyp regions and fine-grained details, improving adaptability to lesions with diverse morphologies while reducing the computational complexity associated with large convolutional kernels.
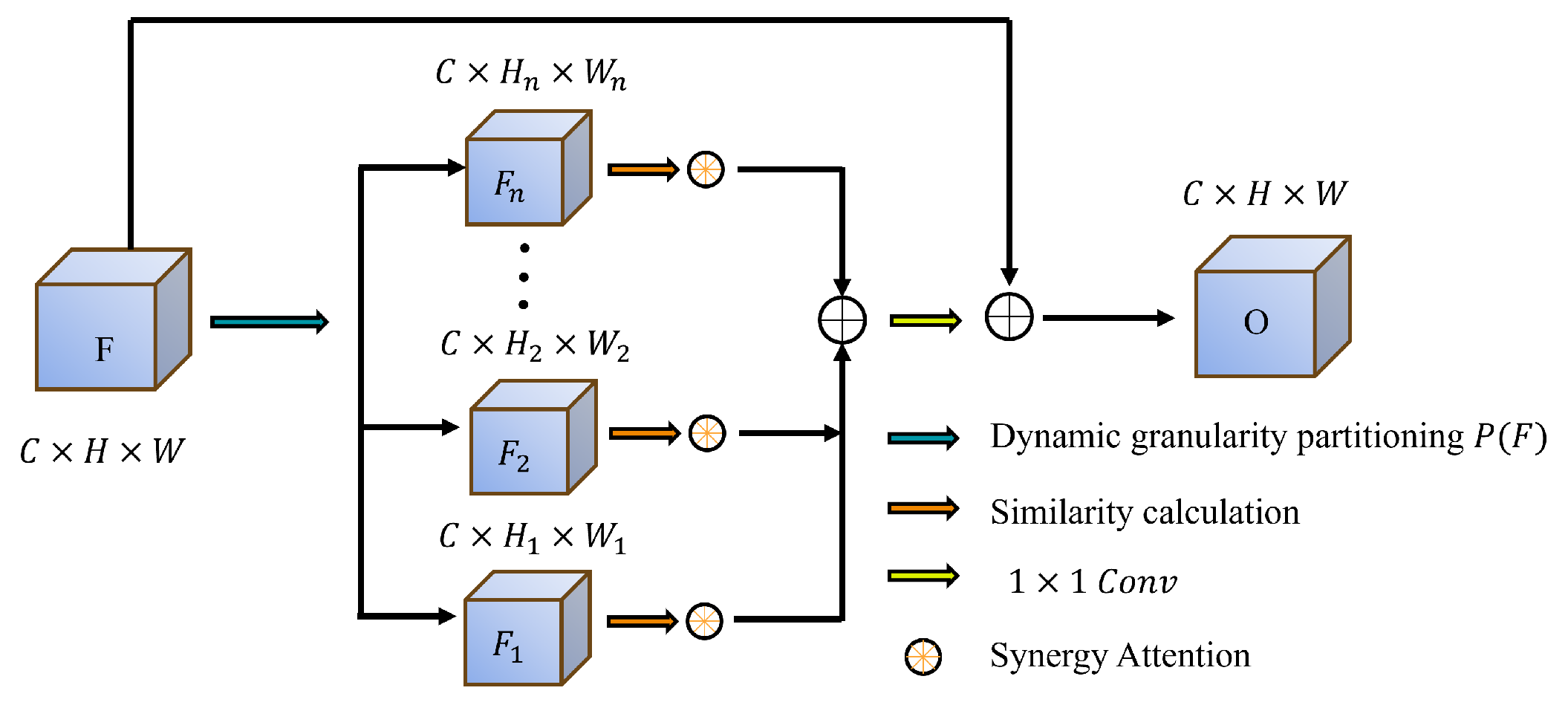
Complex backgrounds and low contrast frequently degrade segmentation performance, leading to inaccurate target region identification. To address this, we propose the DGPS mechanism, which dynamically adjusts the granularity partitioning strategy based on the distribution of polyp region features and integrates local and global information to enhance the distinction between targets and backgrounds. This approach significantly improves segmentation performance in low-contrast scenarios while reducing mis-segmentation errors.
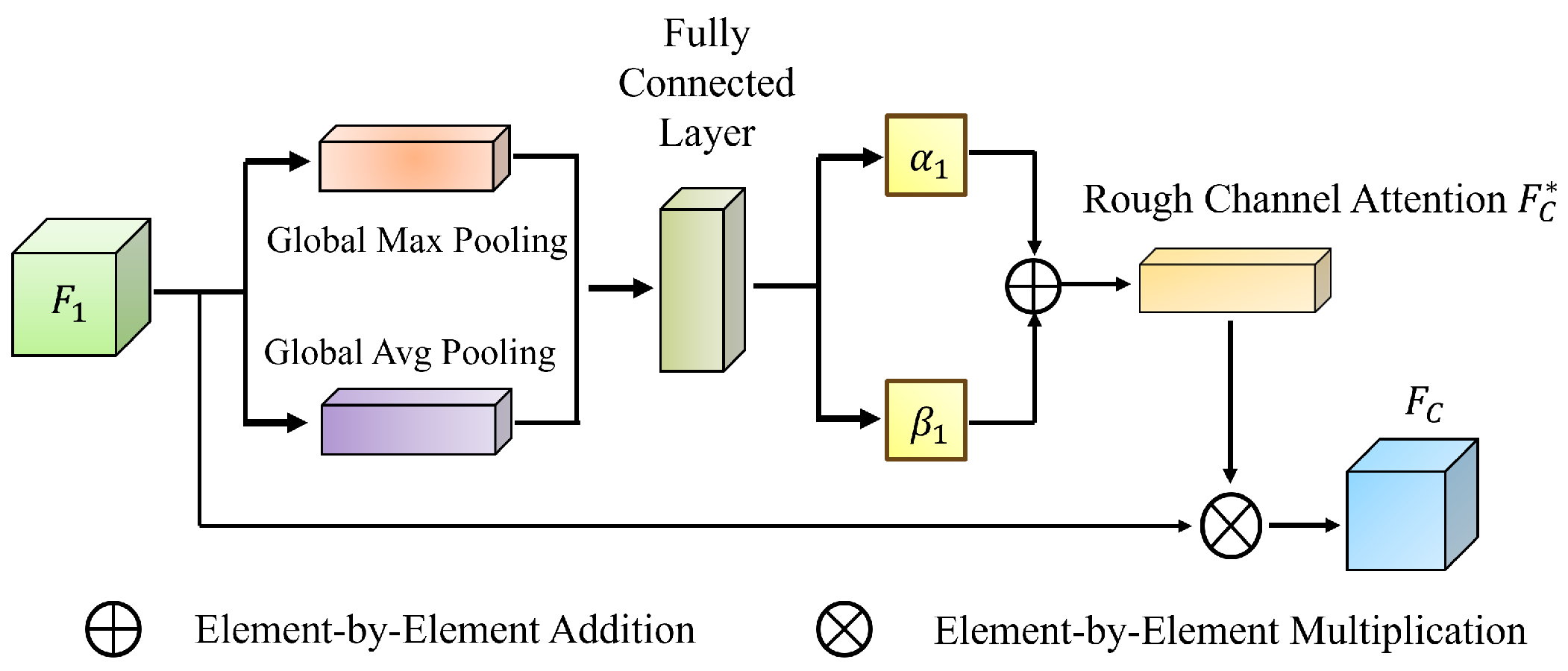
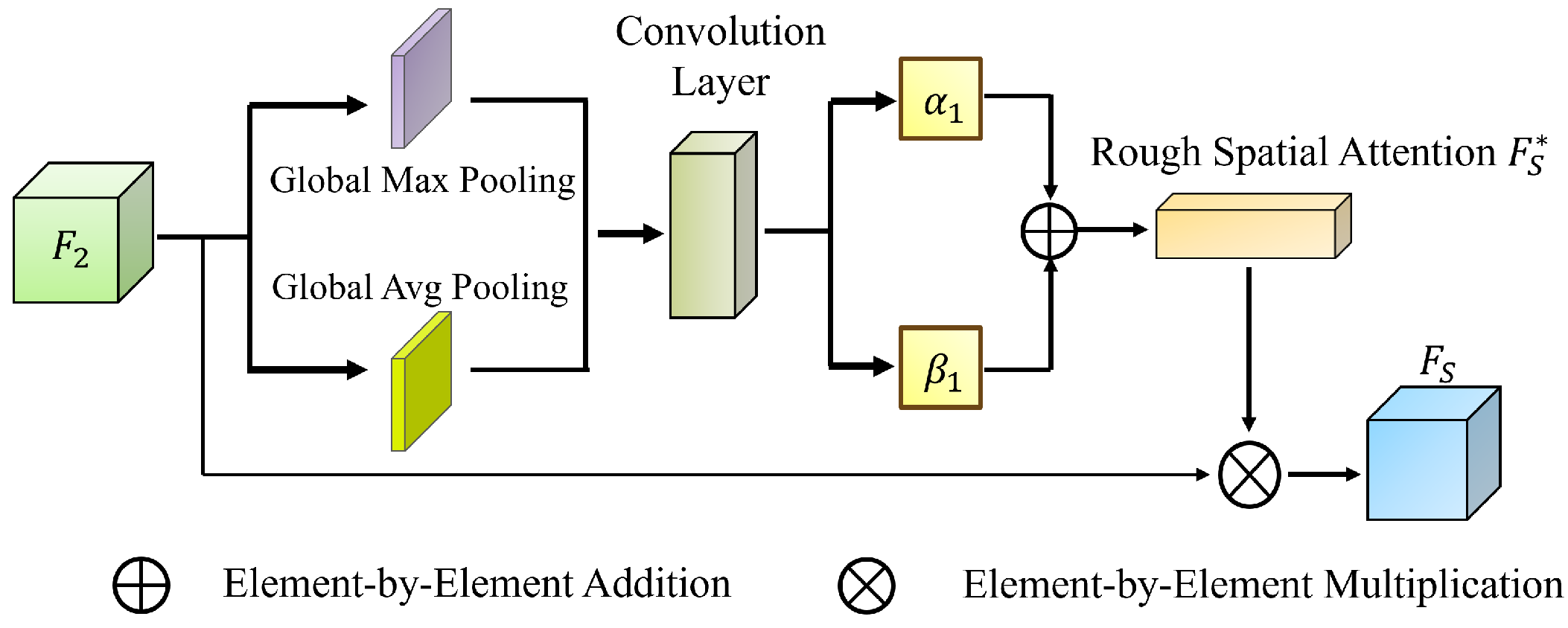
Leveraging rough set theory, we introduce the MGRA mechanism, which employs a coarse-to-fine progressive feature refinement strategy. Initially, coarse-grained features are used for rough localization of the polyp region (upper approximation). Subsequently, fine-grained features iteratively refine boundary details (lower approximation), enhancing the distinction between polyp regions and normal tissues. This strategy improves the network’s ability to handle ambiguous boundaries and reduces computational complexity while ensuring high segmentation accuracy.
The structure of this paper is arranged as follows:
Section 2 reviews related research work.
Section 3 provides a detailed introduction to the architecture of S-MGRAUNet, its core modules, and the experimental setup.
Section 4 analyzes the experimental results and discussion, while also evaluating the model’s performance on benchmark datasets.
Section 5 concludes the study and outlines future research directions.
5. Conclusions
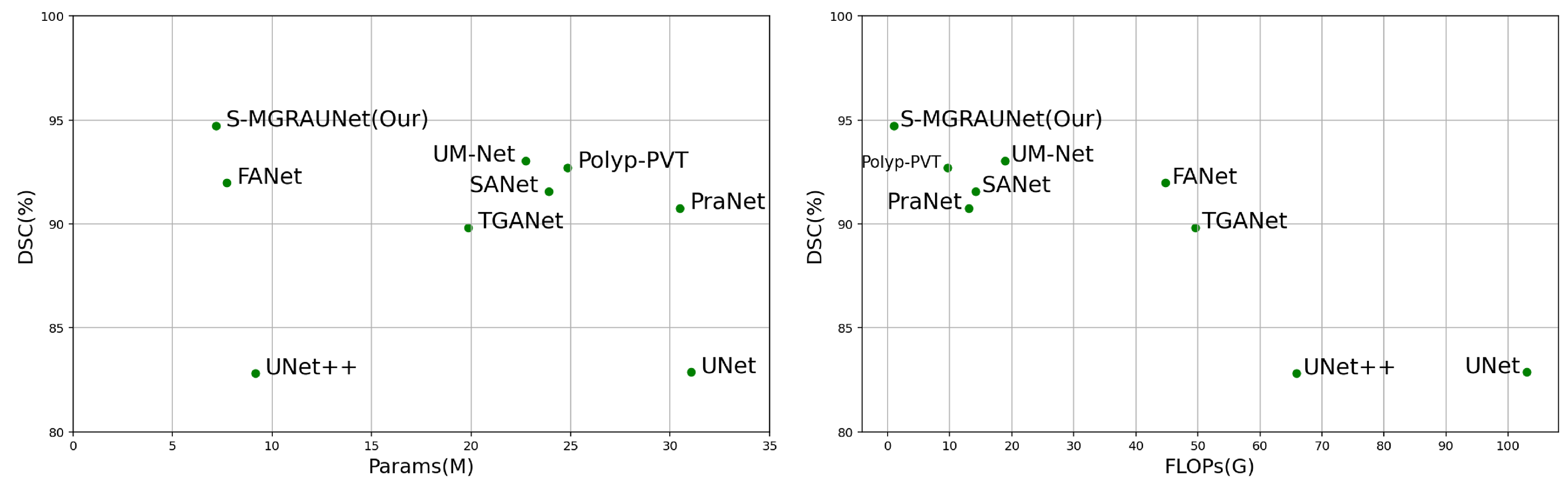
This study proposes the S-MGRAUNet network to improve the accuracy of polyp segmentation while reducing computational complexity. Compared with traditional U-Net and its variants, S-MGRAUNet integrates MGHF, DGPS, and MGRA to achieve efficient feature extraction, contextual modeling, and boundary refinement. Experimental results show that the proposed method performs excellently on multiple benchmark datasets, particularly in boundary refinement and maintaining the integrity of target regions.
Compared with existing methods, traditional deep learning networks for polyp segmentation mainly rely on global or local features, making it difficult to capture multi-scale information simultaneously. As a result, small polyps or polyps with unclear boundaries are prone to over-segmentation or under-segmentation. Although multi-scale fusion and attention mechanisms can improve segmentation accuracy, they often come at the cost of increased computational complexity. In this study, the MGHF module combines small-scale convolutions with strip convolutions, which can approximate large-scale convolutions, reducing complexity while enhancing the representation capability of polyp morphology from a directional perspective. The DGPS mechanism optimizes feature interaction between polyps and the background through dynamic weight allocation, improving the model’s ability to focus on important features and thus enhancing segmentation robustness. At the same time, this mechanism adaptively adjusts weights, reducing computational redundancy caused by fixed weights and further lowering resource consumption. MGRA first utilizes coarse-grained features to quickly locate polyps and then gradually refines their boundaries to improve computational efficiency.
Experimental results indicate that S-MGRAUNet not only improves segmentation quality but also enhances the transparency of network decision-making, providing new insights for computer-aided diagnosis. In the future, we will further investigate the adaptability of this method to large-scale datasets and different modalities to enhance its generalization ability and clinical application value. Additionally, this study integrates multi-granularity knowledge from granular computing and rough set theory with U-Net as the backbone network, providing potential theoretical support for our subsequent research on the interpretability of neural networks.