Dual-Path Convolutional Neural Network with Squeeze-and-Excitation Attention for Lung and Colon Histopathology Classification
Abstract
1. Introduction
- A lightweight, end-to-end dual-path CNN with asymmetric kernel scaling is proposed to enable efficient and effective multi-scale feature extraction from histopathological images.
- A cross-path attention design is introduced by integrating a Squeeze-and-Excitation (SE) block after feature fusion to dynamically recalibrate and enhance discriminative multi-scale representations.
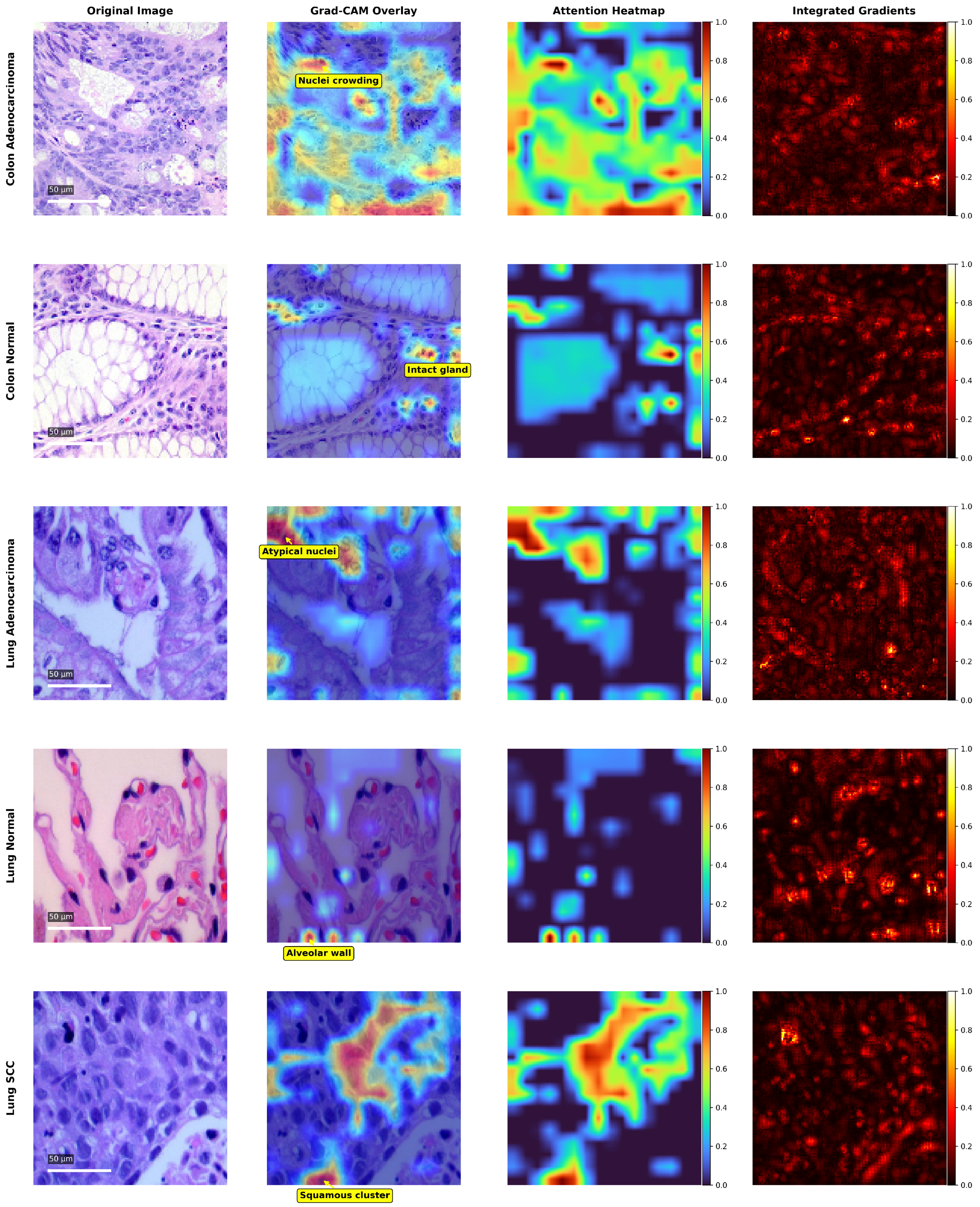
- Multiple explainable AI (XAI) techniques, including Grad-CAM, attention heatmaps, and Integrated Gradients, are incorporated to ensure that the model’s focus aligns with clinically relevant tissue structures.
- A computationally efficient training pipeline is developed to improve convergence stability and mitigate overfitting through adaptive learning strategies and callback mechanisms.
2. Related Work
3. Materials and Methods
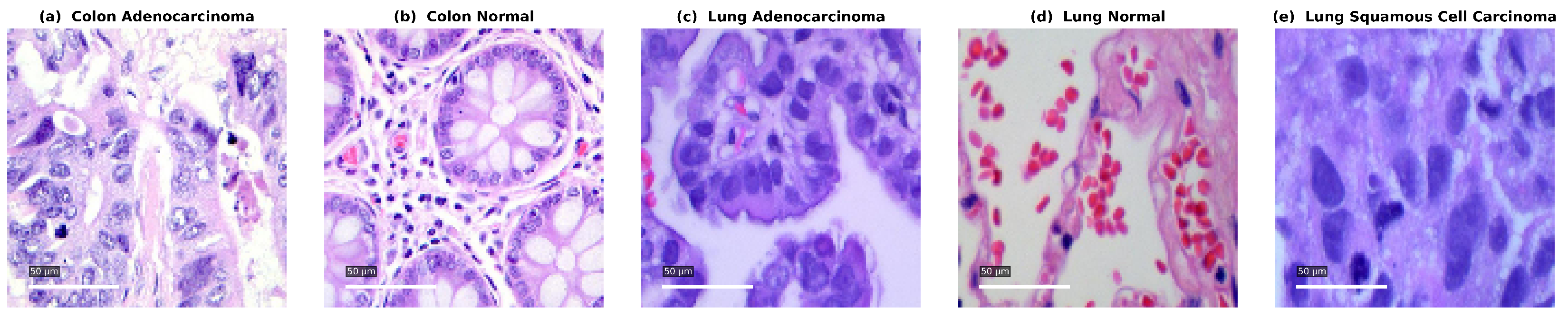
3.1. Dataset and Preprocessing
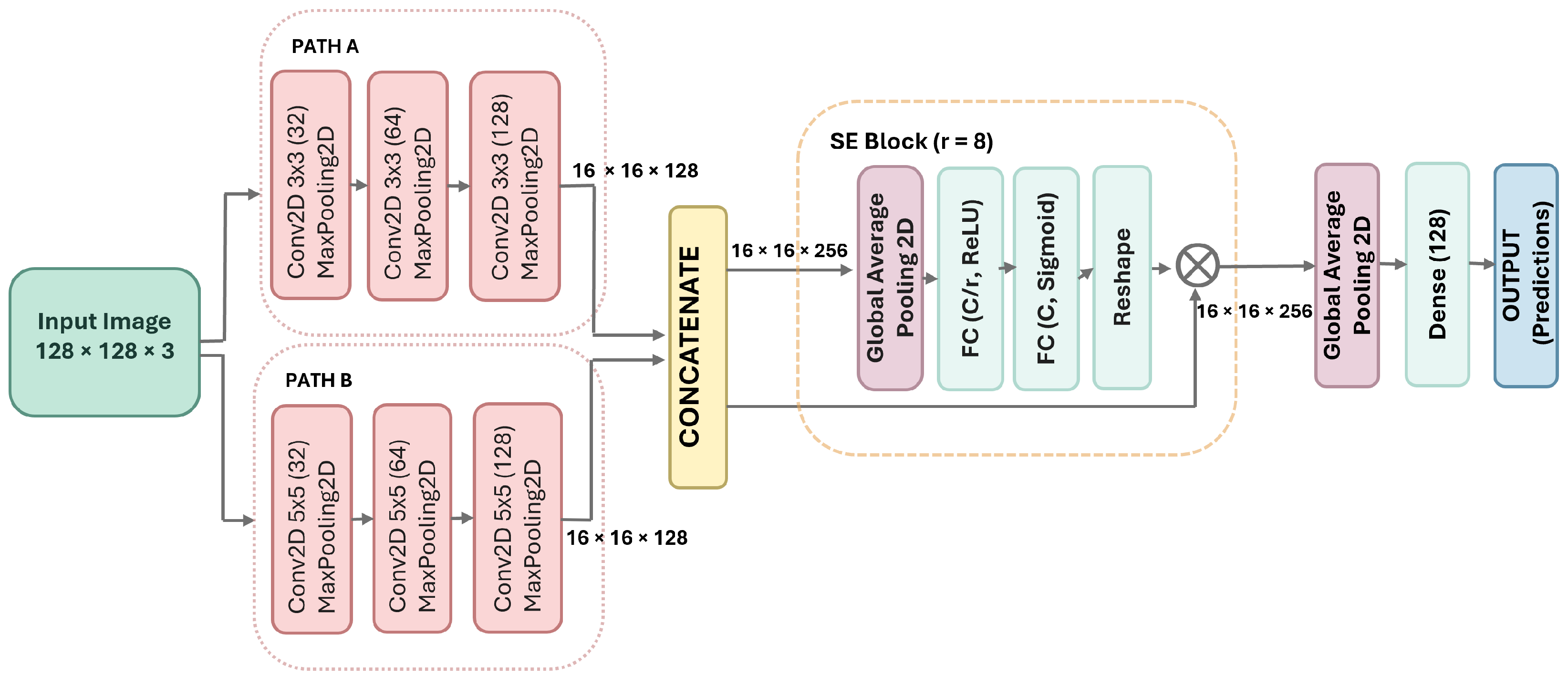
3.2. The DPCSE-Net Architecture
3.2.1. Input
3.2.2. Dual-Path Processing
- Path A employs 3 × 3 convolutional filters in three consecutive blocks with 32, 64, and 128 filters, respectively. Each convolutional block is followed by a MaxPooling2D layer to progressively reduce spatial dimensions while retaining key local features.
- Path B utilizes 5 × 5 convolutional filters in three consecutive blocks with 32, 64, and 128 filters, respectively. Each convolutional block is followed by a MaxPooling2D layer to capture broader spatial context while progressively reducing spatial dimensions.
3.2.3. Feature Concatenation
3.2.4. Squeeze-and-Excitation Block
3.2.5. Classification and Output Layer
3.3. Explainability and Model Interpretation
3.3.1. Gradient-Weighted Class Activation Mapping (Grad-CAM)
3.3.2. SE Attention Heatmaps
3.3.3. Integrated Gradients
3.3.4. Explainability Pipeline Summary
| Algorithm 1 Explainability Pipeline for DPCSE-Net |
Require: Input image x, trained DPCSE-Net model F
|
3.4. Experimental Setup
3.4.1. Training Setup
3.4.2. Training Configuration
4. Results
4.1. Quantitative Evaluation and Ablation Analysis
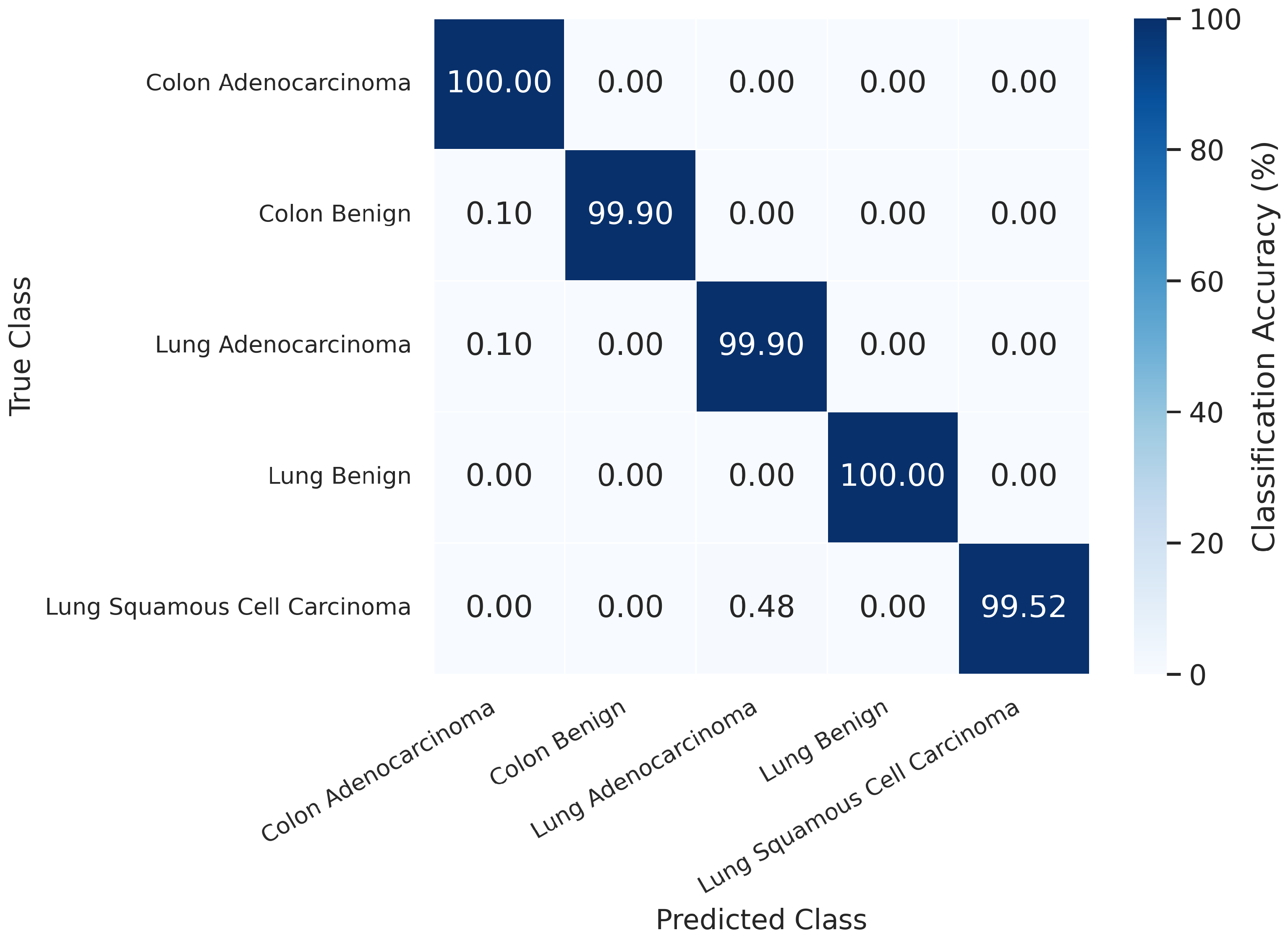
4.1.1. Overall Performance on the LC25000 Dataset
4.1.2. Ablation Study
4.2. Computational Efficiency Analysis
4.3. Comparative Analysis with Existing Methods
4.4. Explainability and Visual Interpretation
5. Discussion
5.1. Summary of Findings
5.2. Ablation and Explainability Insights
5.3. Comparison with Previous Studies
5.4. Limitations
5.5. Clinical Considerations and Future Work
5.6. Overall Implications
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CAD | Computer-Aided Diagnosis |
| CNN | Convolutional Neural Network |
| SE | Squeeze and Excitation |
| XAI | Explainable Artificial Intelligence |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| GAP | Global Average Pooling |
| ReLU | Rectified Linear Unit |
| AUC | Area Under the Curve |
| ROC | Receiver Operating Characteristic |
| TP | True Positive |
| TN | True Negative |
| FP | False Positive |
| FN | False Negative |
| ANN | Artificial Neural Network |
| SVM | Support Vector Machine |
References
- World Health Organization. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. Available online: https://gco.iarc.fr/today/home (accessed on 20 January 2025).
- Bray, F.; Laversanne, M.; Weiderpass, E.; Soerjomataram, I. The ever-increasing importance of cancer as a leading cause of premature death worldwide. Cancer 2021, 127, 3029–3030. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- World Health Organization. Effects of Tobacco on Health; World Health Organization (WHO): Geneva, Switzerland, 2024; Available online: https://www.who.int/europe/news-room/fact-sheets/item/effects-of-tobacco-on-health (accessed on 25 November 2025).
- Centers for Disease Control and Prevention. What Are the Risk Factors for Lung Cancer? U.S. Department of Health and Human Services (CDC): Atlanta, GA, USA, 2024. Available online: https://www.cdc.gov/lung-cancer/risk-factors/index.html (accessed on 25 November 2025).
- Kurishima, K.; Miyazaki, K.; Watanabe, H.; Shiozawa, T.; Ishikawa, H.; Satoh, H.; Hizawa, N. Lung cancer patients with synchronous colon cancer. Mol. Clin. Oncol. 2017, 7, 3029–3030. [Google Scholar] [CrossRef]
- Sánchez-Peralta, L.F.; Bote-Curiel, L.; Picón, A.; Sánchez-Margallo, F.M.; Pagador, J.B. Deep learning to find colorectal polyps in colonoscopy: A systematic literature review. Artif. Intell. Med. 2020, 108, 101923. [Google Scholar] [CrossRef]
- Tummala, S.; Kadry, S.; Nadeem, A.; Rauf, H.T.; Gul, N. An explainable classification method based on complex scaling in histopathology images for lung and colon cancer. Diagnostics 2023, 13, 1594. [Google Scholar] [CrossRef]
- Bhattacharya, A.; Saha, B.; Chattopadhyay, S.; Sarkar, R. Deep feature selection using adaptive β-hill climbing aided whale optimization algorithm for lung and colon cancer detection. Biomed. Signal Process. Control 2023, 83, 104692. [Google Scholar] [CrossRef]
- Provath, M.A.-M.; Deb, K.; Dhar, P.K.; Shimamura, T. Classification of lung and colon cancer histopathological images using global context attention-based convolutional neural network. IEEE Access 2023, 11, 110164–110183. [Google Scholar] [CrossRef]
- Cinar, U.; Cetin Atalay, R.; Cetin, Y.Y. Human Hepatocellular Carcinoma Classification from H&E Stained Histopathology Images with 3D Convolutional Neural Networks and Focal Loss Function. J. Imaging 2023, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.A.; Haque, F.; Sabuj, S.R.; Sarker, H.; Goni, M.O.F.; Rahman, F.; Rashid, M.M. An end-to-end lightweight multi-scale CNN for the classification of lung and colon cancer with XAI integration. Technologies 2024, 12, 56. [Google Scholar] [CrossRef]
- Al-Jabbar, M.; Alshahrani, M.; Senan, E.M.; Ahmed, I.A. Histopathological analysis for detecting lung and colon cancer malignancies using hybrid systems with fused features. Bioengineering 2023, 10, 383. [Google Scholar] [CrossRef]
- Attallah, O.; Aslan, M.F.; Sabancı, K. A framework for lung and colon cancer diagnosis via lightweight deep learning models and transformation methods. Diagnostics 2022, 12, 2926. [Google Scholar] [CrossRef]
- Alsubai, S. Transfer learning based approach for lung and colon cancer detection using local binary pattern features and explainable artificial intelligence (AI) techniques. PeerJ Comput. Sci. 2024, 10, 1996. [Google Scholar] [CrossRef] [PubMed]
- Attallah, O. Lung and colon cancer classification using multiscale deep features integration of compact convolutional neural networks and feature selection. Technologies 2025, 13, 54. [Google Scholar] [CrossRef]
- Degadwala, S.; Oza, P.R. A review on lung and colon combine cancer detection using ML and DL techniques. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2024, 10, 24–35. [Google Scholar] [CrossRef]
- Sloboda, T.; Hudec, L.; Halinkovič, M.; Benesova, W. Attention-Enhanced Unpaired xAI-GANs for Transformation of Histological Stain Images. J. Imaging 2024, 10, 32. [Google Scholar] [CrossRef]
- Ijaz, M.A.; Ashraf, I.; Zahid, U.; Yasin, A.; Ali, S.; Khan, M.A.; AlQahtani, S.A.; Zhang, Y. DS2LC3Net: A decision support system for lung colon cancer classification using fusion of deep neural networks and normal distribution-based gray wolf optimization. ACM Trans. Asian Low-Resour. Lang. Inf. Process. 2023. [Google Scholar] [CrossRef]
- Mangal, S.; Chaurasia, A.; Khajanchi, A. Convolution neural networks for diagnosing colon and lung cancer histopathological images. arXiv 2020, arXiv:2009.03878. [Google Scholar] [CrossRef]
- Hadiyoso, S.; Aulia, S.; Irawati, I.D. Diagnosis of lung and colon cancer based on clinical pathology images using convolutional neural network and CLAHE framework. Int. J. Appl. Sci. Eng. 2023, 20, 2022004. [Google Scholar] [CrossRef]
- Iqbal, S.; Qureshi, A.N.; Alhussein, M.; Aurangzeb, K.; Kadry, S. A novel heteromorphous convolutional neural network for automated assessment of tumors in colon and lung histopathology images. Biomimetics 2023, 8, 370. [Google Scholar] [CrossRef] [PubMed]
- AlGhamdi, R.; Asar, T.O.; Assiri, F.Y.; Mansouri, R.A.; Ragab, M. Al-Biruni earth radius optimization with transfer learning-based histopathological image analysis for lung and colon cancer detection. Cancers 2023, 15, 3300. [Google Scholar] [CrossRef]
- Gowthamy, J.; Ramesh, S. A novel hybrid model for lung and colon cancer detection using pre-trained deep learning and KELM. Expert Syst. Appl. 2024, 252, 124114. [Google Scholar] [CrossRef]
- Mengash, H.A.; Alamgeer, M.; Maashi, M.; Othman, M.; Hamza, M.A.; Ibrahim, S.S.; Zamani, A.S.; Yaseen, I. Leveraging marine predators algorithm with deep learning for lung and colon cancer diagnosis. Cancers 2023, 15, 1591. [Google Scholar] [CrossRef] [PubMed]
- Singh, O.; Singh, K.K. An approach to classify lung and colon cancer of histopathology images using deep feature extraction and an ensemble method. Int. J. Inf. Technol. 2023, 15, 4149–4160. [Google Scholar] [CrossRef]
- Borkowski, A.A.; Bui, M.M.; Thomas, L.B.; Wilson, C.P.; DeLand, L.A.; Mastorides, S.M. Lung and colon cancer histopathological image dataset (LC25000). arXiv 2019, arXiv:1912.12142. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. arXiv 2017, arXiv:1703.01365. [Google Scholar] [CrossRef]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Powers, D.M.W. Evaluation: From precision, recall and F-measure to ROC, informedness, markedness and correlation. arXiv 2020, arXiv:2010.16061. [Google Scholar] [CrossRef]
- Dozat, T. Incorporating Nesterov momentum into Adam. In Proceedings of the 4th International Conference on Learning Representations, Workshop Track, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]

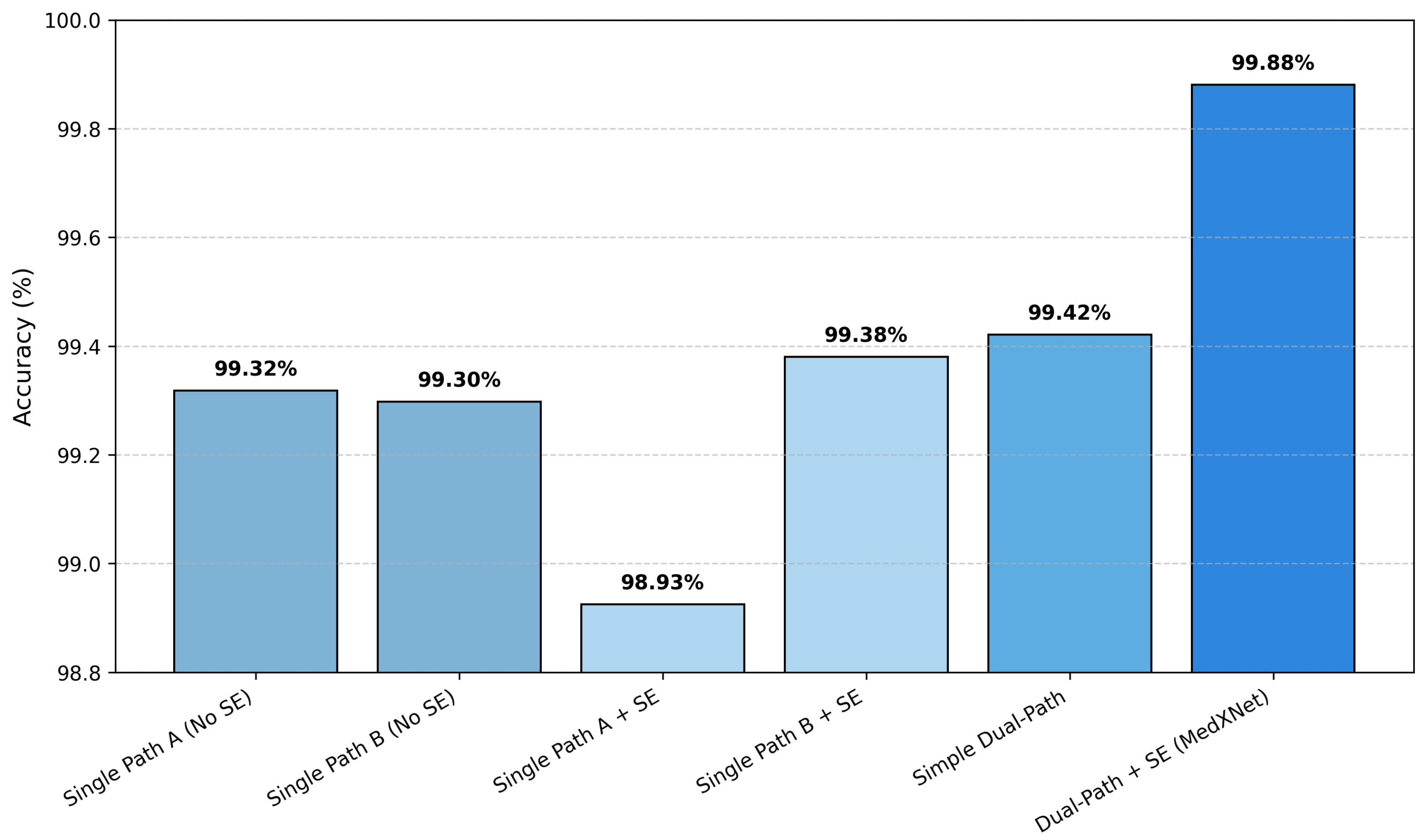
| Model Variant | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| Single Path A (No SE) | 99.32 | 99.32 | 99.32 | 99.32 |
| Single Path B (No SE) | 99.30 | 99.30 | 99.30 | 99.30 |
| Single Path A + SE | 98.93 | 98.95 | 98.93 | 98.92 |
| Single Path B + SE | 99.38 | 99.38 | 99.38 | 99.38 |
| Simple Dual-Path (No SE) | 99.42 | 99.43 | 99.42 | 99.42 |
| Dual-Path + SE (Full DPCSE-Net) | 99.88 | 99.88 | 99.88 | 99.88 |
| Metric | Value | Description |
|---|---|---|
| Total parameters | 287,525 | Trainable only |
| Model size | 1.10 MB | Float32 representation |
| FLOPs per inference | 0.988 GFLOPs | 128 × 128 × 3 input |
| Inference time | 2.41 ms | Per image on GPU |
| Throughput | 415 images/s | Batch size of 32 |
| Functional Block | GFLOPs | Percentage |
|---|---|---|
| Path A | 0.339 | 34.28% |
| Path B | 0.649 | 65.70% |
| Squeeze-and-Excitation | 0.000098 | 0.01% |
| Classifier | 0.000067 | 0.01% |
| Total | 0.988 | 100.00% |
| Authors (Year) | Accuracy (%) | Sensitivity (%) | Specificity (%) | Precision (%) | F1-Score (%) | Model Size/Parameters |
|---|---|---|---|---|---|---|
| Hasan et al. (2024) [12] | 99.20 | 99.36 | 99.16 | 99.16 | – | 1.10 M parameters |
| Al-Jabbar et al. (2023) [13] | 99.64 | 99.85 | 100.00 | 100.00 | – | – |
| Attallah et al. (2022) [14] | 99.60 | 99.60 | 99.90 | 99.60 | 99.60 | – |
| Alsubai (2024) [15] | 99.88 | 99.42 | 99.46 | 99.76 | – | – |
| Attallah (2025) [16] | 99.78 | 99.78 | 99.95 | 99.78 | 99.78 | – |
| DPCSE-Net (Proposed) | 99.88 | 99.88 | 99.88 | 99.88 | 99.88 | 287,525 parameters |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AlShehri, H. Dual-Path Convolutional Neural Network with Squeeze-and-Excitation Attention for Lung and Colon Histopathology Classification. J. Imaging 2025, 11, 448. https://doi.org/10.3390/jimaging11120448
AlShehri H. Dual-Path Convolutional Neural Network with Squeeze-and-Excitation Attention for Lung and Colon Histopathology Classification. Journal of Imaging. 2025; 11(12):448. https://doi.org/10.3390/jimaging11120448
Chicago/Turabian StyleAlShehri, Helala. 2025. "Dual-Path Convolutional Neural Network with Squeeze-and-Excitation Attention for Lung and Colon Histopathology Classification" Journal of Imaging 11, no. 12: 448. https://doi.org/10.3390/jimaging11120448
APA StyleAlShehri, H. (2025). Dual-Path Convolutional Neural Network with Squeeze-and-Excitation Attention for Lung and Colon Histopathology Classification. Journal of Imaging, 11(12), 448. https://doi.org/10.3390/jimaging11120448






