Current Trends and Future Opportunities of AI-Based Analysis in Mesenchymal Stem Cell Imaging: A Scoping Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol
2.2. Research Question
- Population: MSCs of any type, from either animal or human sources;
- Concept: application of AI methods for image processing;
- Context: analysis of cell culture images.
2.3. Search Strategy
2.4. Study Selection and Data Extraction
3. Results
3.1. Search Results
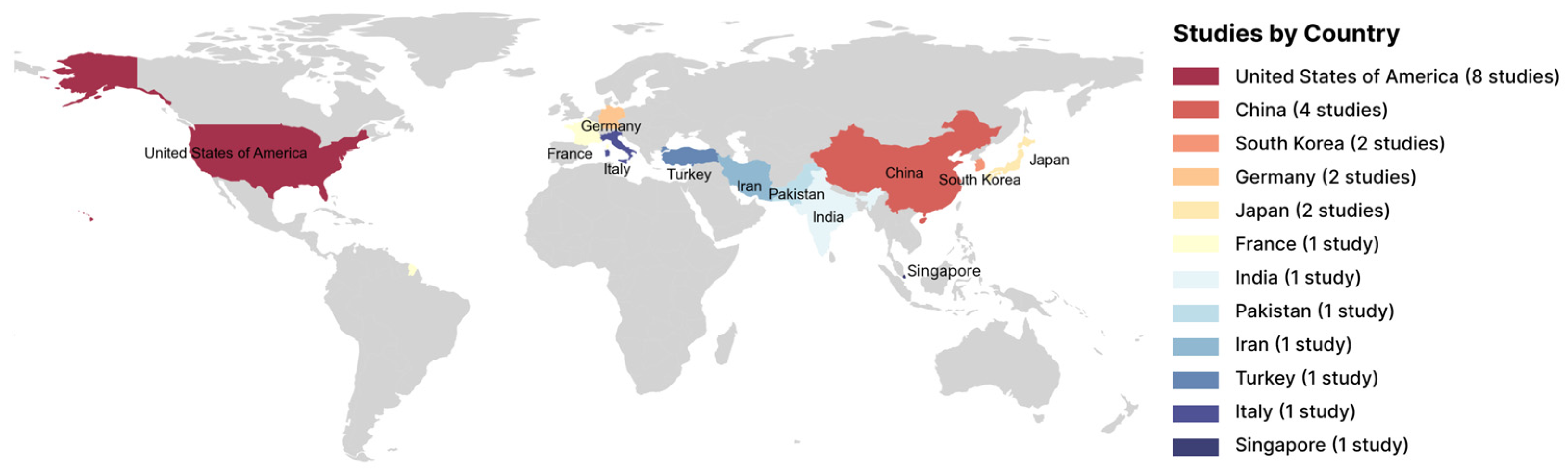
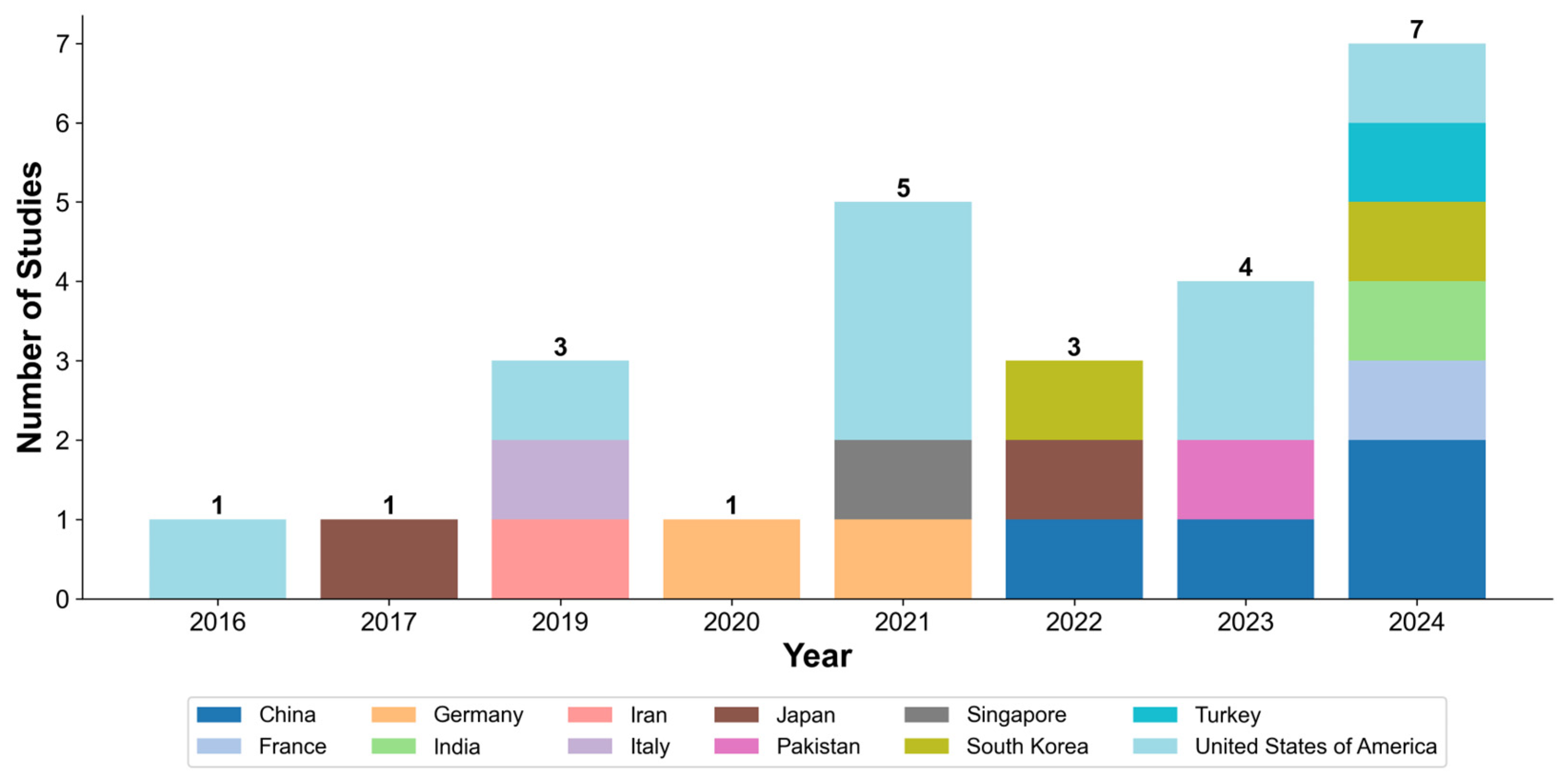
3.2. Characteristics of Included Studies
3.3. Areas of AI Application in MSC Image Analysis
3.3.1. Cell Classification
3.3.2. Cell Segmentation and Counting
3.3.3. Assessment of Differentiation
3.3.4. Analysis of Senescence
3.3.5. Other Areas
3.4. Types of AI Algorithms
3.5. Effectiveness of AI Methods for MSCs Image Analysis
4. Discussion
4.1. Principal Findings and Implications of AI in MSC Analysis
4.2. Key Limitations and Challenges
4.3. Future Perspectives and Research Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| AUC | Area Under the Curve |
| cGAN | Conditional Generative Adversarial Network |
| DAE | Denoising Autoencoder |
| FLIM | Fluorescence Lifetime Imaging Microscopy |
| H-SCNN | Hyperspectral Separable Convolutional Neural Network |
| HWJ | Human Wharton’s Jelly |
| IFN-γ | Interferon-gamma |
| IoU | Intersection over Union |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| LDA | Linear Discriminant Analysis |
| LSVM | Linear Support Vector Machine |
| mAP | Mean Average Precision |
| MRI | Magnetic Resonance Imaging |
| MSCs | Mesenchymal Stem/Stromal Cells |
| PCL | Polycaprolactone |
| R-CNN | Region-Based Convolutional Neural Network |
| RF | Random Forest |
| ROC | Receiver Operating Characteristic |
| SHED | Stem Cells from Human Exfoliated Deciduous Teeth |
| SRS | Stimulated Raman Scattering Microscopy |
| SSL | Self-Supervised Learning |
| SVM | Support Vector Machine |
| VAE | Variational Autoencoder |
| viSNE | Visual Stochastic Neighbor Embedding |
References
- Liu, J.; Gao, J.; Liang, Z.; Gao, C.; Niu, Q.; Wu, F.; Zhang, L. Mesenchymal Stem Cells and Their Microenvironment. Stem Cell Res. Ther. 2022, 13, 429. [Google Scholar] [CrossRef] [PubMed]
- Mei, R.; Wan, Z.; Yang, C.; Shen, X.; Wang, R.; Zhang, H.; Yang, R.; Li, J.; Song, Y.; Su, H. Advances and Clinical Challenges of Mesenchymal Stem Cell Therapy. Front. Immunol. 2024, 15, 1421854. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Rodríguez, P.; Cuenca-Martagón, A.; Castillo-González, J.; Serrano-Martínez, I.; Luque, R.M.; Delgado, M.; González-Rey, E. Novel Therapeutic Opportunities for Neurodegenerative Diseases with Mesenchymal Stem Cells: The Focus on Modulating the Blood-Brain Barrier. Int. J. Mol. Sci. 2023, 24, 14117. [Google Scholar] [CrossRef]
- Jiang, L.; Lu, J.; Chen, Y.; Lyu, K.; Long, L.; Wang, X.; Liu, T.; Li, S. Mesenchymal Stem Cells: An Efficient Cell Therapy for Tendon Repair (Review). Int. J. Mol. Med. 2023, 52, 70. [Google Scholar] [CrossRef]
- Chung, M.-J.; Son, J.-Y.; Park, S.; Park, S.-S.; Hur, K.; Lee, S.-H.; Lee, E.-J.; Park, J.-K.; Hong, I.-H.; Kim, T.-H.; et al. Mesenchymal Stem Cell and MicroRNA Therapy of Musculoskeletal Diseases. Int. J. Stem Cells 2021, 14, 150–167. [Google Scholar] [CrossRef]
- Zaripova, L.N.; Midgley, A.; Christmas, S.E.; Beresford, M.W.; Pain, C.; Baildam, E.M.; Oldershaw, R.A. Mesenchymal Stem Cells in the Pathogenesis and Therapy of Autoimmune and Autoinflammatory Diseases. IJMS 2023, 24, 16040. [Google Scholar] [CrossRef]
- Yong, K.W.; Choi, J.R.; Mohammadi, M.; Mitha, A.P.; Sanati-Nezhad, A.; Sen, A. Mesenchymal Stem Cell Therapy for Ischemic Tissues. Stem Cells Int. 2018, 2018, 1–11. [Google Scholar] [CrossRef]
- Kangari, P.; Salahlou, R.; Vandghanooni, S. Harnessing the Therapeutic Potential of Mesenchymal Stem Cells in Cancer Treatment. Adv. Pharm. Bull. 2024, 14, 574–590. [Google Scholar] [CrossRef]
- Han, X.; Liao, R.; Li, X.; Zhang, C.; Huo, S.; Qin, L.; Xiong, Y.; He, T.; Xiao, G.; Zhang, T. Mesenchymal Stem Cells in Treating Human Diseases: Molecular Mechanisms and Clinical Studies. Sig Transduct. Target. Ther. 2025, 10, 262. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, C.; Zheng, L.; Li, Q.; Ge, J.; Geng, S.; Zhai, M.; Chen, X.; Yuan, H.; Li, Y.; et al. Safety and Efficacy of Umbilical Cord Tissue-Derived Mesenchymal Stem Cells in the Treatment of Patients with Aging Frailty: A Phase I/II Randomized, Double-Blind, Placebo-Controlled Study. Stem Cell Res. Ther. 2024, 15, 122. [Google Scholar] [CrossRef]
- Ivanovski, S.; Han, P.; Peters, O.A.; Sanz, M.; Bartold, P.M. The Therapeutic Use of Dental Mesenchymal Stem Cells in Human Clinical Trials. J. Dent. Res. 2024, 103, 1173–1184. [Google Scholar] [CrossRef]
- Elhadi, M.; Khaled, T.; Faraj, H.; Msherghi, A. Evaluating the Impact of Mesenchymal Stromal Cell Therapy on Mortality and Safety in Acute Respiratory Distress Syndrome Caused by COVID-19: A Meta-Analysis of Clinical Trials. CHEST 2023, 164, A1726–A1727. [Google Scholar] [CrossRef]
- Chugan, G.S.; Lyundup, A.V.; Bondarenko, O.N.; Galstyan, G.R. The Application of Cell Products for the Treatment of Critical Limb Ischemia in Patients with Diabetes Mellitus: A Review of the Literature. Probl. Endokrinol. 2024, 70, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, N.; Kelly, J.J.; Sehl, O.C.; Gevaert, J.J.; Fox, M.S.; Qi, Q.; Foster, P.J.; Thiessen, J.D.; Hicks, J.W.; Scholl, T.J.; et al. Complementary Early-Phase Magnetic Particle Imaging and Late-Phase Positron Emission Tomography Reporter Imaging of Mesenchymal Stem Cells In Vivo. Nanoscale 2023, 15, 3408–3418. [Google Scholar] [CrossRef] [PubMed]
- Perottoni, S.; Neto, N.G.B.; Nitto, C.D.; Dmitriev, R.I.; Raimondi, M.T.; Monaghan, M.G. Intracellular Label-Free Detection of Mesenchymal Stem Cell Metabolism within a Perivascular Niche-on-a-Chip. Lab. Chip 2021, 21, 1395–1408. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wang, C.; Kuddannaya, S.; Zhang, J.; Arifin, D.R.; Han, Z.; Walczak, P.; Liu, G.; Bulte, J.W.M. In Vivo Tracking of Unlabelled Mesenchymal Stromal Cells by Mannose-Weighted Chemical Exchange Saturation Transfer MRI. Nat. Biomed. Eng. 2022, 6, 658–666. [Google Scholar] [CrossRef]
- Oja, S.; Komulainen, P.; Penttilä, A.; Nystedt, J.; Korhonen, M. Automated Image Analysis Detects Aging in Clinical-Grade Mesenchymal Stromal Cell Cultures. Stem Cell Res. Ther. 2018, 9, 6. [Google Scholar] [CrossRef]
- Eggerschwiler, B.; Canepa, D.D.; Pape, H.-C.; Casanova, E.A.; Cinelli, P. Automated Digital Image Quantification of Histological Staining for the Analysis of the Trilineage Differentiation Potential of Mesenchymal Stem Cells. Stem Cell Res. Ther. 2019, 10, 69. [Google Scholar] [CrossRef]
- Wright, K.T.; Griffiths, G.J.; Johnson, W.E.B. A Comparison of High-Content Screening versus Manual Analysis to Assay the Effects of Mesenchymal Stem Cell–Conditioned Medium on Neurite Outgrowth in Vitro. SLAS Discov. 2010, 15, 576–582. [Google Scholar] [CrossRef]
- Mota, S.M.; Rogers, R.E.; Haskell, A.W.; McNeill, E.; Kaunas, R.R.; Gregory, C.A.; Giger, M.L.; Maitland, K.C. Automated Mesenchymal Stem Cell Segmentation and Machine Learning-Based Phenotype Classification Using Morphometric and Textural Analysis. J. Med. Imaging 2021, 8, 014503. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Chandel, P.; Prasad, K.; Chakraborty, U. Machine Learning Aided Single Cell Image Analysis Improves Understanding of Morphometric Heterogeneity of Human Mesenchymal Stem Cells. Methods 2024, 225, 62–73. [Google Scholar] [CrossRef]
- Marzec-Schmidt, K.; Ghosheh, N.; Stahlschmidt, S.R.; Küppers-Munther, B.; Synnergren, J.; Ulfenborg, B. Artificial Intelligence Supports Automated Characterization of Differentiated Human Pluripotent Stem Cells. Stem Cells 2023, 41, 850–861. [Google Scholar] [CrossRef]
- Torro, R.; Díaz-Bello, B.; Arawi, D.E.; Dervanova, K.; Ammer, L.; Dupuy, F.; Chames, P.; Sengupta, K.; Limozin, L. Celldetective: An AI-Enhanced Image Analysis Tool for Unraveling Dynamic Cell Interactions. bioRxiv 2024. [Google Scholar] [CrossRef]
- Issa, J.; Abou Chaar, M.; Kempisty, B.; Gasiorowski, L.; Olszewski, R.; Mozdziak, P.; Dyszkiewicz-Konwińska, M. Artificial-Intelligence-Based Imaging Analysis of Stem Cells: A Systematic Scoping Review. Biology 2022, 11, 1412. [Google Scholar] [CrossRef] [PubMed]
- Guez, J.-S.; Lacroix, P.-Y.; Château, T.; Vial, C. Deep in Situ Microscopy for Real-Time Analysis of Mammalian Cell Populations in Bioreactors. Sci. Rep. 2023, 13, 22045. [Google Scholar] [CrossRef] [PubMed]
- Peters, M.D.J.; Godfrey, C.M.; Khalil, H.; McInerney, P.; Parker, D.; Soares, C.B. Guidance for Conducting Systematic Scoping Reviews. JBI Evid. Implement. 2015, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 7. [Google Scholar] [CrossRef]
- Ouzzani, M.; Hammady, H.; Fedorowicz, Z.; Elmagarmid, A. Rayyan—A Web and Mobile App for Systematic Reviews. Syst. Rev. 2016, 5, 210. [Google Scholar] [CrossRef]
- Chen, D.; Sarkar, S.; Candia, J.; Florczyk, S.J.; Bodhak, S.; Driscoll, M.K.; Simon, C.G.; Dunkers, J.P.; Losert, W. Machine Learning Based Methodology to Identify Cell Shape Phenotypes Associated with Microenvironmental Cues. Biomaterials 2016, 104, 104–118. [Google Scholar] [CrossRef]
- Marklein, R.A.; Klinker, M.W.; Drake, K.A.; Polikowsky, H.G.; Lessey-Morillon, E.C.; Bauer, S.R. Morphological Profiling Using Machine Learning Reveals Emergent Subpopulations of Interferon-γ-Stimulated Mesenchymal Stromal Cells That Predict Immunosuppression. Cytotherapy 2019, 21, 17–31. [Google Scholar] [CrossRef]
- Imboden, S.; Liu, X.; Lee, B.S.; Payne, M.C.; Hsieh, C.-J.; Lin, N.Y.C. Investigating Heterogeneities of Live Mesenchymal Stromal Cells Using AI-Based Label-Free Imaging. Sci. Rep. 2021, 11, 6728. [Google Scholar] [CrossRef]
- Chen, D.; Dunkers, J.P.; Losert, W.; Sarkar, S. Early Time-Point Cell Morphology Classifiers Successfully Predict Human Bone Marrow Stromal Cell Differentiation Modulated by Fiber Density in Nanofiber Scaffolds. Biomaterials 2021, 274, 120812. [Google Scholar] [CrossRef]
- Weber, L.; Lee, B.S.; Imboden, S.; Hsieh, C.-J.; Lin, N.Y.C. Phenotyping Senescent Mesenchymal Stromal Cells Using AI Image Translation. Curr. Res. Biotechnol. 2023, 5, 100120. [Google Scholar] [CrossRef] [PubMed]
- Mai, M.; Luo, S.; Fasciano, S.; Oluwole, T.E.; Ortiz, J.; Pang, Y.; Wang, S. Morphology-Based Deep Learning Approach for Predicting Adipogenic and Osteogenic Differentiation of Human Mesenchymal Stem Cells (hMSCs). Front. Cell Dev. Biol. 2023, 11, 1329840. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.; Zheng, S.; Zhang, H.; Murphy, R.F.; Dahl, K.N.; Zaritsky, A. Image-Based Discrimination of the Early Stages of Mesenchymal Stem Cell Differentiation. Mol. Biol. Cell 2024, 35, ar103. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Huang, N.; Fu, Y.; Liu, K.; Zhang, H.; Li, Y.; Yang, S. Morphology-Based Deep Learning Approach for Predicting Osteogenic Differentiation. Front. Bioeng. Biotechnol. 2022, 9, 802794. [Google Scholar] [CrossRef]
- Kong, Y.; Ao, J.; Chen, Q.; Su, W.; Zhao, Y.; Fei, Y.; Ma, J.; Ji, M.; Mi, L. Evaluating Differentiation Status of Mesenchymal Stem Cells by Label-Free Microscopy System and Machine Learning. Cells 2023, 12, 1524. [Google Scholar] [CrossRef]
- He, L.; Li, M.; Wang, X.; Wu, X.; Yue, G.; Wang, T.; Zhou, Y.; Lei, B.; Zhou, G. Morphology-Based Deep Learning Enables Accurate Detection of Senescence in Mesenchymal Stem Cell Cultures. BMC Biol. 2024, 22, 1. [Google Scholar] [CrossRef]
- Liu, M.; Du, X.; Ju, H.; Liang, X.; Wang, H. Utilization of Convolutional Neural Networks to Analyze Microscopic Images for High-Throughput Screening of Mesenchymal Stem Cells. Open Life Sci. 2024, 19, 20220859. [Google Scholar] [CrossRef]
- Kim, G.; Jeon, J.H.; Park, K.; Kim, S.W.; Kim, D.H.; Lee, S. High Throughput Screening of Mesenchymal Stem Cell Lines Using Deep Learning. Sci. Rep. 2022, 12, 17507. [Google Scholar] [CrossRef]
- Ngo, D.; Lee, J.; Kwon, S.J.; Park, J.H.; Cho, B.H.; Chang, J.W. Application of Deep Neural Networks in the Manufacturing Process of Mesenchymal Stem Cells Therapeutics. Int. J. Stem Cells 2024, 18, 186–193. [Google Scholar] [CrossRef]
- Dursun, G.; Balkrishna Tandale, S.; Eschweiler, J.; Tohidnezhad, M.; Markert, B.; Stoffel, M. Recognition of Tenogenic Differentiation Using Convolutional Neural Network. Curr. Dir. Biomed. Eng. 2020, 6, 200–204. [Google Scholar] [CrossRef]
- Ochs, J.; Biermann, F.; Piotrowski, T.; Erkens, F.; Nießing, B.; Herbst, L.; König, N.; Schmitt, R.H. Fully Automated Cultivation of Adipose-Derived Stem Cells in the StemCellDiscovery—A Robotic Laboratory for Small-Scale, High-Throughput Cell Production Including Deep Learning-Based Confluence Estimation. Processes 2021, 9, 575. [Google Scholar] [CrossRef]
- Tanaka, N.; Yamashita, T.; Sato, A.; Vogel, V.; Tanaka, Y. Simple Agarose Micro-Confinement Array and Machine-Learning-Based Classification for Analyzing the Patterned Differentiation of Mesenchymal Stem Cells. PLoS ONE 2017, 12, e0173647. [Google Scholar] [CrossRef]
- Suyama, T.; Takemoto, Y.; Miyauchi, H.; Kato, Y.; Matsuzaki, Y.; Kato, R. Morphology-Based Noninvasive Early Prediction of Serial-Passage Potency Enhances the Selection of Clone-Derived High-Potency Cell Bank from Mesenchymal Stem Cells. Inflamm. Regen. 2022, 42, 30. [Google Scholar] [CrossRef] [PubMed]
- Halima, I.; Maleki, M.; Frossard, G.; Thomann, C.; Courtial, E.J. Accurate Detection of Cell Deformability Tracking in Hydrodynamic Flow by Coupling Unsupervised and Supervised Learning. Mach. Learn. Appl. 2024, 16, 100538. [Google Scholar] [CrossRef]
- Adnan, N.; Umer, F.; Malik, S. Implementation of Transfer Learning for the Segmentation of Human Mesenchymal Stem—A Validation Study. Tissue Cell 2023, 83, 102149. [Google Scholar] [CrossRef] [PubMed]
- Hassanlou, L.; Meshgini, S.; Alizadeh, E. Evaluating Adipocyte Differentiation of Bone Marrow-Derived Mesenchymal Stem Cells by a Deep Learning Method for Automatic Lipid Droplet Counting. Comput. Biol. Med. 2019, 112, 103365. [Google Scholar] [CrossRef]
- Çelebi, F.; Boyvat, D.; Ayaz-Guner, S.; Tasdemir, K.; Icoz, K. Improved Senescent Cell Segmentation on Bright-field Microscopy Images Exploiting Representation Level Contrastive Learning. Int. J. Imaging Syst. Technol. 2024, 34, e23052. [Google Scholar] [CrossRef]
- D’Acunto, M.; Martinelli, M.; Moroni, D.; Nguyen, N.T.; Szczerbicki, E.; Trawiński, B.; Nguyen, V.D. From Human Mesenchymal Stromal Cells to Osteosarcoma Cells Classification by Deep Learning. J. Intell. Fuzzy Syst. 2019, 37, 7199–7206. [Google Scholar] [CrossRef]
- Zhang, Z.; Leong, K.W.; Vliet, K.V.; Barbastathis, G.; Ravasio, A. Deep Learning for Label-Free Nuclei Detection from Implicit Phase Information of Mesenchymal Stem Cells. Biomed. Opt. Express 2021, 12, 1683–1706. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Han, Z.C. Heterogeneity of Human Mesenchymal Stromal/Stem Cells. In Stem Cells Heterogeneity—Novel Concepts; Birbrair, A., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 165–177. ISBN 978-3-030-11096-3. [Google Scholar]
- Sankar, B.S.; Gilliland, D.; Rincon, J.; Hermjakob, H.; Yan, Y.; Adam, I.; Lemaster, G.; Wang, D.; Watson, K.; Bui, A.; et al. Building an Ethical and Trustworthy Biomedical AI Ecosystem for the Translational and Clinical Integration of Foundation Models. Bioengineering 2024, 11, 984. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, L.; Wang, L.; Yao, D.; Cui, Y. Bayesian Network Analysis of Risk Classification Strategies in the Regulation of Cellular Products. Artif. Intell. Med. 2024, 155, 102937. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-R.; Chien, J.-T.; Lee, C.-Y. Advancements in Clinical Evaluation and Regulatory Frameworks for AI-Driven Software as a Medical Device (SaMD). IEEE Open J. Eng. Med. Biol. 2025, 6, 147–151. [Google Scholar] [CrossRef]
- Hildt, E. What Is the Role of Explainability in Medical Artificial Intelligence? A Case-Based Approach. Bioengineering 2025, 12, 375. [Google Scholar] [CrossRef]
- Alkhanbouli, R.; Matar Abdulla Almadhaani, H.; Alhosani, F.; Simsekler, M.C.E. The Role of Explainable Artificial Intelligence in Disease Prediction: A Systematic Literature Review and Future Research Directions. BMC Med. Inf. Decis. Mak. 2025, 25, 110. [Google Scholar] [CrossRef]
- Overgaard, S.M.; Graham, M.G.; Brereton, T.; Pencina, M.J.; Halamka, J.D.; Vidal, D.E.; Economou-Zavlanos, N.J. Implementing Quality Management Systems to Close the AI Translation Gap and Facilitate Safe, Ethical, and Effective Health AI Solutions. npj Digit. Med. 2023, 6, 218. [Google Scholar] [CrossRef]
- Higgins, D.C. OnRAMP for Regulating Artificial Intelligence in Medical Products. Adv. Intell. Syst. 2021, 3, 2100042. [Google Scholar] [CrossRef]
- Cichosz, S.L. Frameworks for Developing Machine Learning Models. J. Diabetes Sci. Technol. 2023, 17, 862–863. [Google Scholar] [CrossRef]
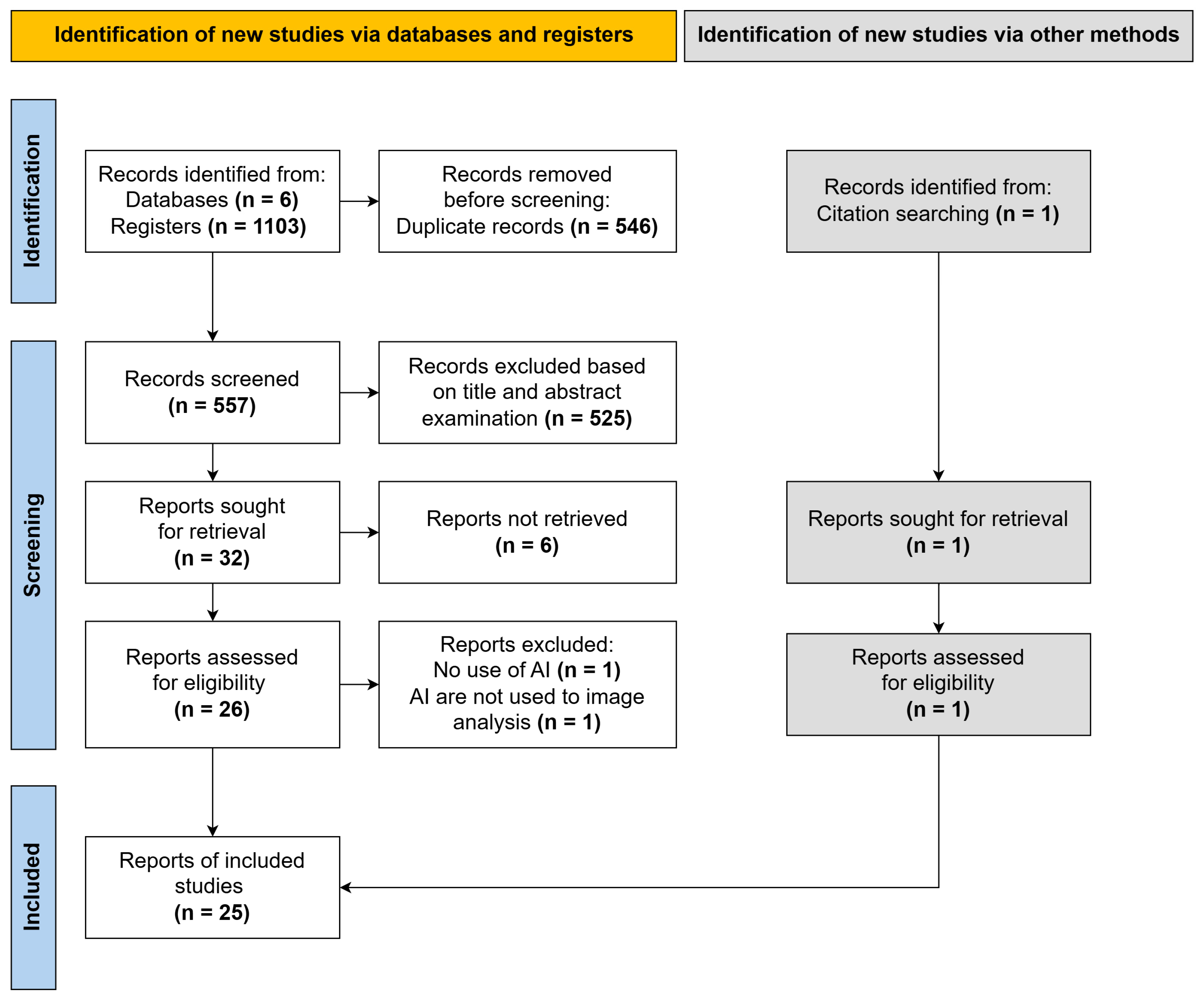


| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Studies on MSCs from animals and humans | Studies involving objects other than MSCs |
| Use of AI methods for MSCs image analysis | Use of AI methods for purposes other than image analysis |
| Studies published in the last 10 years | No use of AI methods |
| Access to full text | Reviews |
| Preprints and conference abstracts | |
| Unavailable full text |
| Authors, Year, Country | Study Objective | Cell Type and Origin | AI Algorithm | Dataset Description | Research Outcomes | Ref. |
|---|---|---|---|---|---|---|
| Chen et al., 2016, USA | Classification of cell morphology on PCL substrates | Human bone marrow MSCs | SVM | Microscopy images of cells on PCL substrates | Identified key morphological indicators; supercell (group of cells) analysis improved accuracy | [29] |
| Tanaka et al., 2017, Japan | Differentiation analysis in agarose microwells | Commercial human bone marrow MSCs | SVM | Annotated microscopy images of cell regions | Achieved 98.2% pixel-level classification accuracy | [44] |
| Marklein et al., 2019, USA | Identification of MSC subpopulations post-IFN-γ stimulation | Commercial bone marrow MSCs | viSNE with LDA | Phase-contrast images of manually segmented cells | Identified subpopulations correlated with T-cell inhibition | [30] |
| Hassanlou et al., 2019, Iran | Automated counting of lipid droplets | Differentiated mouse bone marrow MSCs | Fully convolutional regression network | Cropped microscopy images | Achieved 94% counting accuracy, outperforming manual methods | [48] |
| D’Acunto et al., 2019, Italy | Classification of osteosarcoma vs. MSCs | Human bone marrow MSCs and MG-63 cells | Faster R-CNN with Inception ResNet v2 | Augmented microscopy images of 5 cell classes | Achieved up to 97.5% classification accuracy | [50] |
| Dursun et al., 2020, Germany | Recognition of tenogenic differentiation | Differentiated bone marrow MSCs | VGG16-based CNN | Augmented light microscopy images | Model accuracy of 92.2% | [42] |
| Mota et al., 2021, USA | Segmentation and classification of cell replication speed | Human bone marrow MSCs | Custom algorithm with LSVM, LDA, etc. | Phase-contrast images of segmented cells | Effective for low/mid-density cultures (AUC up to 0.816) | [20] |
| Zhang et al., 2021, Singapore | Detection of cell nuclei in brightfield images | Commercial human MSCs | CNN ensemble | Brightfield images of fixed and live cells | Achieved F1-score of 0.985 on fixed cells | [51] |
| Imboden et al., 2021, USA | Quantitative prediction of marker expression from phase-contrast images | Commercial human bone marrow MSCs | cGAN with U-Net | Paired phase-contrast and immunofluorescence images | Enabled label-free tracking of protein distribution (Corr. Coeff. 0.77) | [31] |
| Ochs et al., 2021, Germany | Automated confluency assessment for quality control | Human adipose tissue MSCs | U-Net | Augmented microscopy images | Achieved F1-score of 0.833 in a high-throughput system | [43] |
| Chen et al., 2021, USA | Prediction of osteogenic differentiation based on morphology | Human bone marrow MSCs | SVM | Synthetic datasets from morphometric data | Correlated morphology with osteogenic potential | [32] |
| Lan et al., 2022, China | Quantitative assessment of osteogenic differentiation | Rat bone marrow MSCs | InceptionV3, VGG16, ResNet50 | Confocal images of stained cells | InceptionV3 achieved AUC of 0.94, outperforming SVM | [36] |
| Suyama et al., 2022, Japan | Noninvasive prediction of high-potency MSC subpopulations | Human bone marrow MSCs | LASSO regression and RF | Time-series morphological data | Predicted cell potency from morphological data; RF/LASSO outperformed | [45] |
| Kim et. al., 2022, South Korea | Identification of MUSE cells based on differentiation potential | Human nasal turbinate-derived MSCs | Transfer learning (DenseNet121, etc.) | Brightfield images validated via immunofluorescence and flow cytometry | DenseNet121 achieved highest AUC (0.975) and accuracy (92.2%) | [40] |
| Weber et al., 2023, USA | Prediction of senescence markers from phase-contrast images | Commercial human adipose and bone marrow MSCs | U-Net-based cGAN | Paired phase-contrast/immunofluorescence images | Strong correlation between predicted and actual senescence markers | [33] |
| Kong et al., 2023, China | Differentiation analysis using FLIM and SRS imaging | Human MSCs | K-means++ clustering on FLIM/SRS data | Single-cell FLIM/SRS images | Successfully tracked differentiation stages; validated by staining | [37] |
| Adnan et al., 2023, Pakistan | Semantic segmentation of MSCs | Commercial human bone marrow MSCs | DeepLab variants | EVICAN dataset (blurred and normal images) | Achieved >99% accuracy; one variant showed better generalizability | [47] |
| Mai et al., 2023, USA | Prediction of differentiation potential from live cell imaging | Human bone marrow MSCs | VGG19, InceptionV3, ResNet18/50 | Time-series images of differentiating cells | ResNet50 achieved >95% accuracy and AUC >0.99 | [34] |
| He et al., 2024, China | Detection of senescent cells | Induced pluripotent stem cell-derived MSCs | Cascade R-CNN with ResNet | Annotated images of SA-β-gal-stained cells | Achieved mAP of 0.81; correlated with senescence markers | [38] |
| Celebi et. al., 2024, Turkey | Segmentation of senescent cells | Commercial human adipose tissue MSCs | Mask R-CNN with SimCLR-based SSL | Images for self-supervised learning and fine-tuning | SSL improved mAP by 8.3%; outperformed U-Net and DeepLabV3 | [49] |
| Mukhopadhyay et al., 2024, India | Classification of SHED vs. HWJ MSCs via imaging flow cytometry | SHED and HWJ MSCs | Custom CNNs and transfer learning | Single-cell brightfield images | Achieved 97.5% accuracy | [21] |
| Halima et al., 2024, France | Cell segmentation and deformability assessment | Human adipose tissue MSCs | Autoencoders (DAE/VAE) and U-Net | Microfluidic images | DAE + U-Net achieved highest precision (81%) | [46] |
| Liu, 2024, China | Functional classification of MSCs via hyperspectral imaging | Commercial human bone marrow MSCs | Hyperspectral separable CNN (H-SCNN) | Hyperspectral images annotated by flow cytometry | H-SCNN achieved 89.6% accuracy, outperforming ResNet/VGG | [39] |
| Hoffman et al., 2024, USA | Determination of stemness and early differentiation | Commercial human bone marrow MSCs | Custom CNN vs. MobileNet | Time-series fluorescent images of actin/chromatin | Achieved up to 90% accuracy with combined actin/chromatin images | [35] |
| Ngo et al., 2024, South Korea | Confluency assessment and anomaly detection | Human Wharton’s jelly MSCs | Ensemble of CNNs and Vision Transformer | Monolayer and multilayer flask images | High accuracy for confluency (AUC 0.958) and anomaly detection | [41] |
| Application Area | Primary Methods | Reported Strengths | Reported Weaknesses/Trade-Offs | Typical Validation Metrics |
|---|---|---|---|---|
| Cell classification | CNN, SVM | CNN: high accuracy, automatic feature extraction. SVM: high interpretability. | CNN: “black-box” nature, requires large datasets. SVM: requires manual feature engineering. | Accuracy, AUC, F1-score |
| Segmentation and counting | U-Net, DeepLab, DAE | U-Net: high precision on clean images. DAE + U-Net: robustness to image noise. | High dependency on large, pixel-level annotated datasets. | Dice coefficient, F1-score, precision, IoU |
| Differentiation assessment | CNN, SVM, k-means | CNN: enables non-invasive prediction on live cells. SVM/k-means: transparent, based on defined features. | SVM/k-means: lower accuracy with subtle morphological changes. | AUC, correlation with biochemical assays |
| Senescence analysis | cGAN, Mask R-CNN | cGAN: “virtual staining” preserves cell viability. R-CNN: Precise detection and segmentation. | Computationally intensive, complex to train, require large datasets. | Correlation with senescence markers, mAP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solopov, M.; Chechekhina, E.; Turchin, V.; Popandopulo, A.; Filimonov, D.; Burtseva, A.; Ishchenko, R. Current Trends and Future Opportunities of AI-Based Analysis in Mesenchymal Stem Cell Imaging: A Scoping Review. J. Imaging 2025, 11, 371. https://doi.org/10.3390/jimaging11100371
Solopov M, Chechekhina E, Turchin V, Popandopulo A, Filimonov D, Burtseva A, Ishchenko R. Current Trends and Future Opportunities of AI-Based Analysis in Mesenchymal Stem Cell Imaging: A Scoping Review. Journal of Imaging. 2025; 11(10):371. https://doi.org/10.3390/jimaging11100371
Chicago/Turabian StyleSolopov, Maksim, Elizaveta Chechekhina, Viktor Turchin, Andrey Popandopulo, Dmitry Filimonov, Anzhelika Burtseva, and Roman Ishchenko. 2025. "Current Trends and Future Opportunities of AI-Based Analysis in Mesenchymal Stem Cell Imaging: A Scoping Review" Journal of Imaging 11, no. 10: 371. https://doi.org/10.3390/jimaging11100371
APA StyleSolopov, M., Chechekhina, E., Turchin, V., Popandopulo, A., Filimonov, D., Burtseva, A., & Ishchenko, R. (2025). Current Trends and Future Opportunities of AI-Based Analysis in Mesenchymal Stem Cell Imaging: A Scoping Review. Journal of Imaging, 11(10), 371. https://doi.org/10.3390/jimaging11100371








