AI Diffusion Models Generate Realistic Synthetic Dental Radiographs Using a Limited Dataset
Abstract
1. Introduction
2. Materials and Methods
2.1. Dental Radiograph Database
2.2. Development of Training Dataset for Model Development
2.3. Clinical Expert Involvement
2.3.1. Expert Panel (EP) of Dentists
2.3.2. Primary Dentist Evaluator
2.4. Survey Instrument
2.5. AI Model Development
2.5.1. Hardware and Software Configuration
2.5.2. Model Architecture and Training Development
2.5.3. SDR Model #1
2.5.4. SDR Model #2
2.5.5. SDR Model #3
2.6. Assessment of Image Generation Model Performance
2.6.1. Expert Evaluation
2.6.2. Distributional Similarity
3. Results
3.1. Training Datasets
3.2. Expert Evaluation of Model Performance
3.3. Objective Analysis of Model Performance
3.3.1. Absolute Performance
3.3.2. Pairwise Comparisons
3.3.3. Overall Patterns
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A2I2 | U.S. Army Artificial Intelligence Institute |
| ADAM | Adaptive Moment Estimation |
| AI | Artificial Intelligence |
| BWRM | Bitewing Right Molar |
| DHA | Defense Health Agency |
| DICOM | Digital Imaging and Communication in Medicine |
| ECIA | Enterprise Clinical Image Archive |
| EP | Expert Panel |
| FID | Fréchet Inception Distance |
| GAN | Generative Adversarial Network |
| GELU | Gaussian Error Linear Unit |
| GPU | Graphic Processing Unit |
| JPEG | Joint Photographic Experts Group |
| KID | Kernel Inception Distance |
| MMD | Minimum Mean Discrepancy |
| MSE | Mean Squared Error |
| SDR | Synthetic Dental Radiograph |
| SiLU | Sigmoid Linear Unit |
| SME | Subject Matter Expert |
| USAISR | U.S. Army Institute of Surgical Research |
| UIC | Unique Identifier Code |
| VAE | Variational Autoencoder |
References
- Koetzier, L.R.; Wu, J.; Mastrodicasa, D.; Lutz, A.; Chung, M.; Koszek, W.A.; Pratap, J.; Chaudhari, A.S.; Rajpurkar, P.; Lungren, M.P.; et al. Generating Synthetic Data for Medical Imaging. Radiology 2024, 312, e232471. [Google Scholar] [CrossRef]
- Zhao, H.; Li, H.; Cheng, L. Chapter 14—Data Augmentation for Medical Image Analysis. In Biomedical Image Synthesis and Simulation; Burgos, N., Svoboda, D., Eds.; The MICCAI Society Book Series; Academic Press: Cambridge, MA, USA, 2022; pp. 279–302. ISBN 978-0-12-824349-7. [Google Scholar]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A Review of Medical Image Data Augmentation Techniques for Deep Learning Applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Sunilkumar, A.P.; Keshari Parida, B.; You, W. Recent Advances in Dental Panoramic X-Ray Synthesis and Its Clinical Applications. IEEE Access 2024, 12, 141032–141051. [Google Scholar] [CrossRef]
- Svoboda, D.; Burgos, N. Chapter 1—Introduction to Medical and Biomedical Image Synthesis. In Biomedical Image Synthesis and Simulation; Burgos, N., Svoboda, D., Eds.; The MICCAI Society Book Series; Academic Press: Cambridge, MA, USA, 2022; pp. 1–3. ISBN 978-0-12-824349-7. [Google Scholar]
- Chen, R.J.; Lu, M.Y.; Chen, T.Y.; Williamson, D.F.K.; Mahmood, F. Synthetic Data in Machine Learning for Medicine and Healthcare. Nat. Biomed. Eng. 2021, 5, 493–497. [Google Scholar] [CrossRef]
- Kokomoto, K.; Okawa, R.; Nakano, K.; Nozaki, K. Intraoral Image Generation by Progressive Growing of Generative Adversarial Network and Evaluation of Generated Image Quality by Dentists. Sci. Rep. 2021, 11, 18517. [Google Scholar] [CrossRef]
- He, H.; Zhao, S.; Xi, Y.; Ho, J.C. MedDiff: Generating Electronic Health Records Using Accelerated Denoising Diffusion Model. arXiv 2023, arXiv:2302.04355. [Google Scholar] [CrossRef]
- Zhang, K.; Khosravi, B.; Vahdati, S.; Faghani, S.; Nugen, F.; Rassoulinejad-Mousavi, S.M.; Moassefi, M.; Jagtap, J.M.M.; Singh, Y.; Rouzrokh, P.; et al. Mitigating Bias in Radiology Machine Learning: 2. Model Development. Radiol. Artif. Intell. 2022, 4, e220010. [Google Scholar] [CrossRef] [PubMed]
- Alaa, A.; Breugel, B.V.; Saveliev, E.S.; van der Schaar, M. How Faithful Is Your Synthetic Data? Sample-Level Metrics for Evaluating and Auditing Generative Models. In Proceedings of the 39th International Conference on Machine Learning, Baltimore, MD, USA, 28 June 2022; pp. 290–306. [Google Scholar]
- Sylolypavan, A.; Sleeman, D.; Wu, H.; Sim, M. The Impact of Inconsistent Human Annotations on AI Driven Clinical Decision Making. NPJ Digit. Med. 2023, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Hegde, S.; Gao, J.; Vasa, R.; Cox, S. Factors Affecting Interpretation of Dental Radiographs. Dentomaxillofacial Radiol. 2023, 52, 20220279. [Google Scholar] [CrossRef] [PubMed]
- Umer, F.; Adnan, N. Generative Artificial Intelligence: Synthetic Datasets in Dentistry. BDJ Open 2024, 10, 13. [Google Scholar] [CrossRef]
- Pedersen, S.; Jain, S.; Chavez, M.; Ladehoff, V.; de Freitas, B.N.; Pauwels, R. Pano-GAN: A Deep Generative Model for Panoramic Dental Radiographs. J. Imaging 2025, 11, 41. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing Medical Imaging Data for Machine Learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Rossum, G.V. Python Software Foundation Python Programming Language 2024; Python Software Foundation: Beaverton, OR, USA, 2024. [Google Scholar]
- SurveyMonkey Inc. SurveyMonkey 2025; SurveyMonkey Inc.: San Mateo, CA, USA, 2025. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Proceedings of the Advances in Neural Information Processing Systems 32 (NeurIPS 2019), Vancouver, BC, Canada, 8–14 December 2019; Volume 32. [Google Scholar]
- R Core Team R: A Language and Environment for Statistical Computing 2023. Available online: https://www.scirp.org/reference/referencespapers?referenceid=3582659 (accessed on 3 June 2025).
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Chong, M.J.; Forsyth, D. Effectively Unbiased FID and Inception Score and Where to Find Them. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 6070–6079. [Google Scholar]
- Konz, N.; Chen, Y.; Gu, H.; Dong, H.; Mazurowski, M.A. Rethinking Perceptual Metrics for Medical Image Translation 2024. arXiv 2024, arXiv:2404.07318. [Google Scholar]
- Li, X.; Ren, Y.; Jin, X.; Lan, C.; Wang, X.; Zeng, W.; Wang, X.; Chen, Z. Diffusion Models for Image Restoration and Enhancement—A Comprehensive Survey. Int. J. Comput. 2023, 1–31. [Google Scholar] [CrossRef]
- Khader, F.; Müller-Franzes, G.; Tayebi Arasteh, S.; Han, T.; Haarburger, C.; Schulze-Hagen, M.; Schad, P.; Engelhardt, S.; Baeßler, B.; Foersch, S.; et al. Denoising Diffusion Probabilistic Models for 3D Medical Image Generation. Sci. Rep. 2023, 13, 7303. [Google Scholar] [CrossRef]
- Ho, J.; Saharia, C.; Chan, W.; Fleet, D.J.; Norouzi, M.; Salimans, T. Cascaded Diffusion Models for High Fidelity Image Generation. J. Mach. Learn. Res. 2022, 23, 1–33. [Google Scholar]
- Karras, T.; Aittala, M.; Lehtinen, J.; Hellsten, J.; Aila, T.; Laine, S. Analyzing and Improving the Training Dynamics of Diffusion Models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 24174–24184. [Google Scholar]
- Liu, J.; Wang, Q.; Fan, H.; Wang, Y.; Tang, Y.; Qu, L. Residual Denoising Diffusion Models. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 2773–2783. [Google Scholar]
- Ho, J.; Jain, A.; Abbeel, P. Denoising Diffusion Probabilistic Models. In Proceedings of the Advances in Neural Information Processing Systems 32 (NeurIPS 2019), Vancouver, BC, Canada, 6–12 December 2020; Volume 33, pp. 6840–6851. [Google Scholar]
- Schmarje, L. Addressing the Challenge of Ambiguous Data in Deep Learning: A Strategy for Creating High-Quality Image Annotations with Human Reliability and Judgement Enhancement. Ph.D. Thesis, Das Institut für Informatik der Christian-Albrechts-Universität zu Kiel, Kiel, Germany, 2024. [Google Scholar] [CrossRef]
- Fukuda, M.; Kotaki, S.; Nozawa, M.; Tsuji, K.; Watanabe, M.; Akiyama, H.; Ariji, Y. An Attempt to Generate Panoramic Radiographs Including Jaw Cysts Using StyleGAN3. Dentomaxillofacial Radiol. 2024, 53, 535–541. [Google Scholar] [CrossRef] [PubMed]
- Lindén, J.; Forsberg, H.; Daneshtalab, M.; Söderquist, I. Evaluating the Robustness of ML Models to Out-of-Distribution Data Through Similarity Analysis. In New Trends in Database and Information Systems; Abelló, A., Vassiliadis, P., Romero, O., Wrembel, R., Bugiotti, F., Gamper, J., Solar, G.V., Zumpano, E., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 348–359. [Google Scholar]
- Kynkäänniemi, T.; Karras, T.; Aittala, M.; Aila, T.; Lehtinen, J. The Role of ImageNet Classes in Fréchet Inception Distance. arXiv 2023, arXiv:2203.06026. [Google Scholar] [CrossRef]
- Kwiatkowski, D.; Dziubich, T. Comparison of Selected Neural Network Models Used for Automatic Liver Tumor Segmentation. In New Trends in Database and Information Systems; Abelló, A., Vassiliadis, P., Romero, O., Wrembel, R., Bugiotti, F., Gamper, J., Vargas Solar, G., Zumpano, E., Eds.; Springer Nature: Cham, Switzerland, 2023; pp. 511–522. [Google Scholar]
- Theis, L.; van den Oord, A.; Bethge, M. A Note on the Evaluation of Generative Models. arXiv 2016, arXiv:1511.01844. [Google Scholar]
- McNeely-White, D.; Beveridge, J.R.; Draper, B.A. Inception and ResNet Features Are (Almost) Equivalent. Cogn. Syst. Res. 2020, 59, 312–318. [Google Scholar] [CrossRef]
- Sismadi, A.; Rianto, S.; Abimanyu, B. Influence of Pixel Size Difference on Imaging Plate Computed Radiography on the Quality of Digital Images in Intraoral Dental Radiography Examination. J. World Sci. 2023, 2, 1822–1829. [Google Scholar] [CrossRef]

| Category | Criteria |
|---|---|
| Inclusion | Contain all teeth present distal to 1st premolar to distal of 2nd molar on both maxillary and mandibular arch on the right side. |
| The entire crown of each tooth is visible. | |
| Alveolar crest is visible interproximal between teeth. | |
| Exclusion | Unable to identify tooth anatomy. |
| Contain any edentulous spaces. | |
| Poor image quality that requires retake of the image. | |
| The extent of overlap of proximal contacts requires retake of the image. | |
| Excessive occlusal plane rotation requires retake of the image. |
| Category | Description | Sample Images | |
|---|---|---|---|
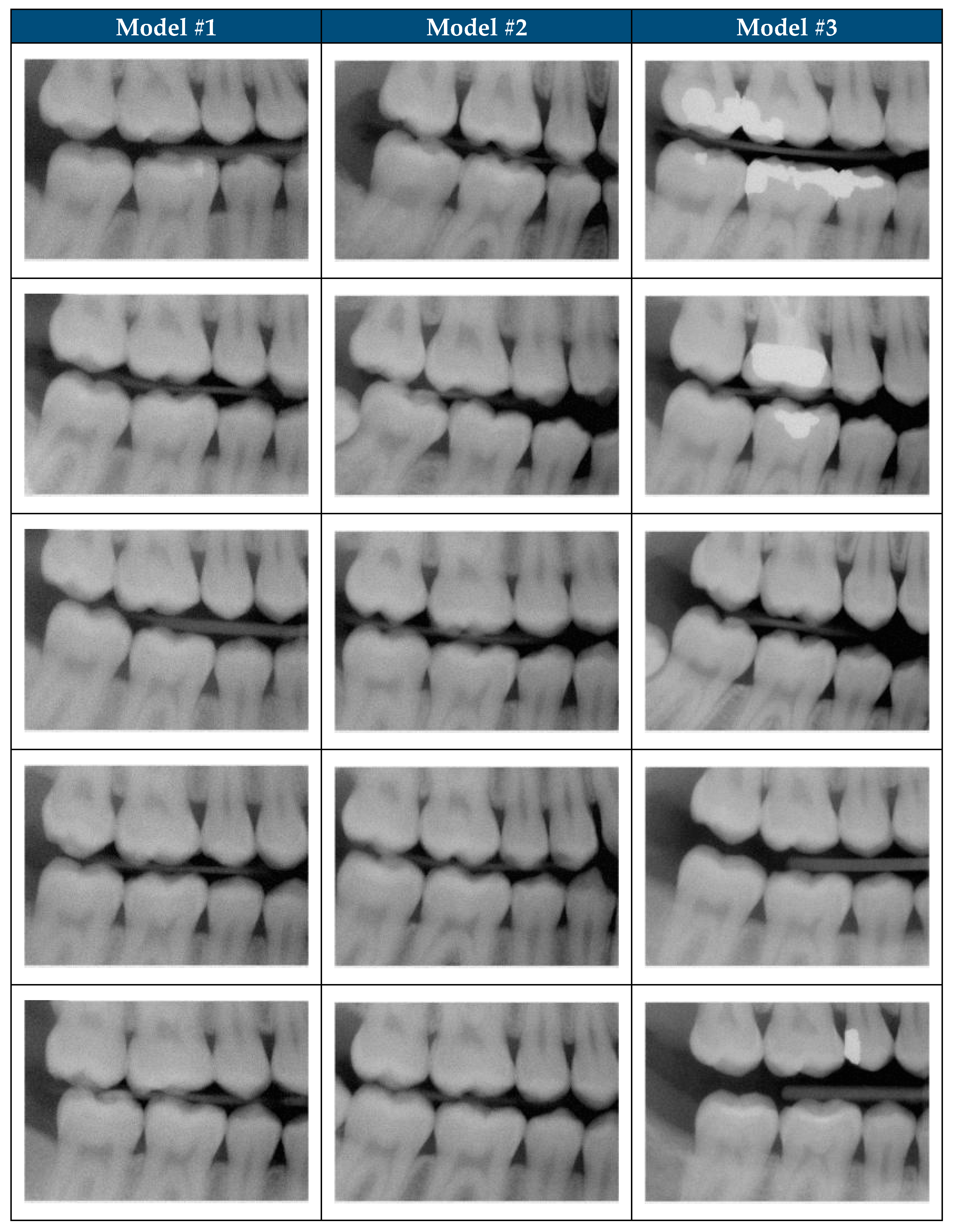
| 1 | The image appears to be a realistic dental radiograph representative of the training data. (Looks real with no anatomic anomalies) |  |  |
| 2 | Image resembles a realistic dental radiograph representative of the training data but contains anatomical hallucinations or abnormalities. (Looks real but tooth count, order or anatomy is unreal) | 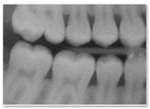 |  |
| 3 | Image is unrealistic but resembles the general appearance of dental radiograph represented in the training data. (Looks like a dental radiograph with features that are obviously fake) |  | 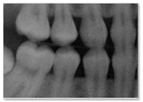 |
| 4 | The image is unrealistic, but portions of the image contain dental-related attributes. (At minimum portions of tooth anatomy are present) | 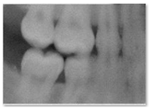 | 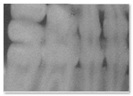 |
| 5 | No recognizable dental-related attributes. |  |  |
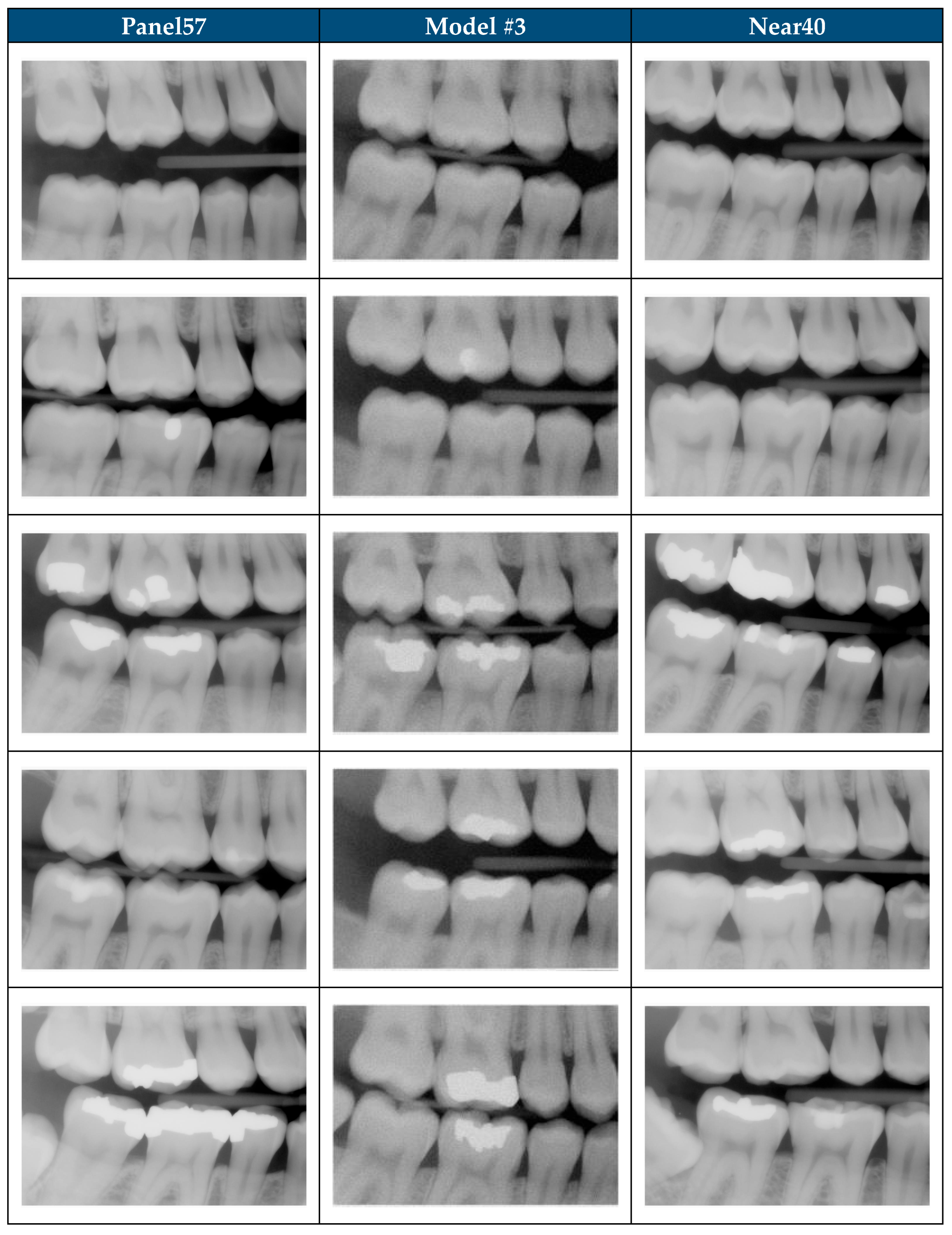
| Subset | Total Images | Selection Method | Training Exposure |
|---|---|---|---|
| All200 | 200 | BWRM radiographs that met image selection criteria | Model #1: all 200. Models #2–#3: Panel57 only |
| Panel57 | 57 | Expert-panel selected subset of All200 | Seen by all 3 models |
| Unseen143 | 143 | Remainder of All200 not in Panel57 | Unseen by Model #2 and Model #3 |
| Shift1000 | 1000 | Clinically acceptable BWRM excluded from model training | No exposure (all 3 models) |
| Near40 | 40 | Algorithmic-matched subset of Shift1000 (closest to All200) | No exposure (all 3 models) |
| Random40 (×5) | 40 | Five independent random subsets from Shift1000 | No exposure (all 3 models) |
| Dataset Description | Number |
|---|---|
| Dental Radiographs (unprocessed) | |
| Total | 10,000 |
| BWRM subset | |
| Total (EP0) | 2226 |
| Clinically Acceptable (EP0) | 1284 |
| Image Selection Criteria satisfied (EP0) | 225 |
| Expert-Informed Curation subset | |
| Total Evaluated (EP) | 100 |
| Include (EP) | 57 |
| Exclude (EP) | 43 |
| EP Agreement (Image Selection Criteria) | |
| Include (4 of 4 EP members) | 36 |
| Include (3 of 4 EP members) | 7 |
| Include (2 of 4 EP members) | 4 |
| Include (1 of 4 EP members) | 10 |
| Exclude (4 of 4 EP members) | 43 |
| Description | Model #1 | Model #1 | Model #1 | Model #2 | Model #3 |
|---|---|---|---|---|---|
| Training Dataset | |||||
| Total number of images | 200 | 200 | 200 | 200 | 200 |
| Number of unique images | 200 | 200 | 200 | 57 | 57 |
| Model Training | |||||
| Epochs | 22,000 | 30,000 | 40,000 | 30,000 | 30,000 |
| Diffusion steps | 300 | 300 | 300 | 300 | 600 |
| Model Performance | |||||
| Total SDR Graded | 500 | 500 | 500 | 500 | 500 |
| Number of Images Scored 1 | 28 | 74 | 9 | 110 | 451 |
| Number of Images Scored 2 | 120 | 51 | 34 | 109 | 42 |
| Number of Images Scored 3 | 88 | 146 | 35 | 134 | 3 |
| Number of Images Scored 4 | 263 | 214 | 379 | 139 | 4 |
| Number of Images Scored 5 | 1 | 15 | 43 | 8 | 0 |
| Average Score | 3.18 | 3.09 | 3.83 | 2.65 | 1.12 |
| Standard Deviation | 0.98 | 1.11 | 0.75 | 1.15 | <0.01 |
| Realistic SDR Generation rate | 6% | 15% | 2% | 22% | 90% |
| Model Refinements | |||||
| Refinement | Training Duration (Epochs) | Training Duration (Epochs) | Training Duration (Epochs) | Expert Panel Refined Dataset | Addition of 300 Diffusion Steps |
| Impact of Refinement on model performance (p-Value) | Worse (<0.05) | Improved (<0.05) | Worse (<0.05) | Improved (<0.05) | Improved (<0.05) |
| Analysis by Subset | Model #1 | Model #2 | Model #3 |
|---|---|---|---|
| All200 | |||
| FID Real vs. Real “floor” (95% CI) | 43.6 (42.2–45.8) | ||
| FID Mean (95% CI) | 221.1 (206.9–235.0) | 182.2 (176.0–189.3) | 114.413 (108.5–120.2) |
| KID Mean (95% CI) | 0.0037 (0.0034–0.0040) | 0.0036 (0.0034–0.0039) | 0.0024 (0.0021–0.0027) |
| Panel57 | |||
| FID Real vs. Real “floor” (95% CI) | 80.0 (75.8–84.5) | ||
| FID Mean (95% CI) | 233.6 (218.9–250.4) | 195.6 (182.9–204.8) | 127.3 (118.0–136.8) |
| KID Mean (95% CI) | 0.0038 (0.0035–0.0042) | 0.0037 (0.0033–0.0040) | 0.0024 (0.0019–0.0028) |
| Unseen143 | |||
| FID Real vs. Real “floor” (95% CI) | 50.6 (48.5–53.2) | ||
| FID Mean (95% CI) | 223.7 (214.4–234.5) | 184.0 (176.5–194.4) | 117.3 (107.2–129.6) |
| KID Mean (95% CI) | 0.0037 (0.0035–0.0039) | 0.0036 (0.0033–0.0039) | 0.0024 (0.0020–0.0030) |
| Shift1000 | |||
| FID Real vs. Real “floor” (95% CI) | 24.5 (24.0–25.1) | ||
| FID Mean (95% CI) | 208.2 (200.4–217.2) | 174.3 (168.7–179.4) | 126.2 (119.1–133.9) |
| KID Mean (95% CI) | 0.0032 (0.0030–0.0036) | 0.0033 (0.0030–0.0035) | 0.0025 (0.0022–0.0028) |
| Near40 | |||
| FID Real vs. Real “floor” (95% CI) | 53.1 (50.4–56.4) | ||
| FID Mean (95% CI) | 231.2 (213.1–248.2) | 197.8 (183.9–209.2) | 129.7 (117.0–144.1) |
| KID Mean (95% CI) | 0.0038 (0.0035–0.0042) | 0.0039 (0.0035–0.0043) | 0.0026 (0.0020–0.0034) |
| Random40_1 | |||
| FID Real vs. Real “floor” (95% CI) | 107.1 (99.8–117.6) | ||
| FID Mean (95% CI) | 239.7 (223.3–253.7) | 204.2 (194.2–213.8) | 145.7 (134.3–159.2) |
| KID Mean (95% CI) | 0.0034 (0.0031–0.0037) | 0.0034 (0.0030–0.0039) | 0.0024 (0.0019–0.0032) |
| Random40_2 | |||
| FID Real vs. Real “floor” (95% CI) | 115.5 (108.6–124.2) | ||
| FID Mean (95% CI) | 241.0 (226.9–253.5) | 204.8 (196.8–213.6) | 161.9 (150.6–174.4) |
| KID Mean (95% CI) | 0.0034 (0.0031–0.0037) | 0.0034 (0.0030–0.0039) | 0.0028 (0.0023–0.0035) |
| Random40_3 | |||
| FID Real vs. Real “floor” (95% CI) | 121.5 (115.2–129.3) | ||
| FID Mean (95% CI) | 236.3 (220.1–250.8) | 200.8 (191.6–209.0) | 149.9 (139.6–162.8) |
| KID Mean (95% CI) | 0.0033 (0.0030–0.0036) | 0.0033 (0.0029–0.0037) | 0.0024 (0.0019–0.0032) |
| Random40_4 | |||
| FID Real vs. Real “floor” (95% CI) | 112.4 (105.7–123.2) | ||
| FID Mean (95% CI) | 228.3 (212.0–244.6) | 194.3 (184.9–203.9) | 143.4 (132.9–155.4) |
| KID Mean (95% CI) | 0.0030 (0.0027–0.0033) | 0.0030 (0.0026–0.0035) | 0.0023 (0.0018–0.0031) |
| Random40_5 | |||
| FID Real vs. Real “floor” (95% CI) | 115.9 (108.1–126.2) | ||
| FID Mean (95% CI) | 232.8 (218.1–248.2) | 195.9 (187.4–204.7) | 151.9 (142.2–162.5) |
| KID Mean (95% CI) | 0.0033 (0.0030–0.0036) | 0.0031 (0.0028–0.0036) | 0.0024 (0.0020–0.0031) |
| Subset | Δ FID Mean * (95% CI) | ||
|---|---|---|---|
| Model #1–Model #2 | Model #2–Model #3 | Model #3–Model #1 | |
| All200 | 38.9 (25.9–55.8) | 67.8 (58.6–79.0) | −106.6 (−118.4–−92.0) |
| Panel57 | 38.0 (19.5–56.1) | 68.3 (55.0–82.6) | −106.4 (−125.3–−90.2) |
| Unseen143 | 39.7 (24.4–53.7) | 66.7 (52.9–77.1) | −106.4 (−119.7–−89.0) |
| Shift1000 | 34.0 (25.6–44.3) | 48.1 (40.4–54.2) | −82.1 (−92.9–−71.8) |
| Near40 | 33.3 (13.1–54.2) | 68.2 (53.3–87.7) | −101.5 (−120.0–−78.0) |
| Random40_1 | 35.5 (16.2–52.8) | 58.5 (43.4 –75.0) | −94.1 (−112.3–−73.3) |
| Random40_2 | 36.1 (18.8–51.3) | 43.0 (27.4–59.1) | −79.1 (−98.7–−60.0) |
| Random40_3 | 35.5 (15.8–52.9) | 50.9 (36.0–66.4) | −86.4 (−104.8–−65.5) |
| Random40_4 | 34.0 (15.1–52.7) | 51.0 (37.1–66.3) | −85.0 (−102.9–−64.7) |
| Random40_5 | 36.9 (20.6–52.4) | 44.0 (31.1–59.0) | −81.0 (−101.0–−60.3) |
| Evaluation Subset | Lowest FID (Best) | Rank Order |
|---|---|---|
| In-distribution Comparison | ||
| All200 | Model #3 | Model #3 > Model #2 > Model #1 |
| Panel57 | Model #3 | Model #3 > Model #2 > Model #1 |
| Unseen143 | Model #3 | Model #3 > Model #2 > Model #1 |
| Out-of-Distribution Comparison | ||
| Shift1000 | Model #3 | Model #3 > Model #2 > Model #1 |
| Near40 | Model #3 | Model #3 > Model #2 > Model #1 |
| Random40_1 | Model #3 | Model #3 > Model #2 > Model #1 |
| Random40_2 | Model #3 | Model #3 > Model #2 > Model #1 |
| Random40_3 | Model #3 | Model #3 > Model #2 > Model #1 |
| Random40_4 | Model #3 | Model #3 > Model #2 > Model #1 |
| Random40_5 | Model #3 | Model #3 > Model #2 > Model #1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirkwood, B.; Choi, B.Y.; Bynum, J.; Salinas, J. AI Diffusion Models Generate Realistic Synthetic Dental Radiographs Using a Limited Dataset. J. Imaging 2025, 11, 356. https://doi.org/10.3390/jimaging11100356
Kirkwood B, Choi BY, Bynum J, Salinas J. AI Diffusion Models Generate Realistic Synthetic Dental Radiographs Using a Limited Dataset. Journal of Imaging. 2025; 11(10):356. https://doi.org/10.3390/jimaging11100356
Chicago/Turabian StyleKirkwood, Brian, Byeong Yeob Choi, James Bynum, and Jose Salinas. 2025. "AI Diffusion Models Generate Realistic Synthetic Dental Radiographs Using a Limited Dataset" Journal of Imaging 11, no. 10: 356. https://doi.org/10.3390/jimaging11100356
APA StyleKirkwood, B., Choi, B. Y., Bynum, J., & Salinas, J. (2025). AI Diffusion Models Generate Realistic Synthetic Dental Radiographs Using a Limited Dataset. Journal of Imaging, 11(10), 356. https://doi.org/10.3390/jimaging11100356









