Automated Lung Cancer Diagnosis Applying Butterworth Filtering, Bi-Level Feature Extraction, and Sparce Convolutional Neural Network to Luna 16 CT Images
Abstract
1. Introduction
- Lack of image quality and resolution: the existing models are significantly influenced by the quality and resolution of the CT scans. Low-quality scans or those with artifacts introduce some noise, thus reducing the model’s effectiveness;
- Feature selection and algorithmic impact: feature selection accuracy depends on the algorithm’s choice and selected features’ number. Assumptions about critical characteristics for lung cancer detection may not always align with the model’s accuracy;
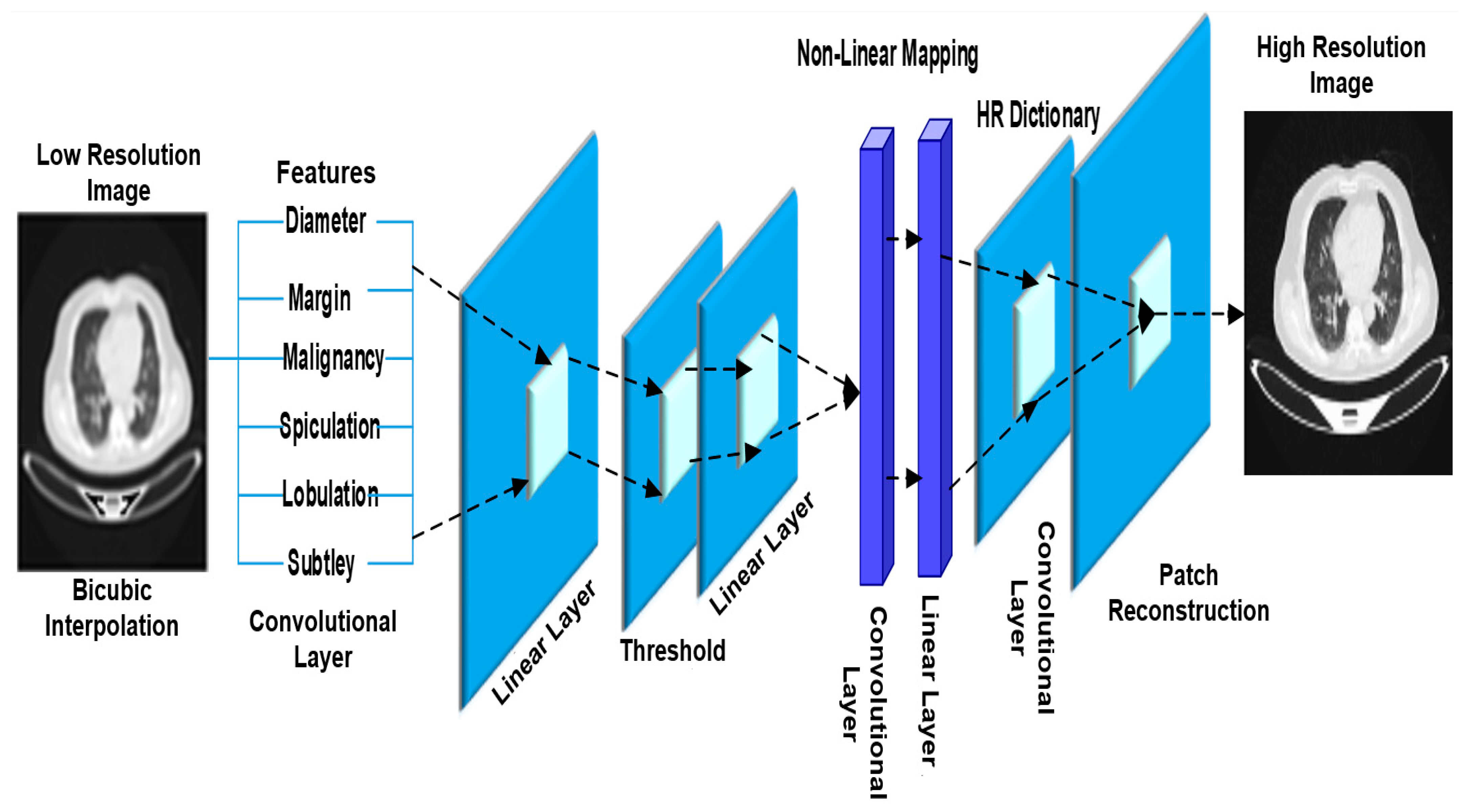
- Low-resolution image generation in the biomedical field: the existing image generation techniques face challenges such as less accuracy and high processing time while struggling with generating high-resolution images essential for the biomedical field;
- Sensitivity and model evaluation: in existing methods, the sensitivity fluctuations at low and high false positive (FP) rates, coupled with the impact of false negative rates on the F-1 score and negative predictive value (NPV), present challenges in achieving robust model evaluation and high performance.
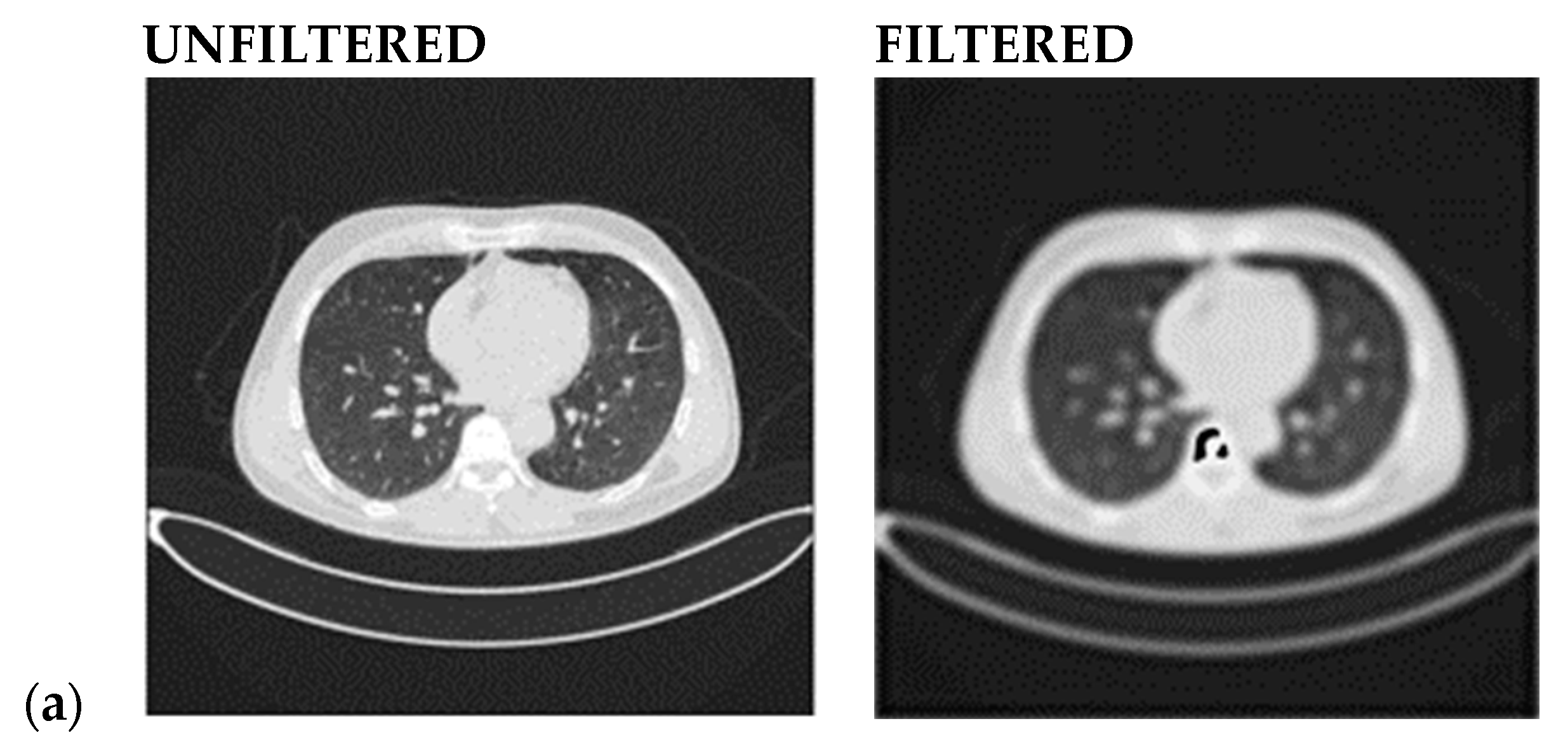
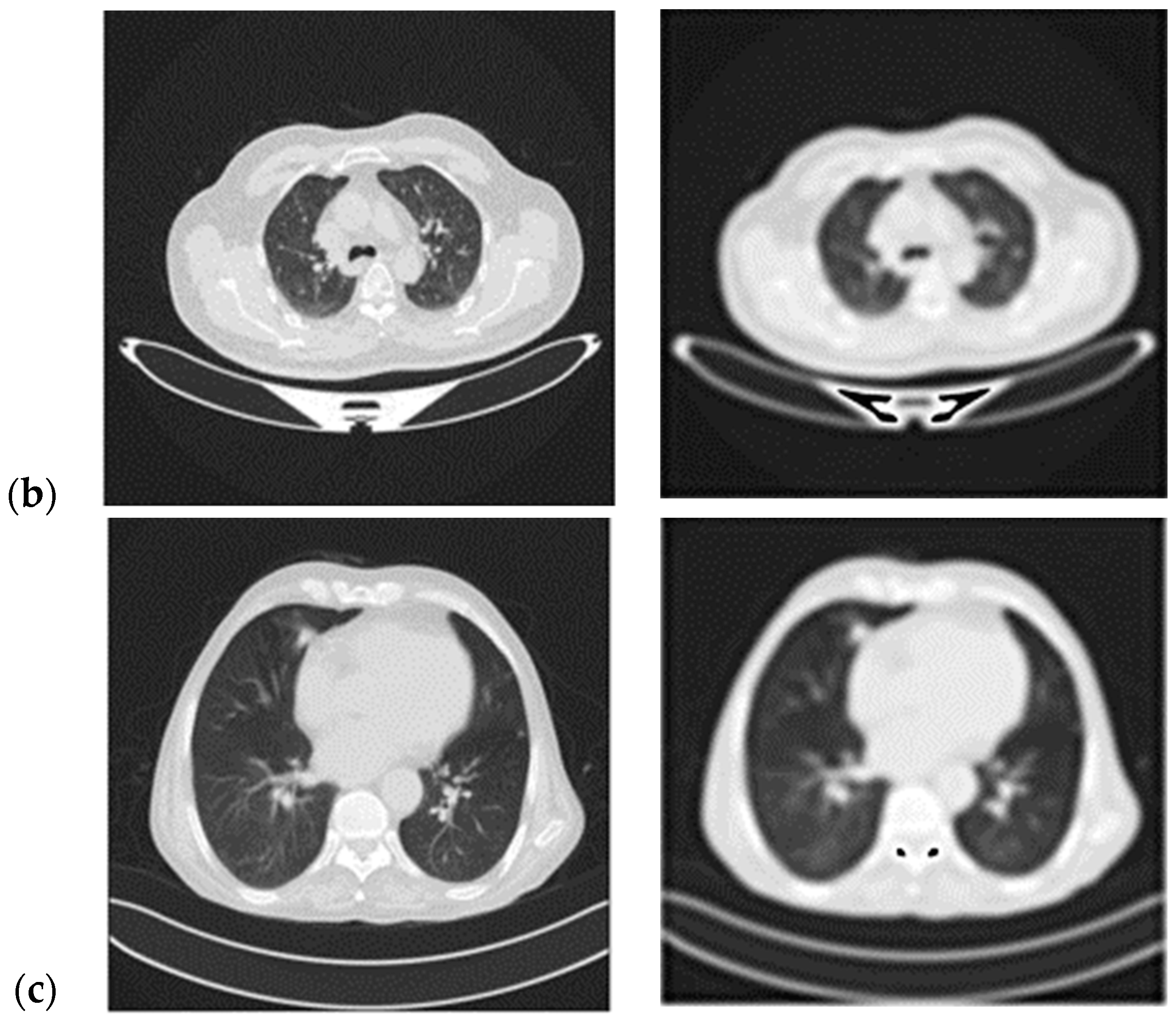
- A Butterworth smooth filter-based pre-processing can be applied to reduce the noise, that could be misinterpreted as features, before utilizing the CNN-based module. The efficacy of CNN models applied to smoothed images is investigated; this methodology provides good results in comparison to literature in which many attempts to segment lesions or increase the lesion quality, but this coincides with keeping noise that impacts the CNN models’ performance (i.e., accuracy) [21,22,23];
- To achieve better feature selection and extraction results, dual CCSA-RF and MIR-GLCM-based methods are performed to select the proper features, thus providing profitable outcomes for the lung-cancer CT image classification;
- To improve the classification accuracy, the Sparse Convolutional Neural Network with a Probabilistic Neural Network (SCNN-PNN)-based classification is applied to remove the redundant features from the attention module and provide high classification accuracy with less complexity.
Literature Survey
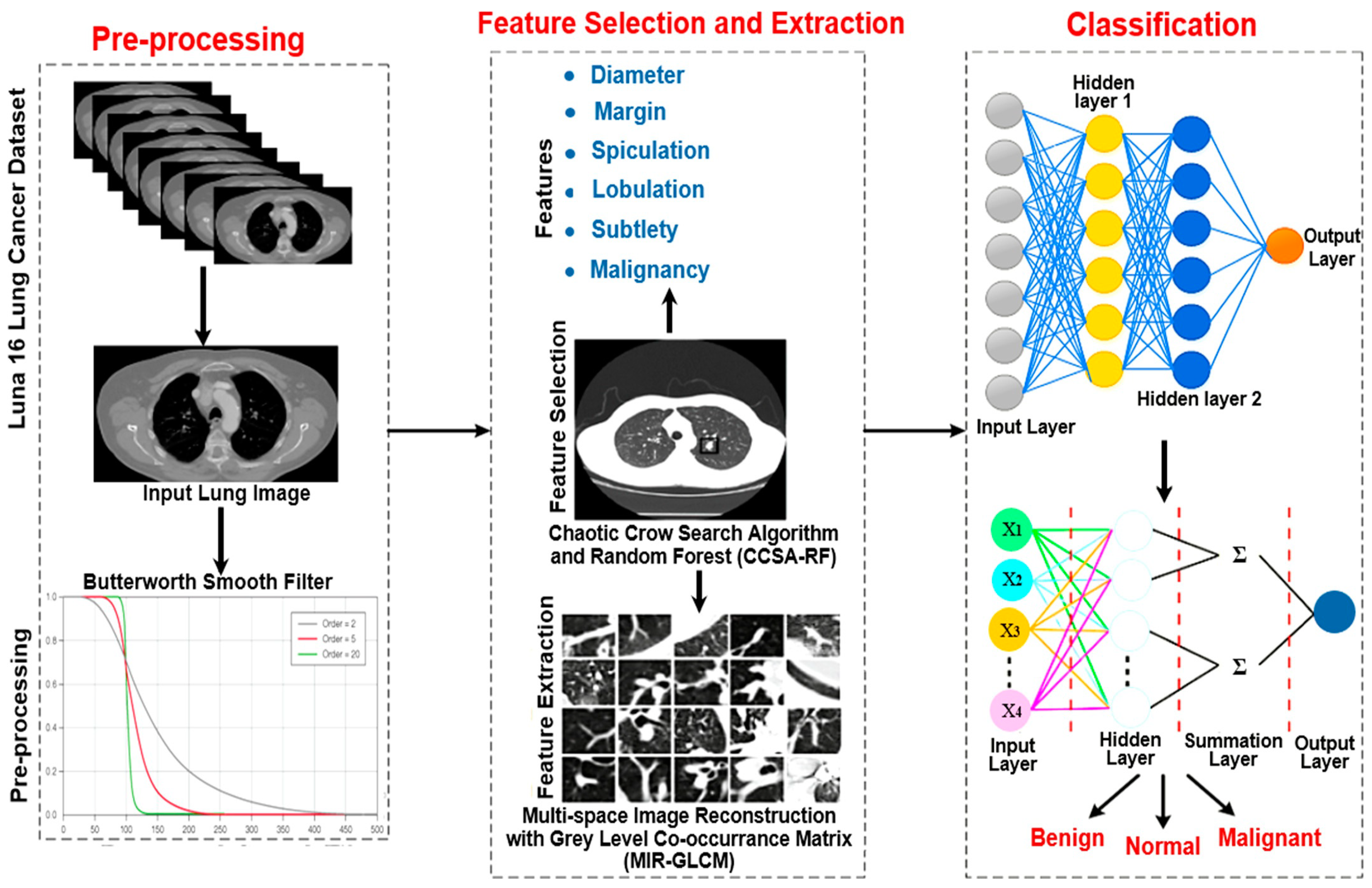
2. Materials and Methods
2.1. Problem Statement and Research Solutions
- The quality and resolution of CT scans can considerably influence the model’s performance. Scans that are low quality or contain artifacts may cause noise in the model, limiting its usefulness;
- It should be noted that the assumption that the chosen parameters are vital for lung cancer diagnosis is not always correct. Additional features or variables can help identify lung cancer but are not included in the model;
- The method used and the number of features chosen can affect the accuracy of the feature selection approach.
- The Lung Cancer Deep Convolutional generative adversarial networks (LDCGAN) are unable to produce high-resolution images, crucial for the biomedical field;
- The ensemble model requires a lot of time and computing resources to train.
2.2. Proposed Work
- Smooth filter-based pre-processing;
- Bi-level-based feature selection;
- Feature extraction;
- Severity classification of the lung tumor.
2.2.1. Smooth Filter-Based Pre-Processing
- Step 1: provide the input image;
- Step 2: Convert the input image to an array;
- Step 3: Use Fast Fourier Transform (FFT) to change the Fourier frequency of the resulting array;
- Step 4: Use Equation (1) to calculate the convolution function for the smooth filter;
- Step 5: Arrange the convolution function provided in Step 4 with the array created in Step 3;
- Step 6: Inverse FFT is used to calculate the magnitude;
- Step 7: Transform the resultant array to an image;
- Step 8: Perform all the procedures for the BHPF; to create the convolution function for the BHPF, Equation (2) is used instead of Equation (1);
- Step 9: Calculate the PSNR (Peak Signal-to-Noise Ratio) value.
2.2.2. Bi-Level-Based Feature Selection
- Parameter Initialization
- Fitness Function
- Position Updating
- Termination Criteria
| Algorithm 1: Chaotic crow search algorithm |
| Initialize the crow position α at random Calculate the fitness function F(α) for each crow Start the search by initializing the crow Q’s memory // Initialization of the counter Set τ: = 1 Repeat For l = 1 to H do Get the value of Chaotic map ς If ς_x ≥ AP^(x,τ) then α^(l,τ+1) = α^(l,τ) + ς_l × L_F × (Q^(x,τ) − α^(l,τ)) Else α^(l,τ+1) = A random position in the search space End if // Ensure viability of the new position If (v(α^(l,τ+1)) ≥ r) then α^(l,τ+1) = 1 Else α^(l,τ+1) = 0 End if End for Verify the viability of α^(l,τ+1) Analyze the crow’s new location F(α^(l,τ+1)) Update the crow’s memory using Q^(l,τ+1) Set τ = τ + 1 Until (τ < τmax) // Produce the best solution Q |
- Random Forest classifier
- Step 1: a random forest is a collection of decision tree algorithms used together as an ensemble. This technique extends the bootstrap aggregation (bagging) concept applied to decision trees, making it suitable for classification and regression tasks. Multiple decision trees are generated in bagging, each built from a distinct bootstrap sample of the training data.
- Step 2: once the random forest trees and classifiers have been built (Equation (7)), the predictions may be formed by running the test data through the rules of each decision tree to forecast the outcome, which is then stored.
| Algorithm 2: Developed RF model |
| Input: Dataset {(Y_j, X_j)|j = 1 to N, X_j ∈ γ^M, X ∈ {1, 2, …, c}} Output: Random Forest classifier For k = 1 to K do: Create bagged subset U_k from the whole dataset U While stopping criteria are not satisfied do: Select randomly mtry features For each feature in mtry do Calculate reduction in node impurity Select the feature that reduces impurity the most Split the nodes into two child nodes End For Construct the K trees to form the RF classifier |
- Step 3: collect the votes from several decision trees and decide the test object’s final classification. In other words, the final prediction made by the ensemble model is presented by Equation (8) [43], which defines the trees of the developed RF classifier:where is the predicted outcome of the kth individual model (in this case, a decision tree ) within the ensemble (K is the total number of models in the ensemble).
- Step 4: the prediction errors allow for the adjustment and validation of the random forest classifier during training. To achieve this goal, one can utilize the technique of Bagging, which involves training the model using random sub-sets of the training data in order to reduce the variance. Additionally, the Out-of-bag (OOB) error estimate can be calculated by averaging the errors obtained from predictions made by the trees that were not trained on their respective bootstrap samples (Equation (9)). This allows for the random forest classifier to be fine-tuned and verified during the training process [43].
- By altering the matching predictor feature values for shadow characteristics, create the extended dataset of two dimensions;
- Construct an RF model from and calculate R replicates of raw importance scores to use RF for predictor features and shadows. To build the comparison sample elements, extract the greatest significance score of each duplicate;
- To determine the weight of each predictor feature, take the R importance values and calculate the p-value using the Wilcoxon test;
- Ignore uninformative characteristics if there is a significance level threshold ;
- Divide the remaining features into two subsets, and (as in Algorithm 3);
- Sample the replacement-to-generate-bagged samples from the training set H;
- For each grow a CART tree as follows:
- For every node, choose a subspace of features at random and apart from and . Then, utilize the subspace features as candidates to divide each node;
- Every tree is grown non-deterministically, without pruning, until the minimum node size is reached;
- Given a , use step (1) to predict the response value.
| Algorithm 3: Random Forest |
| Input: H and an RF training dataset: number of replicates and threshold. Output: and Let For r to R do Build RF model from to produce and Set For i to M do Compute the Wilcoxon rank- sum test with and Compute values for each feature if then Set Compute statistic to get value for i to do if then Return |
2.2.3. Feature Extraction
- Contrast
- Correlation
- Energy
- Homogeneity
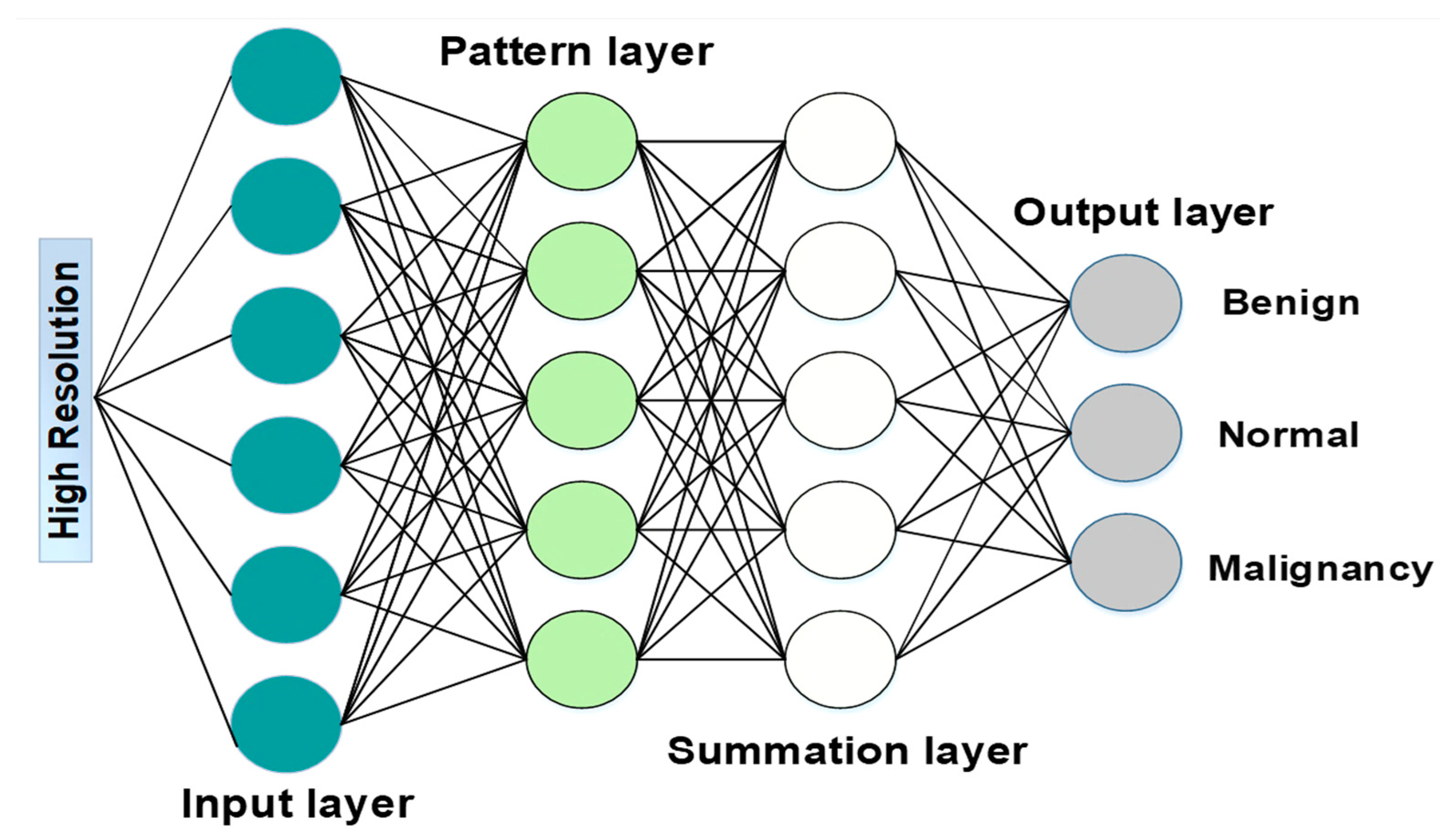
2.2.4. Lung Tumor Severity Classification
- Sparse Convolutional Neural Network (SCNN)
- Restricted Linear Unit
- Loss Function
- Input: read the input units , = 1, 2, …, and connect them to all pattern units.
- Pattern: create a pattern unit (which is either or unit) with weight vector:
- Summation: if goes to class , connect pattern unit to the summation unit , otherwise to unit . The weight used by the summation unit for class B is calculated according to Equation (37), where , is several training patterns in class .
- Decision: if the entire input to the decision unit is positive, the input vector is categorized as belonging to the class .
3. Results
3.1. Simulation Setup
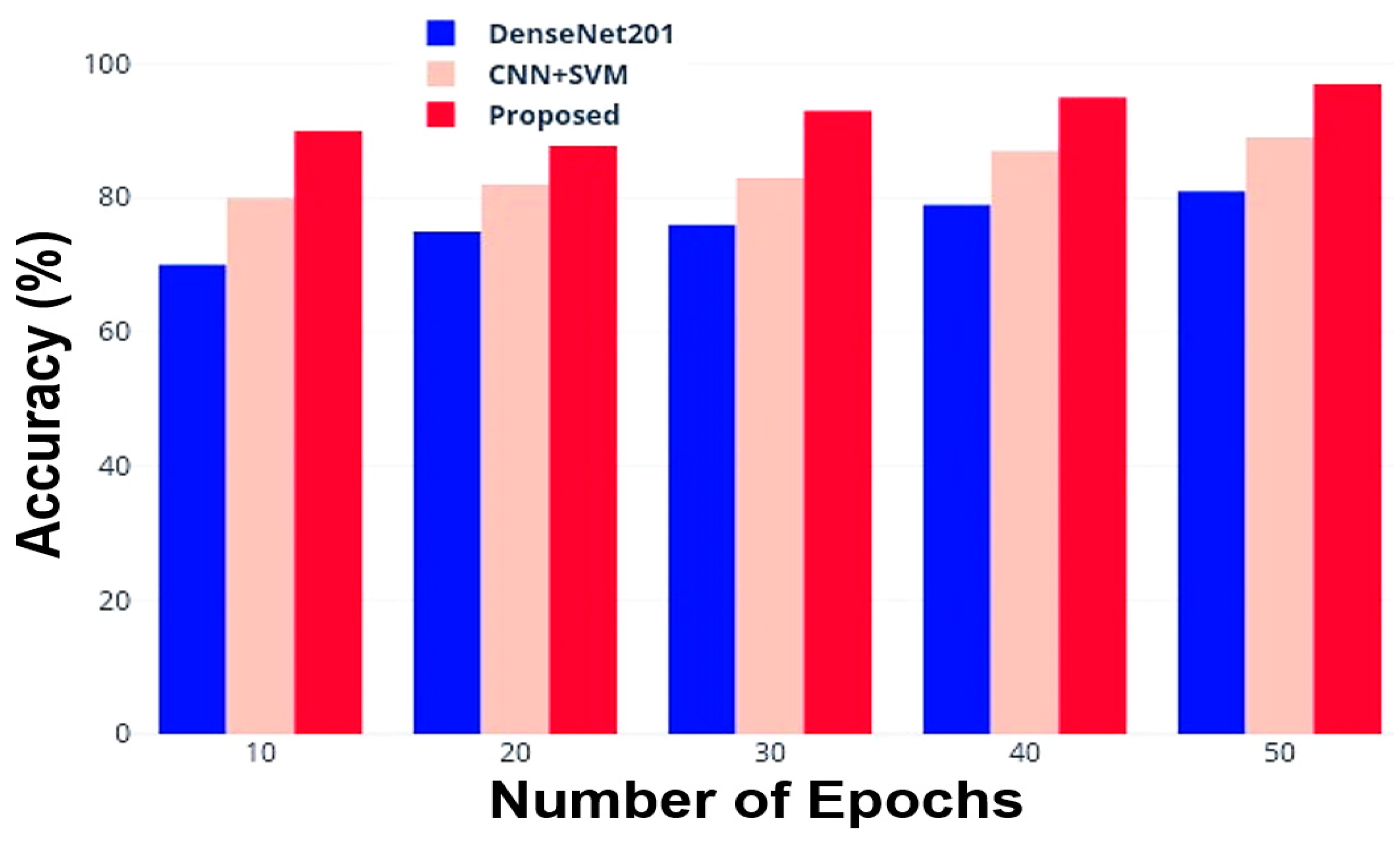
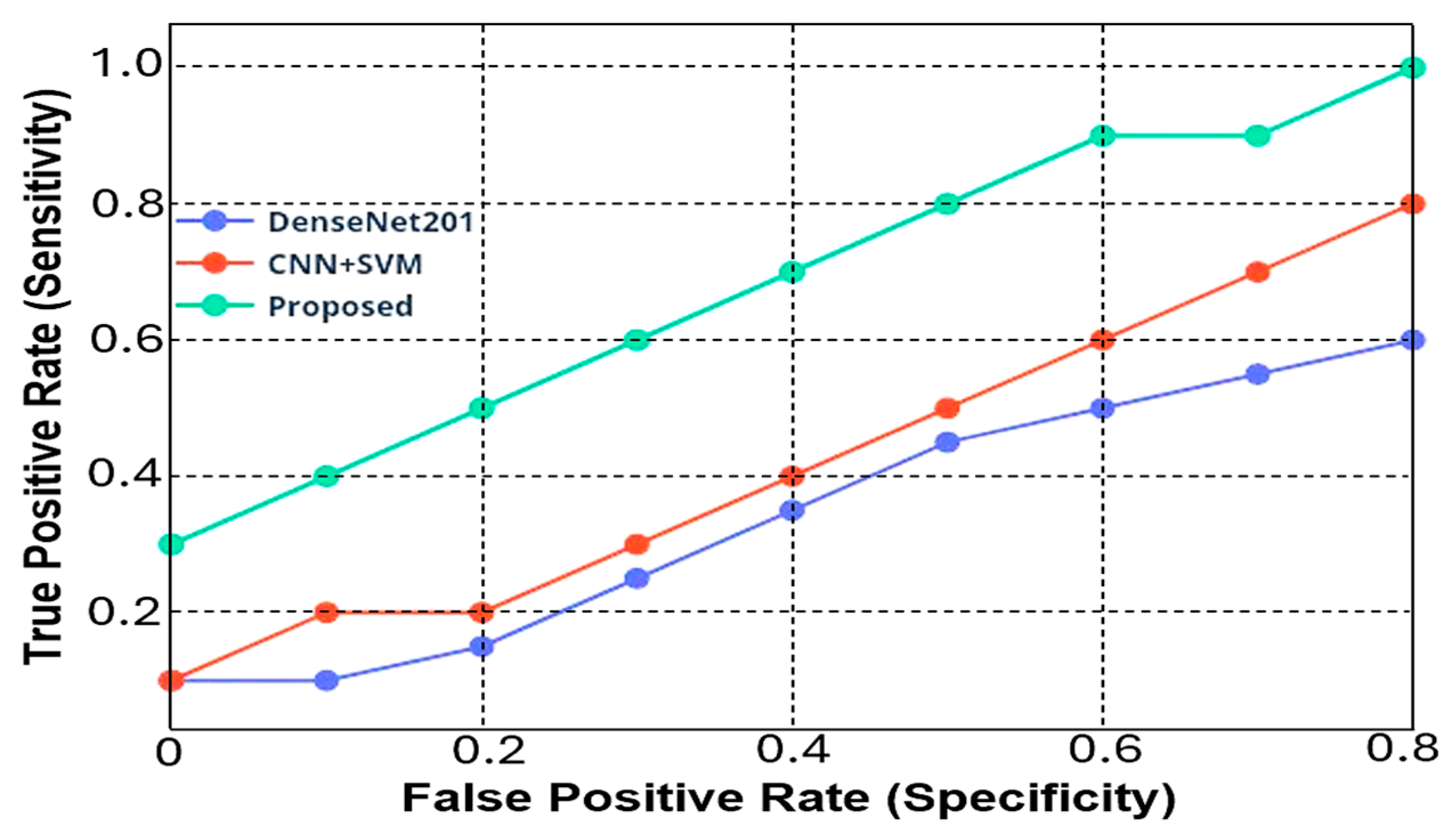
3.2. Comparative Analysis with Other DL Models
- Accuracy
- Precision
- F1-Score
- Sensitivity and Specificity
4. Results Discussion
Performance Comparison with the Scientific Literature
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chiu, H.Y.; Chao, H.S.; Chen, Y.M. Application of Artificial Intelligence in Lung Cancer. Cancers 2022, 14, 1370. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, C.; Liu, X.; Sun, W.; Li, L.; Gao, N.; Zhang, Y.; Yang, X.; Zhang, J.; Wang, H.; et al. Artificial intelligence-assisted system for precision diagnosis of PD-L1 expression in non-small cell lung cancer. Mod. Pathol. 2022, 35, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Khenkina, N.; Toto-Brocchi, M.; Martinenghi, C.; Papa, S.; Carrafiello, G. Artificial Intelligence in Lung Cancer Imaging: Unfolding the Future. Diagnostics 2022, 12, 2644. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Lin, L.; Wu, C.F.; Li, C.F.; Xu, R.H.; Sun, Y. Artificial intelligence for assisting cancer diagnosis and treatment in the era of precision medicine. Cancer Commun. 2021, 41, 1100–1115. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, A.G.; Tsao, M.S.; Beasley, M.B.; Borczuk, A.C.; Brambilla, E.; Cooper, W.A.; Dacic, S.; Jain, D.; Kerr, K.M.; Lantuejoul, S.; et al. The 2021 WHO Classification of Lung Tumors: Impact of Advances Since 2015. J. Thorac. Oncol. 2022, 17, 362–387. [Google Scholar] [CrossRef] [PubMed]
- Raza, R.; Zulfiqar, F.; Khan, M.O.; Arif, M.; Alvi, A.; Iftikhar, M.A.; Alam, T. Lung-EffNet: Lung cancer classification using EfficientNet from CT-scan images. Eng. Appl. Artif. Intell. 2023, 126 Pt B, 106902. [Google Scholar] [CrossRef]
- Montagne, F.; Guisier, F.; Venissac, N.; Baste, J.M. The Role of Surgery in Lung Cancer Treatment: Present Indications and Future Perspectives—State of the Art. Cancers 2021, 13, 3711. [Google Scholar] [CrossRef] [PubMed]
- Johnson, M.; Albizri, A.; Simsek, S. Artificial intelligence in healthcare operations to enhance treatment outcomes: A framework to predict lung cancer prognosis. Ann. Oper. Res. 2022, 308, 275–305. [Google Scholar] [CrossRef]
- Rajasekar, V.; Vaishnnave, M.P.; Premkumar, S.; Sarveshwaran, V.; Rangaraaj, V. Lung cancer disease prediction with CT scan and histopathological images feature analysis using deep learning techniques. Results Eng. 2023, 18, 101111. [Google Scholar] [CrossRef]
- Al-Naami, B.; Badr, B.; Rawash, Y.; Abu Owida, H.; De Fazio, R.; Visconti, P. Social Media Devices’ Influence on User Neck Pain during the COVID-19 Pandemic: Collaborating Vertebral-GLCM Extracted Features with a Decision Tree. J. Imaging 2023, 9, 14. [Google Scholar] [CrossRef]
- Al-Naami, B.; Fraihat, H.; Al-Nabulsi, J.; Gharaibeh, N.Y.; Visconti, P.; Al-Hinnawi, A.R. Assessment of Dual Tree Complex Wavelet Transform to improve SNR in collaboration with Neuro-Fuzzy System for Heart Sound Identification. Electronics 2022, 11, 938. [Google Scholar] [CrossRef]
- Boddu, R.S.K.; Karmakar, P.; Bhaumik, A.; Nassa, V.K.; Vandana; Bhattacharya, S. Analyzing the impact of machine learning and artificial intelligence and its effect on management of lung cancer detection in covid-19 pandemic. Mater. Today Proc. 2022, 56, 2213–2216. [Google Scholar] [CrossRef] [PubMed]
- Shafi, I.; Din, S.; Khan, A.; De La Torre Díez, I.; Casanova, R.d.J.P.; Pifarre, K.T.; Ashraf, I. An effective method for lung cancer diagnosis from CT scan using deep learning-based Support Vector Network. Cancers 2022, 14, 5457. [Google Scholar] [CrossRef] [PubMed]
- Thanoon, M.A.; Zulkifley, M.A.; Mohd Zainuri, M.A.A.; Abdani, S.R. A Review of Deep Learning Techniques for Lung Cancer Screening and Diagnosis Based on CT Images. Diagnostics 2023, 13, 2617. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Thakkar, H.K.; Mallick, P.K.; Tiwari, P.; Alamri, A. A sustainable IoHT based computationally intelligent healthcare monitoring system for lung cancer risk detection. Sustain. Cities Soc. 2021, 72, 103079. [Google Scholar] [CrossRef]
- Manafi-Farid, R.; Askari, E.; Shiri, I.; Pirich, C.; Asadi, M.; Khateri, M.; Beheshti, M. [18F] FDG-PET/CT Radiomics and artificial intelligence in lung cancer: Technical aspects and potential clinical applications. Semin. Nucl. Med. 2022, 52, 759–780. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.; Bhushan, M. Machine learning and deep learning approach for medical image analysis: Diagnosis to detection. Multimed. Tools Appl. 2023, 82, 26731–26769. [Google Scholar] [CrossRef]
- Shen, L.; Fu, H.; Tao, G.; Liu, X.; Yuan, Z.; Ye, X. Pre-immunotherapy contrast-enhanced CT texture-based classification: A useful approach to non-small cell lung cancer immunotherapy efficacy prediction. Front. Oncol. 2021, 11, 591106. [Google Scholar] [CrossRef] [PubMed]
- Karthika, M.S.; Hurikumar, R.; Nair, A.R. Evaluation and Exploration of Machine Learning and Convolutional Neural Network Classifiers in Detection of Lung Cancer from Microarray Gene—A Paradigm Shift. Bioengineering 2023, 10, 933. [Google Scholar] [CrossRef]
- Puttagunta, M.; Ravi, S. Medical image analysis based on deep learning approach. Multimed. Tools Appl. 2021, 80, 24365–24398. [Google Scholar] [CrossRef]
- Najarian, K.; Splinter, R. Biomedical Signal and Image Processing, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2012; ISBN 9781439870334. [Google Scholar] [CrossRef]
- Fave, X.; Zhang, L.; Yang, J.; Mackin, D.; Balter, P.; Gomez, D.; Followill, D.; Jones, A.K.; Stingo, F.; Court, L.E. Impact of image preprocessing on the volume dependence and prognostic potential of radiomics features in non-small cell lung cancer. Transl. Cancer Res. 2016, 5, 349–363. [Google Scholar] [CrossRef]
- Rajasenbagam, T.; Jeyanthi, S. Image Enhancement Filtering Techniques to Enhancect Images of Lung Cancer. Int. J. Innov. Technol. Explor. Eng. (IJITEE) 2020, 9, 241–248. [Google Scholar] [CrossRef]
- Silva, F.; Pereira, T.; Neves, I.; Morgado, J.; Freitas, C.; Malafaia, M.; Oliveira, H.P. Towards machine learning-aided lung cancer clinical routines: Approaches and open challenges. J. Pers. Med. 2022, 12, 480. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, A.E.; Dua, D. Lung Nodule Detection via Optimized Convolutional Neural Network: Impact of Improved Moth Flame Algorithm. Sens. Imaging 2023, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Said, Y.; Alsheikhy, A.A.; Shawly, T.; Lahza, H. Medical image segmentation for lung cancer diagnosis based on deep learning architectures. Diagnostics 2023, 13, 546. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, A.; Rabea, S.; Bhattacharjee, A.; Elkaeed, E.B.; Murugan, R.; Selim, H.M.R.M.; Sahu, R.K.; Shazly, G.A.; Bekhit, M.M. A multi-class deep learning model for early lung cancer and chronic kidney disease detection using computed tomography images. Front. Oncol. 2023, 13, 1193746. [Google Scholar] [CrossRef] [PubMed]
- Riaz, Z.; Khan, B.; Abdullah, S.; Khan, S.; Islam, M.S. Lung tumor image segmentation from computer tomography images using MobileNetV2 and transfer learning. Bioengineering 2023, 10, 981. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhang, H.; Yao, Y.; Shen, Q. Attention-Guided Feature Extraction and Multiscale Feature Fusion 3D ResNet for Automated Pulmonary Nodule Detection. IEEE Access 2022, 10, 61530–61543. [Google Scholar] [CrossRef]
- Kasinathan, G.; Jayakumar, S. Cloud-based lung tumor detection and stage classification using deep learning techniques. BioMed Res. Int. 2022, 2022, 4185835. [Google Scholar] [CrossRef]
- Mithun, S.; Jha, A.K.; Sherkhane, U.B.; Jaiswar, V.; Purandare, N.C.; Dekker, A.; Puts, S.; Bermejo, I.; Rangarajan, V.; Zegers, C.M.L.; et al. Clinical Concept-Based Radiology Reports Classification Pipeline for Lung Carcinoma. J. Digit. Imaging 2023, 36, 812–826. [Google Scholar] [CrossRef]
- Almas, B.; Sathesh, K.; Rajesekaran, S. A Deep Analysis of Google Net and AlexNet for Lung Cancer Detection. Int. J. Eng. Adv. Technol. (IJEAT) 2019, 9, 395–399. [Google Scholar] [CrossRef]
- Kumar, V.; Bakariya, B. Classification of malignant lung cancer using deep learning. J. Med. Eng. Technol. 2021, 45, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Lanjewar, M.G.; Panchbhai, K.G.; Charanarur, P. Lung cancer detection from CT scans using modified DenseNet with feature selection methods and ML classifiers. Expert Syst. Appl. 2023, 224, 119961. [Google Scholar] [CrossRef]
- Pandit, B.R.; Alsadoon, A.; Prasad, P.W.C.; Al Aloussi, S.; Rashid, T.A.; Alsadoon, O.H.; Jerew, O.D. Deep learning neural network for lung cancer classification: Enhanced optimization function. Multimed. Tools Appl. 2023, 82, 6605–6624. [Google Scholar] [CrossRef]
- Ren, Z.; Zhang, Y.; Wang, S. LCDAE: Data augmented ensemble framework for lung cancer classification. Technol. Cancer Res. Treat. 2022, 21, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Zhang, Y.; Wang, S. A hybrid framework for lung cancer classification. Electronics 2022, 11, 1614. [Google Scholar] [CrossRef]
- Siddiqui, E.A.; Chaurasia, V.; Shandilya, M. Detection and classification of lung cancer computed tomography images using a novel improved deep belief network with Gabor filters. Chemom. Intell. Lab. Syst. 2023, 235, 104763. [Google Scholar] [CrossRef]
- van Rikxoort, E.M.B.; de Hoop, B.; Viergever, M.A.; Prokop, M.; van Ginneken, B. Automatic lung segmentation from thoracic computed tomography scans using a hybrid approach with error detection. Med. Phys. 2009, 4236, 2934–2947. [Google Scholar] [CrossRef] [PubMed]
- Armato, S.G., 3rd; McLennan, G.; Bidaut, L.; McNitt-Gray, M.F.; Meyer, C.R.; Reeves, A.P.; Zhao, B.; Aberle, D.R.; Henschke, C.I.; Hoffman, E.A.; et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): A completed reference database of lung nodules on CT scans. Med. Phys. 2011, 38, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.; Setio, A.A.A.; Traverso, A.; van Ginneken, B. LUNA16—Grand Challenge. Available online: https://luna16.grand-challenge.org/Data/ (accessed on 13 April 2024).
- Al-Naami, B.; Fraihat, H.; Owida, H.A.; Al-Hamad, K.; De Fazio, R.; Visconti, P. Automated Detection of Left Bundle Branch Block from ECG signal utilizing the Maximal Overlap Discrete Wavelet Transform with ANFIS. Computers 2022, 11, 93. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Almazloum, A.A.; Al-Hinnawi, A.-R.; De Fazio, R.; Visconti, P. Assessment of Multi-Layer Perceptron Neural Network for Pulmonary Function Test’s diagnosis using ATS and ERS Respiratory Standard Parameters. Computers 2022, 11, 130. [Google Scholar] [CrossRef]
- Shu, X.; Yang, Y.; Liu, J.; Chang, X.; Wu, B. BDAL: Balanced Distribution Active Learning for MRI Cardiac Multistructures Segmentation. IEEE Trans. Ind. Inform. 2024, 20, 6099–6108. [Google Scholar] [CrossRef]
- Shu, X.; Yuan, D. Local Variance-driven Level Set Model with Application to Segment Medical Images. In Proceedings of the 2023 International Conference on Cyber-Physical Social Intelligence (ICCSI), Xi’an, China, 20–23 October 2023; pp. 435–440. [Google Scholar] [CrossRef]


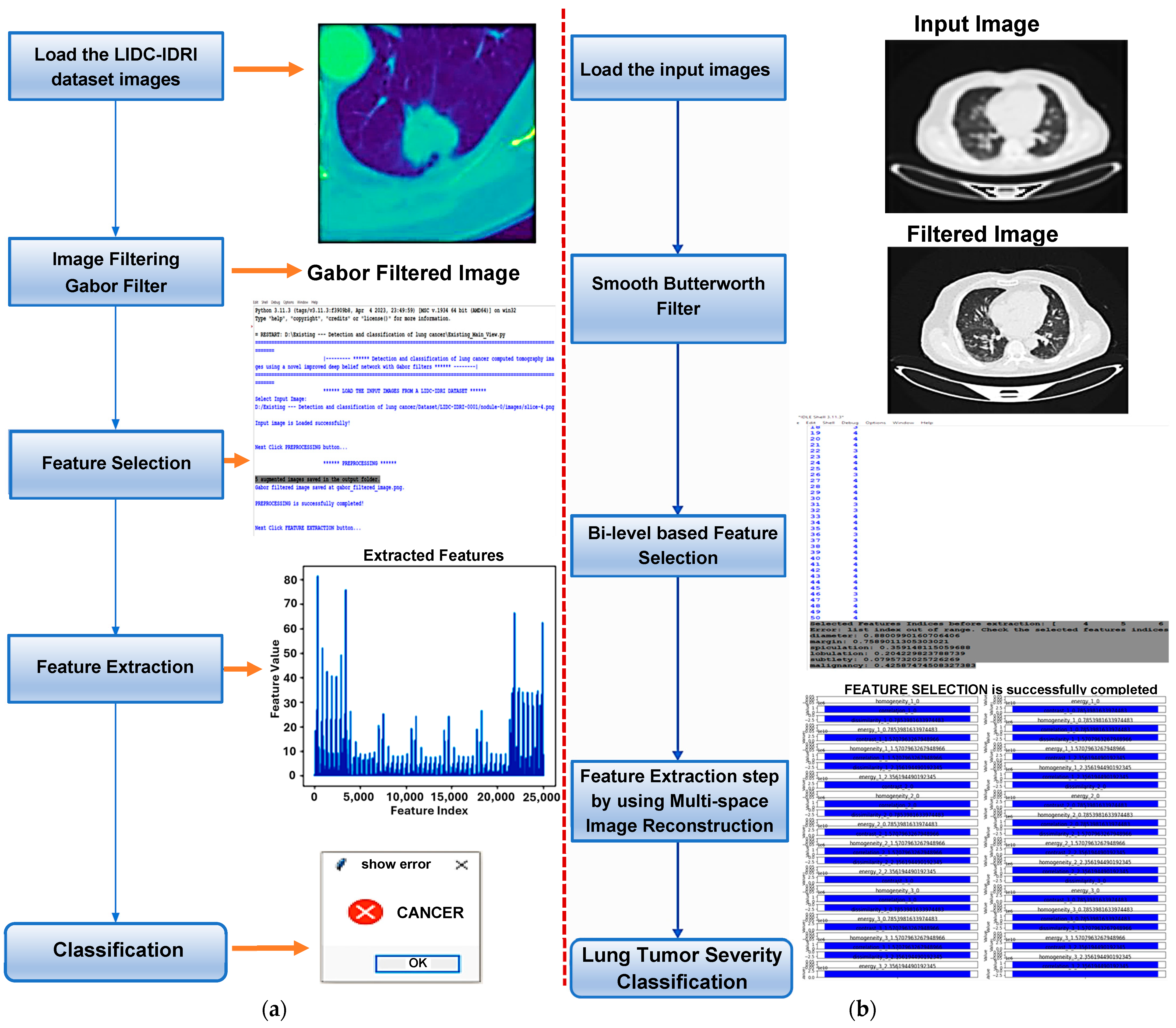
| Work | Objective | The Method or AI Approach | Limitations |
|---|---|---|---|
| A. Sebastian et al. [25] | Detection of Lung Nodules via Enhanced Convolutional Neural Network | Improved Moth Flame Optimization | The model calculation time is high (low computational speed). The model intends to utilize visualization tools to comprehend the networks’ classification components and see whether they match the radiologists’ diagnoses. |
| Y. Said et al. [26] | Deep Learning-Based Medical Image Segmentation for Lung Cancer Diagnosis. | UNETR network, self-supervised network | The model requires high computational demands and a powerful GPU; this can be overcome by using powerful local machines or cloud platforms. |
| A. Bhattacharjee et al. [27] | Computed tomography images for the early diagnosis of chronic renal disease and lung cancer. | Xception framework | The Malignant, Normal, and Benign (MNoB) model did not outperform the Normal, Cyst, Tumour, and Stone (NCTS) model despite promising findings due to insufficient training data. |
| Z. Riaz et al. [28] | Segmentation of Lung Tumour Images from Computer Tomography Images. | MobileNetV2 and UNET | The segmentation was tested only on the challenge validation dataset; to assess its resilience, it has to be tested on other medical image segmentation tasks. The segmentation results were not completely processed. |
| G. Zhang et al. [29] | Automatic Pulmonary Nodule Detection and Attention-Guided Feature Extraction. | R-CNN-based attention-embedded model, ResNet | A low false-positive rate does not imply sensitivity. Focal loss reduces the information imbalance but cannot eliminate it. Even after data quality improvements, the model needs new data as the network deepens to expand its capability. |
| G. Kasinathan et al. [30] | Cloud-Based Identification and Stage Categorization of Lung Tumours. | Deep neural model | Cloud system’s memory constraints limit the analysis to no more than 65 records at once, making it inefficient for large hospitals. |
| S. Mithun et al. [31] | Clinical Concept-Based Radiology Reports Lung Cancer Classification Pipeline. | ML algorithms | The designed pipeline is able to extract data only from HIS but is customizable for other institutions. The rule-based model’s ontology has been customized only using their data. Some false negatives remain unidentified after mapping most of the disease diagnostic keywords. |
| V. Kumar et al. [33] | Categorization of lung cancer that is malignant. | Median filter, Gaussian filter, Gabor filter, and watershed algorithm. | Overfitting occurs when DL algorithms memorize training data instead of capturing patterns. Overfitting may occur when the model’s capability exceeds the training data. Overfitting can be reduced by data augmentation, dropout, and regularisation, but not eliminated. |
| M. Lanjewar, et al. [34] | Detecting lung cancer in CT images and feature selection. | DenseNet framework | The quality and representativeness of the training data can influence the model performance. Demographic imbalances or imaging procedure changes can affect model performance and generalizability. |
| B.R. Pandit et al. [35] | To improve prediction accuracy and save processing time, use multi-space images. | Convolution filter, autoencoder model based on convolutional neural network | Image quality affects MIR’s ability to reconstruct CT images. Image quality issues such as noise, artifacts, and resolution can affect MIR performance and classification accuracy. |
| Z. Ren et al. [36] | Data-augmented ensemble framework for lung cancer categorization. | Deep Convolutional GAN | LCDAE framework performance can depend on pre-processing, hyper-parameters, and model setups. Small changes in these elements could affect the model’s performance and generalizability. |
| Z. Ren et al. [37] | To identify early-stage lung cancer. | Deep convolutional GAN, transfer learning model | LCGANT may require a lot of computing resources, limiting its usability in resource-constrained applications. |
| E. Saddiqui et al. [38] | Lung cancer detection and categorization using computed tomography images. | Deep Belief Network, Gaussian-Bernoulli, support vector machine. | Increased sensitivity for high false positive (FP) rates often follows lower sensitivity for low FP rates. |
| Variables | Value |
|---|---|
| H | 30 |
| AP | 0.1 |
| 2 | |
| Lower bound | 0 |
| Upper bound | 1 |
| 50 | |
| Same as the total number of features in the original dataset |
| Software Specifications | Operating System (OS) | Windows 10 (64-bit) |
| Tool | Python Ver. 3.9.6 | |
| Hardware Specifications | RAM | 4 GB |
| Hard Disk | 500 GB |
| Number of Epochs | Accuracy (%) | ||
|---|---|---|---|
| DenseNet201 | CNN + SVM | Proposed Method | |
| 10 | 70.0 | 80.0 | 90.0 |
| 20 | 75.5 | 82.0 | 91.5 |
| 30 | 76.5 | 83.5 | 93.0 |
| 40 | 79.0 | 87.0 | 95.5 |
| 50 | 81.5 | 89.0 | 97.5 |
| Number of Epochs | Precision (%) | ||
|---|---|---|---|
| DenseNet201 | CNN + SVM | Proposed Method | |
| 10 | 72.0 | 81.0 | 90.0 |
| 20 | 75.0 | 82.5 | 92.0 |
| 30 | 79.0 | 84.5 | 93.5 |
| 40 | 80.5 | 85.5 | 94.5 |
| 50 | 85.0 | 87.0 | 95.5 |
| Number of Epochs | F1-Score (%) | ||
|---|---|---|---|
| DenseNet201 | CNN + SVM | Proposed Method | |
| 10 | 76.0 | 77.0 | 91.0 |
| 20 | 77.0 | 78.0 | 92.0 |
| 30 | 78.5 | 79.5 | 93.5 |
| 40 | 79.5 | 80.5 | 95.0 |
| 50 | 80.5 | 81.5 | 96.5 |
| Scientific Work | Dataset | Accuracy [%] | Precision [%] | Sensitivity (Recall) [%] |
|---|---|---|---|---|
| F. Silva, et al. [24] | LIDC-IDRI | 92.5 | 92.5 | 92.5 |
| A. E. Sebastian, et al. [25] | Decathlon 2018 Challenge | 98.8 | N.A. | 96.8 |
| Y. Said, et al. [26] | Local | 98.3 | 99.3 | 98.0 |
| A. Bhattacharjee, et al. [27] | Decathlon 2018 Challenge | N.A. | 93.2 | 86.0 |
| Z. Riaz, et al. [28] | LUNA 16 | 91.2 | N.A. | 97.7 |
| G. Zhang, et al. [29] | LIDC-IDRI | 97.1 | N.A. | 95.9 |
| S. Mithun, et al. [31] | Local | 91.0 | N.A. | 94.0 |
| V. Kumar, et al. [33] | Local | Up to 100 (97 Avg.) | Up to 100 (95 Avg.) | N.A. |
| Z. Ren, et al. [36] | LIDC-IDRI, LUNA16, TCIA | 99.4 | N.A. | 98.5 |
| E.A. Siddiqui, et al. [38] | LUNA 16 | 99.1% | N.A. | 98.0% |
| Proposed hybrid approach | LUNA16 | 97.5 | 95.5 | 99.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharaibeh, N.Y.; De Fazio, R.; Al-Naami, B.; Al-Hinnawi, A.-R.; Visconti, P. Automated Lung Cancer Diagnosis Applying Butterworth Filtering, Bi-Level Feature Extraction, and Sparce Convolutional Neural Network to Luna 16 CT Images. J. Imaging 2024, 10, 168. https://doi.org/10.3390/jimaging10070168
Gharaibeh NY, De Fazio R, Al-Naami B, Al-Hinnawi A-R, Visconti P. Automated Lung Cancer Diagnosis Applying Butterworth Filtering, Bi-Level Feature Extraction, and Sparce Convolutional Neural Network to Luna 16 CT Images. Journal of Imaging. 2024; 10(7):168. https://doi.org/10.3390/jimaging10070168
Chicago/Turabian StyleGharaibeh, Nasr Y., Roberto De Fazio, Bassam Al-Naami, Abdel-Razzak Al-Hinnawi, and Paolo Visconti. 2024. "Automated Lung Cancer Diagnosis Applying Butterworth Filtering, Bi-Level Feature Extraction, and Sparce Convolutional Neural Network to Luna 16 CT Images" Journal of Imaging 10, no. 7: 168. https://doi.org/10.3390/jimaging10070168
APA StyleGharaibeh, N. Y., De Fazio, R., Al-Naami, B., Al-Hinnawi, A.-R., & Visconti, P. (2024). Automated Lung Cancer Diagnosis Applying Butterworth Filtering, Bi-Level Feature Extraction, and Sparce Convolutional Neural Network to Luna 16 CT Images. Journal of Imaging, 10(7), 168. https://doi.org/10.3390/jimaging10070168













