A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging
Abstract
:1. Introduction
- 1.
- Comprehensive Survey: We offer a thorough survey of innovative approaches for interpreting and visualizing DL models in MI, including a broad range of techniques aimed at enhancing model transparency and trust.
- 2.
- Methodological Review: We provide an in-depth review of current methodologies, focusing on post-hoc visualization techniques such as perturbation-based, gradient-based, decomposition-based, trainable attention (TA)-based methods, and vision transformers (ViT). We evaluate each method’s effectiveness and applicability in MI.
- 3.
- Clinical Relevance: We emphasize the importance of interpretability techniques in clinical settings, demonstrating how they lead to more reliable and actionable insights from DL models, thus supporting better decision-making in healthcare.
- 4.
- Future Directions: We outline future research directions in model interpretability and visualization, highlighting the need for more robust and scalable techniques that can handle the complexity of DL models while ensuring practical utility in medical applications.
2. Research Methodology
- What innovative methods exist for interpreting and visualizing deep learning models in medical imaging?
- How effective are post-hoc visualization techniques (perturbation-based, gradient-based, decomposition-based, TA-based, and ViT) in improving model transparency?
- What is the clinical relevance of interpretability techniques for actionable insights from deep learning models in healthcare?
- What are the future research directions for model interpretability and visualization in medical applications?
3. Interpreting Model Design and Workflow
- 1.
- Autoencoders for Learning Latent Representations
- 2.
- Visualizing High-Dimensional Latent Data in a Two-Dimensional Space
- 3.
- Visualizing Filters and Activations in Feature Maps
3.1. Autoencoders for Learning Latent Representations
3.2. Visualizing High-Dimensional Latent Data in a Two-Dimensional Space
3.3. Visualizing Filters and Activations in Feature Maps
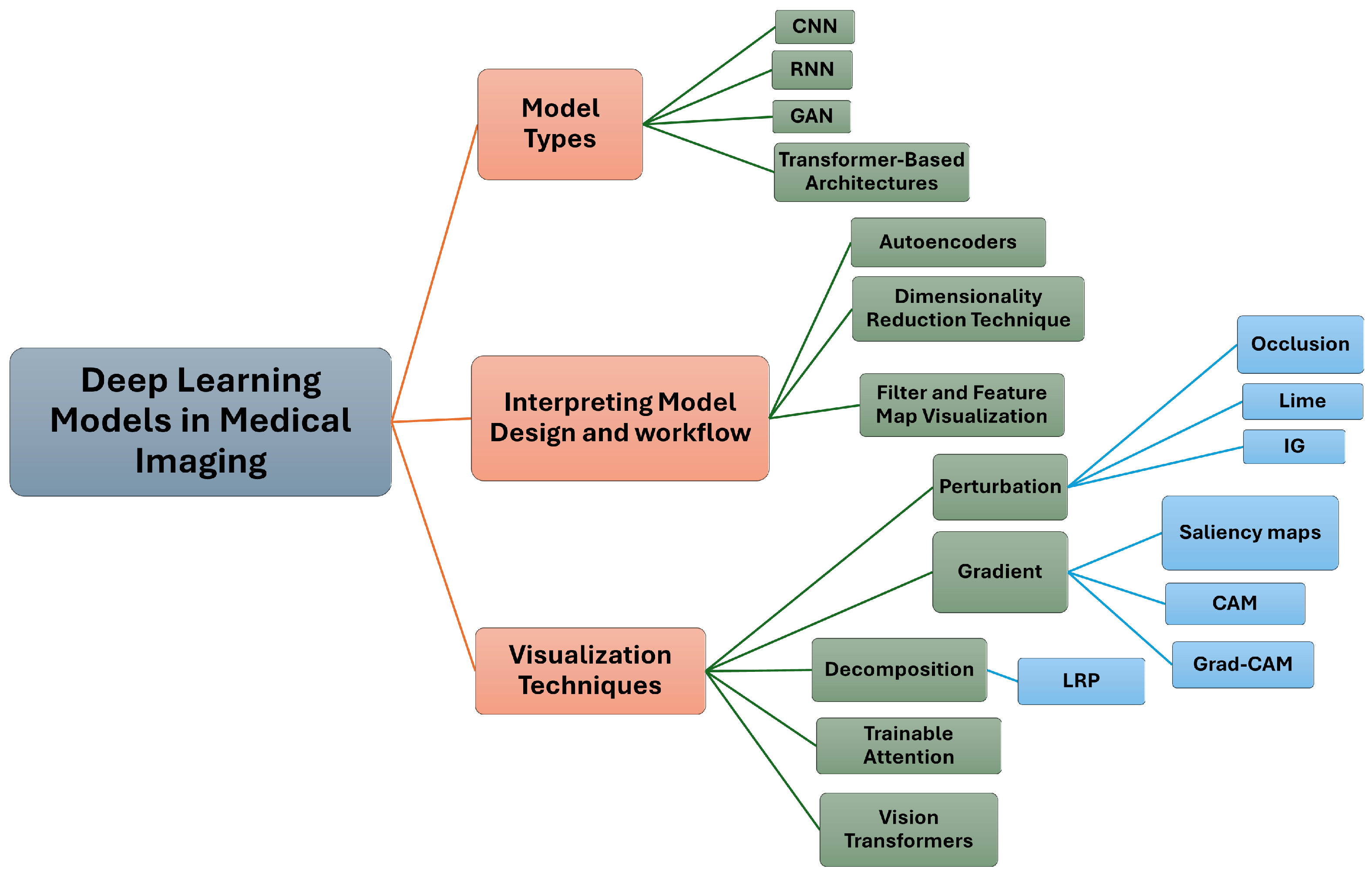
4. Deep Learning Models in Medical Imaging
Transformer-Based Architectures
5. Interpretation and Visualization Techniques
5.1. Perturbation-Based Methods
5.1.1. Occlusion
5.1.2. Local Interpretable Model-Agnostic Explanations (LIME)
5.1.3. Integrated Gradients
5.2. Gradient-Based Methods
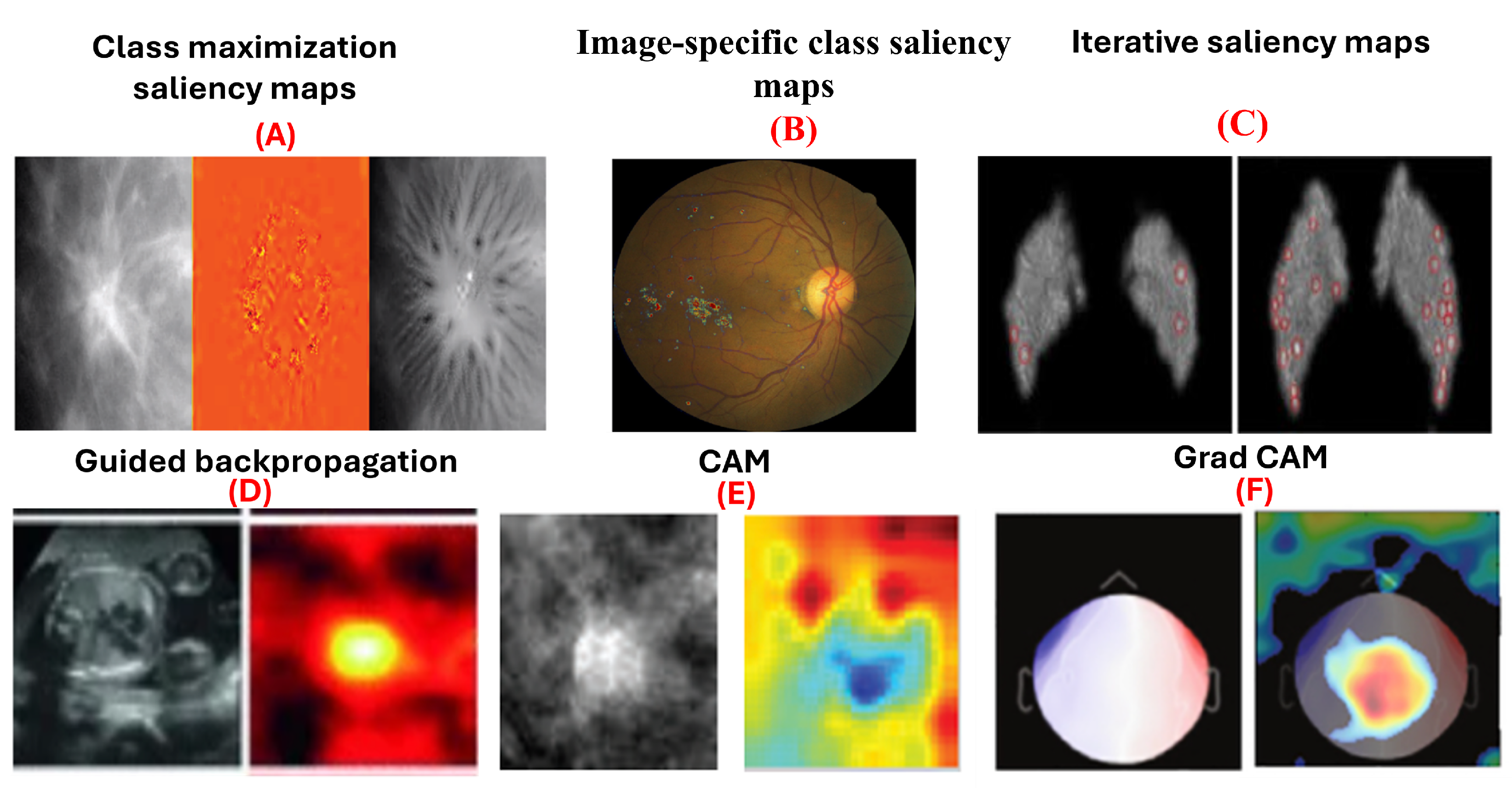
5.2.1. Saliency Maps
5.2.2. Guided Backpropagation
5.2.3. Class Activation Maps (CAM)
5.2.4. Grad-CAM
5.3. Decomposition-Based Methods
Layer-Wise Relevance Propagation (LRP)
5.4. Trainable Attention Models
5.5. Vision Transformers
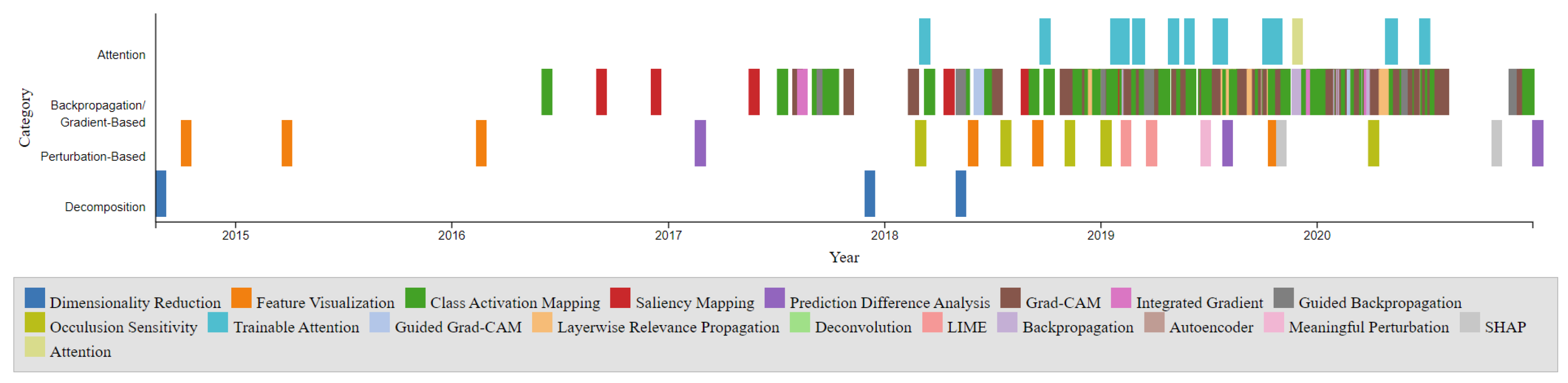
6. Comparison of Different Interpretation Methods
6.1. Categorization by Visualization Technique
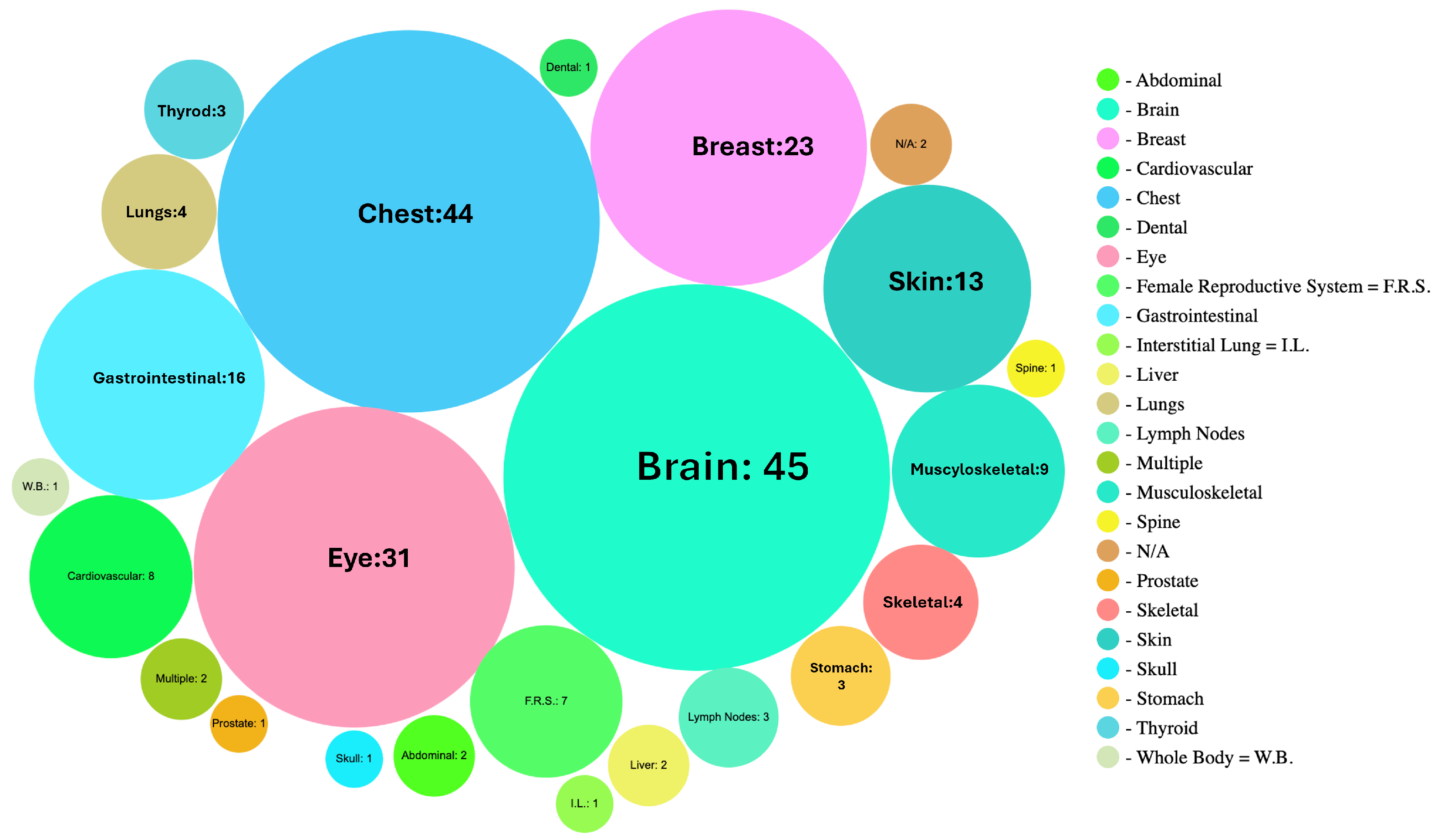
6.2. Categorization by Body Parts, Modality, and Accuracy
6.3. Categorization by Task
7. Current Challenges and Future Directions
7.1. Current Challenges
7.2. Future Directions
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Neikirk, K.; Lopez, E.G.; Marshall, A.G.; Alghanem, A.; Krystofiak, E.; Kula, B.; Smith, N.; Shao, J.; Katti, P.; Hinton, A.O., Jr. Call to action to properly utilize electron microscopy to measure organelles to monitor disease. Eur. J. Cell Biol. 2023, 102, 151365. [Google Scholar] [CrossRef] [PubMed]
- Galaz-Montoya, J.G. The advent of preventive high-resolution structural histopathology by artificial-intelligence-powered cryogenic electron tomography. Front. Mol. Biosci. 2024, 11, 1390858. [Google Scholar] [CrossRef] [PubMed]
- Banerji, S.; Mitra, S. Deep learning in histopathology: A review. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2022, 12, e1439. [Google Scholar] [CrossRef]
- Niazi, M.K.K.; Parwani, A.V.; Gurcan, M.N. Digital pathology and artificial intelligence. Lancet Oncol. 2019, 20, e253–e261. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Wu, F.; Peng, J.; Bao, Y.; Chen, F.; Kong, D. Automatic abdominal multi-organ segmentation using deep convolutional neural network and time-implicit level sets. Int. J. Comput. Assist. Radiol. Surg. 2017, 12, 399–411. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Roth, H.R.; Lu, L.; Farag, A.; Shin, H.C.; Liu, J.; Turkbey, E.B.; Summers, R.M. Deeporgan: Multi-level deep convolutional networks for automated pancreas segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part I 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 556–564. [Google Scholar]
- Gao, Y.; Alison Noble, J. Detection and characterization of the fetal heartbeat in free-hand ultrasound sweeps with weakly-supervised two-streams convolutional networks. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part II 20. Springer: Berlin/Heidelberg, Germany, 2017; pp. 305–313. [Google Scholar]
- Roth, H.R.; Lu, L.; Liu, J.; Yao, J.; Seff, A.; Cherry, K.; Kim, L.; Summers, R.M. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans. Med. Imaging 2015, 35, 1170–1181. [Google Scholar] [CrossRef]
- Kim, S.T.; Lee, J.H.; Lee, H.; Ro, Y.M. Visually interpretable deep network for diagnosis of breast masses on mammograms. Phys. Med. Biol. 2018, 63, 235025. [Google Scholar] [CrossRef]
- Yang, X.; Do Yang, J.; Hwang, H.P.; Yu, H.C.; Ahn, S.; Kim, B.W.; You, H. Segmentation of liver and vessels from CT images and classification of liver segments for preoperative liver surgical planning in living donor liver transplantation. Comput. Methods Programs Biomed. 2018, 158, 41–52. [Google Scholar] [CrossRef]
- Chen, X.; Shi, B. Deep mask for X-ray based heart disease classification. arXiv 2018, arXiv:1808.08277. [Google Scholar]
- Yi, D.; Sawyer, R.L.; Cohn III, D.; Dunnmon, J.; Lam, C.; Xiao, X.; Rubin, D. Optimizing and visualizing deep learning for benign/malignant classification in breast tumors. arXiv 2017, arXiv:1705.06362. [Google Scholar]
- Hengstler, M.; Enkel, E.; Duelli, S. Applied artificial intelligence and trust—The case of autonomous vehicles and medical assistance devices. Technol. Forecast. Soc. Chang. 2016, 105, 105–120. [Google Scholar] [CrossRef]
- Nundy, S.; Montgomery, T.; Wachter, R.M. Promoting trust between patients and physicians in the era of artificial intelligence. JAMA 2019, 322, 497–498. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Jia, X.; Ren, L.; Cai, J. Clinical implementation of AI technologies will require interpretable AI models. Med. Phys. 2020, 47, 1–4. [Google Scholar] [CrossRef]
- Reyes, M.; Meier, R.; Pereira, S.; Silva, C.A.; Dahlweid, F.M.; Tengg-Kobligk, H.v.; Summers, R.M.; Wiest, R. On the interpretability of artificial intelligence in radiology: Challenges and opportunities. Radiol. Artif. Intell. 2020, 2, e190043. [Google Scholar] [CrossRef]
- Gastounioti, A.; Kontos, D. Is it time to get rid of black boxes and cultivate trust in AI? Radiol. Artif. Intell. 2020, 2, e200088. [Google Scholar] [CrossRef]
- Guo, R.; Wei, J.; Sun, L.; Yu, B.; Chang, G.; Liu, D.; Zhang, S.; Yao, Z.; Xu, M.; Bu, L. A survey on advancements in image-text multimodal models: From general techniques to biomedical implementations. Comput. Biol. Med. 2024, 178, 108709. [Google Scholar] [CrossRef]
- Rasool, N.; Bhat, J.I. Brain tumour detection using machine and deep learning: A systematic review. Multimed. Tools Appl. 2024, 1–54. [Google Scholar] [CrossRef]
- Huff, D.T.; Weisman, A.J.; Jeraj, R. Interpretation and visualization techniques for deep learning models in medical imaging. Phys. Med. Biol. 2021, 66, 04TR01. [Google Scholar] [CrossRef]
- Hohman, F.; Kahng, M.; Pienta, R.; Chau, D.H. Visual analytics in deep learning: An interrogative survey for the next frontiers. IEEE Trans. Vis. Comput. Graph. 2018, 25, 2674–2693. [Google Scholar] [CrossRef] [PubMed]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Vincent, P.; Larochelle, H.; Bengio, Y.; Manzagol, P.A. Extracting and composing robust features with denoising autoencoders. In Proceedings of the 25th International Conference on Machine Learning, Helsinki, Finland, 5–9 July 2008; pp. 1096–1103. [Google Scholar]
- Kiran, B.R.; Thomas, D.M.; Parakkal, R. An overview of deep learning based methods for unsupervised and semi-supervised anomaly detection in videos. J. Imaging 2018, 4, 36. [Google Scholar] [CrossRef]
- Theis, L.; Shi, W.; Cunningham, A.; Huszár, F. Lossy image compression with compressive autoencoders. In Proceedings of the International Conference on Learning Representations, Virtually, 25–29 April 2022. [Google Scholar]
- Tschannen, M.; Bachem, O.; Lucic, M. Recent advances in autoencoder-based representation learning. arXiv 2018, arXiv:1812.05069. [Google Scholar]
- Uzunova, H.; Ehrhardt, J.; Kepp, T.; Handels, H. Interpretable explanations of black box classifiers applied on medical images by meaningful perturbations using variational autoencoders. In Proceedings of the Medical Imaging 2019: Image Processing, San Diego, CA, USA, 19–21 February 2019; SPIE: Bellingham, DC, USA, 2019; Volume 10949, pp. 264–271. [Google Scholar]
- Chen, X.; You, S.; Tezcan, K.C.; Konukoglu, E. Unsupervised lesion detection via image restoration with a normative prior. Med. Image Anal. 2020, 64, 101713. [Google Scholar] [CrossRef]
- Hou, L.; Nguyen, V.; Kanevsky, A.B.; Samaras, D.; Kurc, T.M.; Zhao, T.; Gupta, R.R.; Gao, Y.; Chen, W.; Foran, D.; et al. Sparse autoencoder for unsupervised nucleus detection and representation in histopathology images. Pattern Recognit. 2019, 86, 188–200. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Plis, S.M.; Hjelm, D.R.; Salakhutdinov, R.; Allen, E.A.; Bockholt, H.J.; Long, J.D.; Johnson, H.J.; Paulsen, J.S.; Turner, J.A.; Calhoun, V.D. Deep learning for neuroimaging: A validation study. Front. Neurosci. 2014, 8, 229. [Google Scholar] [CrossRef]
- Stoyanov, D.; Taylor, Z.; Kia, S.M.; Oguz, I.; Reyes, M.; Martel, A.; Maier-Hein, L.; Marquand, A.F.; Duchesnay, E.; Löfstedt, T.; et al. Understanding and Interpreting Machine Learning in Medical Image Computing Applications; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Yu, Z.; Tan, E.L.; Ni, D.; Qin, J.; Chen, S.; Li, S.; Lei, B.; Wang, T. A deep convolutional neural network-based framework for automatic fetal facial standard plane recognition. IEEE J. Biomed. Health Inform. 2017, 22, 874–885. [Google Scholar] [CrossRef]
- Zhang, F.; Li, Z.; Zhang, B.; Du, H.; Wang, B.; Zhang, X. Multi-modal deep learning model for auxiliary diagnosis of Alzheimer’s disease. Neurocomputing 2019, 361, 185–195. [Google Scholar] [CrossRef]
- Al’Aref, S.J.; Anchouche, K.; Singh, G.; Slomka, P.J.; Kolli, K.K.; Kumar, A.; Pandey, M.; Maliakal, G.; Van Rosendael, A.R.; Beecy, A.N.; et al. Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur. Heart J. 2019, 40, 1975–1986. [Google Scholar] [CrossRef]
- Nie, D.; Trullo, R.; Lian, J.; Petitjean, C.; Ruan, S.; Wang, Q.; Shen, D. Medical image synthesis with context-aware generative adversarial networks. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part III 20. Springer: Berlin/Heidelberg, Germany, 2017; pp. 417–425. [Google Scholar]
- Frid-Adar, M.; Diamant, I.; Klang, E.; Amitai, M.; Goldberger, J.; Greenspan, H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018, 321, 321–331. [Google Scholar] [CrossRef]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [PubMed]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Al-Hammuri, K.; Gebali, F.; Kanan, A.; Chelvan, I.T. Vision transformer architecture and applications in digital health: A tutorial and survey. Vis. Comput. Ind. Biomed. Art 2023, 6, 14. [Google Scholar] [CrossRef]
- Lecler, A.; Duron, L.; Soyer, P. Revolutionizing radiology with GPT-based models: Current applications, future possibilities and limitations of ChatGPT. Diagn. Interv. Imaging 2023, 104, 269–274. [Google Scholar] [CrossRef]
- Ivanovs, M.; Kadikis, R.; Ozols, K. Perturbation-based methods for explaining deep neural networks: A survey. Pattern Recognit. Lett. 2021, 150, 228–234. [Google Scholar] [CrossRef]
- Papanastasopoulos, Z.; Samala, R.K.; Chan, H.P.; Hadjiiski, L.; Paramagul, C.; Helvie, M.A.; Neal, C.H. Explainable AI for medical imaging: Deep-learning CNN ensemble for classification of estrogen receptor status from breast MRI. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 16–19 February 2020; SPIE: Bellingham, DC, USA, 2020; Volume 11314, pp. 228–235. [Google Scholar]
- Sayres, R.; Taly, A.; Rahimy, E.; Blumer, K.; Coz, D.; Hammel, N.; Webster, D.R. Using a deep learning algorithm and integrated gradients explanation to assist grading for diabetic retinopathy. Ophthalmology 2019, 126, 552–564. [Google Scholar] [CrossRef]
- Sundararajan, M.; Taly, A.; Yan, Q. Axiomatic attribution for deep networks. In Proceedings of the International Conference on Machine Learning, PMLR, Sydney, Australia, 6–11 August 2017; pp. 3319–3328. [Google Scholar]
- Rajaraman, S.; Candemir, S.; Thoma, G.; Antani, S. Visualizing and explaining deep learning predictions for pneumonia detection in pediatric chest radiographs. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis, San Diego, CA, USA, 17–20 February 2019; SPIE: Bellingham, DC, USA, 2019; Volume 10950, pp. 200–211. [Google Scholar]
- Malhi, A.; Kampik, T.; Pannu, H.; Madhikermi, M.; Främling, K. Explaining machine learning-based classifications of in-vivo gastral images. In Proceedings of the 2019 Digital Image Computing: Techniques and Applications (DICTA), Perth, Australia, 2–4 December 2019; pp. 1–7. [Google Scholar]
- Dubost, F.; Adams, H.; Bortsova, G.; Ikram, M.A.; Niessen, W.; Vernooij, M.; De Bruijne, M. 3D regression neural network for the quantification of enlarged perivascular spaces in brain MRI. Med. Image Anal. 2019, 51, 89–100. [Google Scholar] [CrossRef]
- Shahamat, H.; Abadeh, M.S. Brain MRI analysis using a deep learning based evolutionary approach. Neural Netw. 2020, 126, 218–234. [Google Scholar] [CrossRef]
- Gecer, B.; Aksoy, S.; Mercan, E.; Shapiro, L.G.; Weaver, D.L.; Elmore, J.G. Detection and classification of cancer in whole slide breast histopathology images using deep convolutional networks. Pattern Recognit. 2018, 84, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Seah, J.C.; Tang, J.S.; Kitchen, A.; Gaillard, F.; Dixon, A.F. Chest radiographs in congestive heart failure: Visualizing neural network learning. Radiology 2019, 290, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the Computer Vision–ECCV 2014: 13th European Conference, Zurich, Switzerland, 6–12 September 2014; Proceedings, Part I 13. Springer: Berlin/Heidelberg, Germany, 2014; pp. 818–833. [Google Scholar]
- Liang, Y.; Li, S.; Yan, C.; Li, M.; Jiang, C. Explaining the black-box model: A survey of local interpretation methods for deep neural networks. Neurocomputing 2021, 419, 168–182. [Google Scholar] [CrossRef]
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should i trust you?” Explaining the predictions of any classifier. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–16 August 2016; pp. 1135–1144. [Google Scholar]
- Xu, X.; Li, C.; Lan, X.; Fan, X.; Lv, X.; Ye, X.; Wu, T. A lightweight and robust framework for circulating genetically abnormal cells (CACs) identification using 4-color fluorescence in situ hybridization (FISH) image and deep refined learning. J. Digit. Imaging 2023, 36, 1687–1700. [Google Scholar] [CrossRef]
- Garg, P.; Davenport, E.; Murugesan, G.; Wagner, B.; Whitlow, C.; Maldjian, J.; Montillo, A. Using convolutional neural networks to automatically detect eye-blink artifacts in magnetoencephalography without resorting to electrooculography. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, 11–13 September 2017; Proceedings, Part III 20. Springer: Berlin/Heidelberg, Germany, 2017; pp. 374–381. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013. [Google Scholar]
- Dubost, F.; Bortsova, G.; Adams, H.; Ikram, A.; Niessen, W.J.; Vernooij, M.; De Bruijne, M. Gp-unet: Lesion detection from weak labels with a 3d regression network. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Quebec City, QC, Canada, 11–13 September 2017; pp. 214–221. [Google Scholar]
- Lévy, D.; Jain, A. Breast mass classification from mammograms using deep convolutional neural networks. arXiv 2016. [Google Scholar]
- Rayan, J.C.; Reddy, N.; Kan, J.H.; Zhang, W.; Annapragada, A. Binomial classification of pediatric elbow fractures using a deep learning multiview approach emulating radiologist decision making. Radiol. Artif. Intell. 2019, 1, e180015. [Google Scholar] [CrossRef]
- Liefers, B.; González-Gonzalo, C.; Klaver, C.; van Ginneken, B.; Sánchez, C.I. Dense segmentation in selected dimensions: Application to retinal optical coherence tomography. In Proceedings of the International Conference on Medical Imaging with Deep Learning (MIDL), London, UK, 8–10 July 2019; PMLR. pp. 337–346. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2014. [Google Scholar]
- Böhle, M.; Eitel, F.; Weygandt, M.; Ritter, K. Layer-wise relevance propagation for explaining deep neural network decisions in MRI-based Alzheimer’s disease classification. Front. Aging Neurosci. 2019, 11, 456892. [Google Scholar] [CrossRef]
- Dubost, F.; Yilmaz, P.; Adams, H.; Bortsova, G.; Ikram, M.A.; Niessen, W.; Vernooij, M.; de Bruijne, M. Enlarged perivascular spaces in brain MRI: Automated quantification in four regions. Neuroimage 2019, 185, 534–544. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liang, X.; Jiang, Z.; Nguchu, B.A.; Zhou, Y.; Wang, Y.; Wang, H.; Li, Y.; Zhu, Y.; Wu, F.; et al. Decoding and mapping task states of the human brain via deep learning. Hum. Brain Mapp. 2020, 41, 1505–1519. [Google Scholar] [CrossRef] [PubMed]
- Gessert, N.; Latus, S.; Abdelwahed, Y.S.; Leistner, D.M.; Lutz, M.; Schlaefer, A. Bioresorbable scaffold visualization in IVOCT images using CNNs and weakly supervised localization. In Proceedings of the Medical Imaging 2019: Image Processing, San Diego, CA, USA, 19–21 February 2019; SPIE: Bellingham, DC, USA, 2019; Volume 10949, pp. 606–612. [Google Scholar]
- Wickstrøm, K.; Kampffmeyer, M.; Jenssen, R. Uncertainty and interpretability in convolutional neural networks for semantic segmentation of colorectal polyps. Med. Image Anal. 2020, 60, 101619. [Google Scholar] [CrossRef] [PubMed]
- Jamaludin, A.; Kadir, T.; Zisserman, A. SpineNet: Automated classification and evidence visualization in spinal MRIs. Med. Image Anal. 2017, 41, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013. [Google Scholar]
- Feng, X.; Lipton, Z.C.; Yang, J.; Small, S.A.; Provenzano, F.A.; Initiative, A.D.N.; Initiative, F.L.D.N. Estimating brain age based on a uniform healthy population with deep learning and structural magnetic resonance imaging. Neurobiol. Aging 2020, 91, 15–25. [Google Scholar] [CrossRef]
- Zhao, G.; Zhou, B.; Wang, K.; Jiang, R.; Xu, M. Respond-CAM: Analyzing deep models for 3D imaging data by visualizations. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part I. Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 485–492. [Google Scholar]
- Oquab, M.; Bottou, L.; Laptev, I.; Sivic, J. Learning and transferring mid-level image representations using convolutional neural networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; pp. 1717–1724. [Google Scholar]
- Woerl, A.C.; Eckstein, M.; Geiger, J.; Wagner, D.C.; Daher, T.; Stenzel, P.; Fernandez, A.; Hartmann, A.; Wand, M.; Roth, W.; et al. Deep learning predicts molecular subtype of muscle-invasive bladder cancer from conventional histopathological slides. Eur. Urol. 2020, 78, 256–264. [Google Scholar] [CrossRef]
- Ahmad, A.; Sarkar, S.; Shah, A.; Gore, S.; Santosh, V.; Saini, J.; Ingalhalikar, M. Predictive and discriminative localization of IDH genotype in high grade gliomas using deep convolutional neural nets. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 372–375. [Google Scholar]
- Shinde, S.; Prasad, S.; Saboo, Y.; Kaushick, R.; Saini, J.; Pal, P.K.; Ingalhalikar, M. Predictive markers for Parkinson’s disease using deep neural nets on neuromelanin sensitive MRI. Neuroimage Clin. 2019, 22, 101748. [Google Scholar] [CrossRef]
- Chakraborty, S.; Aich, S.; Kim, H.C. Detection of Parkinson’s disease from 3T T1 weighted MRI scans using 3D convolutional neural network. Diagnostics 2020, 10, 402. [Google Scholar] [CrossRef]
- Choi, H.; Kim, Y.K.; Yoon, E.J.; Lee, J.Y.; Lee, D.S.; Initiative, A.D.N. Cognitive signature of brain FDG PET based on deep learning: Domain transfer from Alzheimer’s disease to Parkinson’s disease. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 403–412. [Google Scholar] [CrossRef]
- Huang, Z.; Zhu, X.; Ding, M.; Zhang, X. Medical image classification using a light-weighted hybrid neural network based on PCANet and DenseNet. IEEE Access 2020, 8, 24697–24712. [Google Scholar] [CrossRef]
- Kim, C.; Kim, W.H.; Kim, H.J.; Kim, J. Weakly-supervised US breast tumor characterization and localization with a box convolution network. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 16–19 February 2020; SPIE: Bellingham, DC, USA, 2020; Volume 11314, pp. 298–304. [Google Scholar]
- Luo, L.; Chen, H.; Wang, X.; Dou, Q.; Lin, H.; Zhou, J.; Li, G.; Heng, P.A. Deep angular embedding and feature correlation attention for breast MRI cancer analysis. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part IV 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 504–512. [Google Scholar]
- Yi, P.H.; Lin, A.; Wei, J.; Yu, A.C.; Sair, H.I.; Hui, F.K.; Hager, G.D.; Harvey, S.C. Deep-learning-based semantic labeling for 2D mammography and comparison of complexity for machine learning tasks. J. Digit. Imaging 2019, 32, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Nishikawa, R.M. Detecting mammographically occult cancer in women with dense breasts using deep convolutional neural network and Radon Cumulative Distribution Transform. J. Med. Imaging 2019, 6, 044502. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, L.; Chen, Y.; Pi, Y.; Chen, Y.; Lv, Q.; Yi, Z. Automated diagnosis of breast ultrasonography images using deep neural networks. Med. Image Anal. 2019, 52, 185–198. [Google Scholar] [CrossRef]
- Xi, P.; Guan, H.; Shu, C.; Borgeat, L.; Goubran, R. An integrated approach for medical abnormality detection using deep patch convolutional neural networks. Vis. Comput. 2020, 36, 1869–1882. [Google Scholar] [CrossRef]
- Zhou, L.Q.; Wu, X.L.; Huang, S.Y.; Wu, G.G.; Ye, H.R.; Wei, Q.; Bao, L.Y.; Deng, Y.B.; Li, X.R.; Cui, X.W.; et al. Lymph node metastasis prediction from primary breast cancer US images using deep learning. Radiology 2020, 294, 19–28. [Google Scholar] [CrossRef]
- Dunnmon, J.A.; Yi, D.; Langlotz, C.P.; Ré, C.; Rubin, D.L.; Lungren, M.P. Assessment of convolutional neural networks for automated classification of chest radiographs. Radiology 2019, 290, 537–544. [Google Scholar] [CrossRef]
- Huang, Z.; Fu, D. Diagnose chest pathology in X-ray images by learning multi-attention convolutional neural network. In Proceedings of the 2019 IEEE 8th Joint International Information Technology and Artificial Intelligence Conference (ITAIC), Chongqing, China, 24–26 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 294–299. [Google Scholar]
- Khakzar, A.; Albarqouni, S.; Navab, N. Learning interpretable features via adversarially robust optimization. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part VI 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 793–800. [Google Scholar]
- Kumar, D.; Sankar, V.; Clausi, D.; Taylor, G.W.; Wong, A. Sisc: End-to-end interpretable discovery radiomics-driven lung cancer prediction via stacked interpretable sequencing cells. IEEE Access 2019, 7, 145444–145454. [Google Scholar] [CrossRef]
- Lei, Y.; Tian, Y.; Shan, H.; Zhang, J.; Wang, G.; Kalra, M.K. Shape and margin-aware lung nodule classification in low-dose CT images via soft activation mapping. Med. Image Anal. 2020, 60, 101628. [Google Scholar] [CrossRef]
- Tang, Y.X.; Tang, Y.B.; Peng, Y.; Yan, K.; Bagheri, M.; Redd, B.A.; Brandon, C.J.; Lu, Z.; Han, M.; Xiao, J.; et al. Automated abnormality classification of chest radiographs using deep convolutional neural networks. NPJ Digit. Med. 2020, 3, 70. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, X.; Huang, S. KGZNet: Knowledge-guided deep zoom neural networks for thoracic disease classification. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1396–1401. [Google Scholar]
- Yi, P.H.; Kim, T.K.; Yu, A.C.; Bennett, B.; Eng, J.; Lin, C.T. Can AI outperform a junior resident? Comparison of deep neural network to first-year radiology residents for identification of pneumothorax. Emerg. Radiol. 2020, 27, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, L.; Nan, Y.; Jin, F.; Wang, Q.; Pu, J. SDFN: Segmentation-based deep fusion network for thoracic disease classification in chest X-ray images. Comput. Med. Imaging Graph. 2019, 75, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Kasukurthi, N.; Pande, H. Deep learning for weak supervision of diabetic retinopathy abnormalities. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 573–577. [Google Scholar]
- Liao, W.; Zou, B.; Zhao, R.; Chen, Y.; He, Z.; Zhou, M. Clinical interpretable deep learning model for glaucoma diagnosis. IEEE J. Biomed. Health Inform. 2019, 24, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Perdomo, O.; Rios, H.; Rodríguez, F.J.; Otálora, S.; Meriaudeau, F.; Müller, H.; González, F.A. Classification of diabetes-related retinal diseases using a deep learning approach in optical coherence tomography. Comput. Methods Programs Biomed. 2019, 178, 181–189. [Google Scholar] [CrossRef]
- Shen, Y.; Sheng, B.; Fang, R.; Li, H.; Dai, L.; Stolte, S.; Qin, J.; Jia, W.; Shen, D. Domain-invariant interpretable fundus image quality assessment. Med. Image Anal. 2020, 61, 101654. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Ran, A.R.; Luo, L.; Chan, P.P.; Tham, C.C.; Chang, R.T.; Mannil, S.S.; Cheung, C.Y.; Heng, P.A. Towards multi-center glaucoma OCT image screening with semi-supervised joint structure and function multi-task learning. Med. Image Anal. 2020, 63, 101695. [Google Scholar] [CrossRef]
- Jiang, H.; Yang, K.; Gao, M.; Zhang, D.; Ma, H.; Qian, W. An interpretable ensemble deep learning model for diabetic retinopathy disease classification. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 2045–2048. [Google Scholar]
- Tu, Z.; Gao, S.; Zhou, K.; Chen, X.; Fu, H.; Gu, Z.; Cheng, J.; Yu, Z.; Liu, J. SUNet: A lesion regularized model for simultaneous diabetic retinopathy and diabetic macular edema grading. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1378–1382. [Google Scholar]
- Kumar, D.; Taylor, G.W.; Wong, A. Discovery radiomics with CLEAR-DR: Interpretable computer aided diagnosis of diabetic retinopathy. IEEE Access 2019, 7, 25891–25896. [Google Scholar] [CrossRef]
- Liu, C.; Han, X.; Li, Z.; Ha, J.; Peng, G.; Meng, W.; He, M. A self-adaptive deep learning method for automated eye laterality detection based on color fundus photography. PLoS ONE 2019, 14, e0222025. [Google Scholar] [CrossRef]
- Narayanan, B.N.; Hardie, R.C.; De Silva, M.S.; Kueterman, N.K. Hybrid machine learning architecture for automated detection and grading of retinal images for diabetic retinopathy. J. Med. Imaging 2020, 7, 034501. [Google Scholar] [CrossRef]
- Everson, M.; Herrera, L.G.P.; Li, W.; Luengo, I.M.; Ahmad, O.; Banks, M.; Magee, C.; Alzoubaidi, D.; Hsu, H.; Graham, D.; et al. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United Eur. Gastroenterol. J. 2019, 7, 297–306. [Google Scholar] [CrossRef]
- García-Peraza-Herrera, L.C.; Everson, M.; Lovat, L.; Wang, H.P.; Wang, W.L.; Haidry, R.; Stoyanov, D.; Ourselin, S.; Vercauteren, T. Intrapapillary capillary loop classification in magnification endoscopy: Open dataset and baseline methodology. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Xing, Y.; Zhang, L.; Gao, H.; Zhang, H. Deep convolutional neural network for ulcer recognition in wireless capsule endoscopy: Experimental feasibility and optimization. Comput. Math. Methods Med. 2019, 2019, 7546215. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Xu, J.; Xie, J.; Cai, C.; Lu, H. Prior-aware CNN with multi-task learning for colon images analysis. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 254–257. [Google Scholar]
- Heinemann, F.; Birk, G.; Stierstorfer, B. Deep learning enables pathologist-like scoring of NASH models. Sci. Rep. 2019, 9, 18454. [Google Scholar] [CrossRef] [PubMed]
- Kiani, A.; Uyumazturk, B.; Rajpurkar, P.; Wang, A.; Gao, R.; Jones, E.; Yu, Y.; Langlotz, C.P.; Ball, R.L.; Montine, T.J.; et al. Impact of a deep learning assistant on the histopathologic classification of liver cancer. NPJ Digit. Med. 2020, 3, 23. [Google Scholar] [CrossRef]
- Chang, G.H.; Felson, D.T.; Qiu, S.; Guermazi, A.; Capellini, T.D.; Kolachalama, V.B. Assessment of knee pain from MR imaging using a convolutional Siamese network. Eur. Radiol. 2020, 30, 3538–3548. [Google Scholar] [CrossRef]
- Yi, P.H.; Kim, T.K.; Wei, J.; Shin, J.; Hui, F.K.; Sair, H.I.; Hager, G.D.; Fritz, J. Automated semantic labeling of pediatric musculoskeletal radiographs using deep learning. Pediatr. Radiol. 2019, 49, 1066–1070. [Google Scholar] [CrossRef]
- Li, W.; Zhuang, J.; Wang, R.; Zhang, J.; Zheng, W.S. Fusing metadata and dermoscopy images for skin disease diagnosis. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1996–2000. [Google Scholar]
- Xie, Y.; Zhang, J.; Xia, Y.; Shen, C. A mutual bootstrapping model for automated skin lesion segmentation and classification. IEEE Trans. Med Imaging 2020, 39, 2482–2493. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, K.J.; Sunwoo, L.; Choi, D.; Nam, C.M.; Cho, J.; Kim, J.; Bae, Y.J.; Yoo, R.E.; Choi, B.S.; et al. Deep learning in diagnosis of maxillary sinusitis using conventional radiography. Investig. Radiol. 2019, 54, 7–15. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, L.; Zhu, M.; Qi, X.; Yi, Z. Automatic diagnosis for thyroid nodules in ultrasound images by deep neural networks. Med. Image Anal. 2020, 61, 101665. [Google Scholar] [CrossRef]
- Huang, Y.; Chung, A.C. Evidence localization for pathology images using weakly supervised learning. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part I 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 613–621. [Google Scholar]
- Kim, I.; Rajaraman, S.; Antani, S. Visual interpretation of convolutional neural network predictions in classifying medical image modalities. Diagnostics 2019, 9, 38. [Google Scholar] [CrossRef]
- Tang, C. Discovering Unknown Diseases with Explainable Automated Medical Imaging. In Proceedings of the Medical Image Understanding and Analysis: 24th Annual Conference, MIUA 2020, Oxford, UK, 15–17 July 2020; Proceedings 24. Springer: Berlin/Heidelberg, Germany, 2020; pp. 346–358. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Hilbert, A.; Ramos, L.A.; van Os, H.J.; Olabarriaga, S.D.; Tolhuisen, M.L.; Wermer, M.J.; Marquering, H.A. Data-efficient deep learning of radiological image data for outcome prediction after endovascular treatment of patients with acute ischemic stroke. Comput. Biol. Med. 2019, 115, 103516. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Ye, J.C. Understanding graph isomorphism network for rs-fMRI functional connectivity analysis. Front. Neurosci. 2020, 14, 630. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Zhang, X.; Zhao, F.; Lou, J.; Wang, L.; Xu, X.; Li, G. Multi-branch deformable convolutional neural network with label distribution learning for fetal brain age prediction. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 424–427. [Google Scholar]
- Natekar, P.; Kori, A.; Krishnamurthi, G. Demystifying brain tumor segmentation networks: Interpretability and uncertainty analysis. Front. Comput. Neurosci. 2020, 14, 6. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Proceedings of the Understanding and Interpreting Machine Learning in Medical Image Computing Applications: First International Workshops, MLCN 2018, DLF 2018, and iMIMIC 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 16–20 September 2018; Proceedings 1. Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 106–114. [Google Scholar]
- Pominova, M.; Artemov, A.; Sharaev, M.; Kondrateva, E.; Bernstein, A.; Burnaev, E. Voxelwise 3D convolutional and recurrent neural networks for epilepsy and depression diagnostics from structural and functional MRI data. In Proceedings of the 2018 IEEE International Conference on Data Mining Workshops (ICDMW), Singapore, 17–20 November 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 299–307. [Google Scholar]
- Xie, B.; Lei, T.; Wang, N.; Cai, H.; Xian, J.; He, M.; Xie, H. Computer-aided diagnosis for fetal brain ultrasound images using deep convolutional neural networks. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1303–1312. [Google Scholar] [CrossRef]
- El Adoui, M.; Drisis, S.; Benjelloun, M. Multi-input deep learning architecture for predicting breast tumor response to chemotherapy using quantitative MR images. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 1491–1500. [Google Scholar] [CrossRef]
- Obikane, S.; Aoki, Y. Weakly supervised domain adaptation with point supervision in histopathological image segmentation. In Proceedings of the Pattern Recognition: ACPR 2019 Workshops, Auckland, New Zealand, 26 November 2019; Proceedings 5. Springer: Singapore, 2020; pp. 127–140. [Google Scholar]
- Candemir, S.; White, R.D.; Demirer, M.; Gupta, V.; Bigelow, M.T.; Prevedello, L.M.; Erdal, B.S. Automated coronary artery atherosclerosis detection and weakly supervised localization on coronary CT angiography with a deep 3-dimensional convolutional neural network. Comput. Med Imaging Graph. 2020, 83, 101721. [Google Scholar] [CrossRef]
- Cong, C.; Kato, Y.; Vasconcellos, H.D.; Lima, J.; Venkatesh, B. Automated stenosis detection and classification in X-ray angiography using deep neural network. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 1301–1308. [Google Scholar]
- Huo, Y.; Terry, J.G.; Wang, J.; Nath, V.; Bermudez, C.; Bao, S.; Landman, B.A. Coronary calcium detection using 3D attention identical dual deep network based on weakly supervised learning. In Proceedings of the Medical Imaging 2019: Image Processing, San Diego, CA, USA, 19–21 February 2019; SPIE: Bellingham, DC, USA, 2019; Volume 10949, pp. 308–315. [Google Scholar]
- Patra, A.; Noble, J.A. Incremental learning of fetal heart anatomies using interpretable saliency maps. In Proceedings of the Medical Image Understanding and Analysis: 23rd Conference, MIUA 2019, Liverpool, UK, 24–26 July 2019; Proceedings 23. Springer International Publishing: Berlin/Heidelberg, Germany, 2020; pp. 129–141. [Google Scholar]
- Brunese, L.; Mercaldo, F.; Reginelli, A.; Santone, A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput. Methods Programs Biomed. 2020, 196, 105608. [Google Scholar] [CrossRef]
- Chen, B.; Li, J.; Lu, G.; Zhang, D. Lesion location attention guided network for multi-label thoracic disease classification in chest X-rays. IEEE J. Biomed. Health Inform. 2019, 24, 2016–2027. [Google Scholar] [CrossRef]
- He, J.; Shang, L.; Ji, H.; Zhang, X. Deep learning features for lung adenocarcinoma classification with tissue pathology images. In Proceedings of the Neural Information Processing: 24th International Conference, ICONIP 2017, Guangzhou, China, 14–18 November 2017; Proceedings, Part IV. Springer International Publishing: Berlin/Heidelberg, Germany, 2017; Volume 24, pp. 742–751. [Google Scholar]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Aerts, H.J. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef]
- Humphries, S.M.; Notary, A.M.; Centeno, J.P.; Strand, M.J.; Crapo, J.D.; Silverman, E.K.; For the Genetic Epidemiology of COPD (COPDGene) Investigators. Deep learning enables automatic classification of emphysema pattern at CT. Radiology 2020, 294, 434–444. [Google Scholar] [CrossRef]
- Ko, H.; Chung, H.; Kang, W.S.; Kim, K.W.; Shin, Y.; Kang, S.J.; Lee, J. COVID-19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: Model development and validation. J. Med Internet Res. 2020, 22, e19569. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, T.; Rahman, M.A.; Fattah, S.A. CovXNet: A multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020, 122, 103869. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Schabath, M.; Gillies, R.; Hall, L.; Goldgof, D. Convolutional Neural Network ensembles for accurate lung nodule malignancy prediction 2 years in the future. Comput. Biol. Med. 2020, 122, 103882. [Google Scholar] [CrossRef]
- Philbrick, K.A.; Yoshida, K.; Inoue, D.; Akkus, Z.; Kline, T.L.; Weston, A.D.; Erickson, B.J. What does deep learning see? Insights from a classifier trained to predict contrast enhancement phase from CT images. Am. J. Roentgenol. 2018, 211, 1184–1193. [Google Scholar] [CrossRef] [PubMed]
- Qin, R.; Wang, Z.; Jiang, L.; Qiao, K.; Hai, J.; Chen, J.; Yan, B. Fine-Grained Lung Cancer Classification from PET and CT Images Based on Multidimensional Attention Mechanism. Complexity 2020, 2020, 6153657. [Google Scholar] [CrossRef]
- Teramoto, A.; Yamada, A.; Kiriyama, Y.; Tsukamoto, T.; Yan, K.; Zhang, L.; Fujita, H. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inform. Med. Unlocked 2019, 16, 100205. [Google Scholar] [CrossRef]
- Xu, R.; Cong, Z.; Ye, X.; Hirano, Y.; Kido, S.; Gyobu, T.; Tomiyama, N. Pulmonary textures classification via a multi-scale attention network. IEEE J. Biomed. Health Inform. 2019, 24, 2041–2052. [Google Scholar] [CrossRef]
- Vila-Blanco, N.; Carreira, M.J.; Varas-Quintana, P.; Balsa-Castro, C.; Tomas, I. Deep neural networks for chronological age estimation from OPG images. IEEE Trans. Med Imaging 2020, 39, 2374–2384. [Google Scholar] [CrossRef]
- Kim, M.; Han, J.C.; Hyun, S.H.; Janssens, O.; Van Hoecke, S.; Kee, C.; De Neve, W. Medinoid: Computer-aided diagnosis and localization of glaucoma using deep learning. Appl. Sci. 2019, 9, 3064. [Google Scholar] [CrossRef]
- Martins, J.; Cardoso, J.S.; Soares, F. Offline computer-aided diagnosis for Glaucoma detection using fundus images targeted at mobile devices. Comput. Methods Programs Biomed. 2020, 192, 105341. [Google Scholar] [CrossRef]
- Meng, Q.; Hashimoto, Y.; Satoh, S. How to extract more information with less burden: Fundus image classification and retinal disease localization with ophthalmologist intervention. IEEE J. Biomed. Health Inform. 2020, 24, 3351–3361. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Fan, D.; Lv, B.; Wang, M.; Zhou, Q.; Lv, C.; Xie, G.; Wang, L. OCT image quality evaluation based on deep and shallow features fusion network. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1561–1564. [Google Scholar]
- Zhang, R.; Tan, S.; Wang, R.; Manivannan, S.; Chen, J.; Lin, H.; Zheng, W.S. Biomarker localization by combining CNN classifier and generative adversarial network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part I 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 209–217. [Google Scholar]
- Chen, X.; Lin, L.; Liang, D.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.H.; Chen, Y.W.; Tong, R.; Wu, J. A dual-attention dilated residual network for liver lesion classification and localization on CT images. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 235–239. [Google Scholar]
- Itoh, H.; Lu, Z.; Mori, Y.; Misawa, M.; Oda, M.; Kudo, S.e.; Mori, K. Visualising decision-reasoning regions in computer-aided pathological pattern diagnosis of endoscytoscopic images based on CNN weights analysis. In Proceedings of the Medical Imaging 2020: Computer-Aided Diagnosis, Houston, TX, USA, 6–19 February 2020; SPIE: Bellingham, DC, USA, 2020; Volume 11314, pp. 761–768. [Google Scholar]
- Korbar, B.; Olofson, A.M.; Miraflor, A.P.; Nicka, C.M.; Suriawinata, M.A.; Torresani, L.; Suriawinata, A.A.; Hassanpour, S. Looking under the hood: Deep neural network visualization to interpret whole-slide image analysis outcomes for colorectal polyps. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 69–75. [Google Scholar]
- Kowsari, K.; Sali, R.; Ehsan, L.; Adorno, W.; Ali, A.; Moore, S.; Amadi, B.; Kelly, P.; Syed, S.; Brown, D. Hmic: Hierarchical medical image classification, a deep learning approach. Information 2020, 11, 318. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cui, Y.; Shi, G.; Zhao, J.; Yang, X.; Qiang, Y.; Du, Q.; Ma, Y.; Kazihise, N.G.F. Multi-branch cross attention model for prediction of KRAS mutation in rectal cancer with t2-weighted MRI. Appl. Intell. 2020, 50, 2352–2369. [Google Scholar] [CrossRef]
- Cheng, C.T.; Ho, T.Y.; Lee, T.Y.; Chang, C.C.; Chou, C.C.; Chen, C.C.; Chung, I.; Liao, C.H. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur. Radiol. 2019, 29, 5469–5477. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Demirer, M.; Bigelow, M.; Sarah, M.Y.; Joseph, S.Y.; Prevedello, L.M.; White, R.D.; Erdal, B.S. Using transfer learning and class activation maps supporting detection and localization of femoral fractures on anteroposterior radiographs. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1526–1529. [Google Scholar]
- Zhang, B.; Tan, J.; Cho, K.; Chang, G.; Deniz, C.M. Attention-based cnn for kl grade classification: Data from the osteoarthritis initiative. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 731–735. [Google Scholar]
- von Schacky, C.E.; Sohn, J.H.; Liu, F.; Ozhinsky, E.; Jungmann, P.M.; Nardo, L.; Posadzy, M.; Foreman, S.C.; Nevitt, M.C.; Link, T.M.; et al. Development and validation of a multitask deep learning model for severity grading of hip osteoarthritis features on radiographs. Radiology 2020, 295, 136–145. [Google Scholar] [CrossRef]
- Lee, J.H.; Ha, E.J.; Kim, D.; Jung, Y.J.; Heo, S.; Jang, Y.H.; An, S.H.; Lee, K. Application of deep learning to the diagnosis of cervical lymph node metastasis from thyroid cancer with CT: External validation and clinical utility for resident training. Eur. Radiol. 2020, 30, 3066–3072. [Google Scholar] [CrossRef]
- Langner, T.; Wikström, J.; Bjerner, T.; Ahlström, H.; Kullberg, J. Identifying morphological indicators of aging with neural networks on large-scale whole-body MRI. IEEE Trans. Med. Imaging 2019, 39, 1430–1437. [Google Scholar] [CrossRef]
- Li, C.; Yao, G.; Xu, X.; Yang, L.; Zhang, Y.; Wu, T.; Sun, J. DCSegNet: Deep learning framework based on divide-and-conquer method for liver segmentation. IEEE Access 2020, 8, 146838–146846. [Google Scholar] [CrossRef]
- Mohamed Musthafa, M.; Mahesh, T.R.; Vinoth Kumar, V.; Guluwadi, S. Enhancing brain tumor detection in MRI images through explainable AI using Grad-CAM with Resnet 50. BMC Med. Imaging 2024, 24, 107. [Google Scholar]
- Wang, C.W.; Khalil, M.A.; Lin, Y.J.; Lee, Y.C.; Chao, T.K. Detection of erbb2 and cen17 signals in fluorescent in situ hybridization and dual in situ hybridization for guiding breast cancer her2 target therapy. Artif. Intell. Med. 2023, 141, 102568. [Google Scholar] [CrossRef]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.R.; Samek, W. On pixel-wise explanations for non-linear classifier decisions by layer-wise relevance propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef]
- Montavon, G.; Lapuschkin, S.; Binder, A.; Samek, W.; Müller, K.R. Explaining nonlinear classification decisions with deep taylor decomposition. Pattern Recognit. 2017, 65, 211–222. [Google Scholar] [CrossRef]
- Samek, W.; Binder, A.; Montavon, G.; Lapuschkin, S.; Müller, K.R. Evaluating the visualization of what a deep neural network has learned. IEEE Trans. Neural Netw. Learn. Syst. 2016, 28, 2660–2673. [Google Scholar] [CrossRef] [PubMed]
- Kohlbrenner, M.; Bauer, A.; Nakajima, S.; Binder, A.; Samek, W.; Lapuschkin, S. Towards best practice in explaining neural network decisions with LRP. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–7. [Google Scholar]
- Arquilla, K.; Gajera, I.D.; Darling, M.; Bhati, D.; Singh, A.; Guercio, A. Exploring Fine-Grained Feature Analysis for Bird Species Classification using Layer-wise Relevance Propagation. In Proceedings of the 2024 IEEE World AI IoT Congress (AIIoT), Melbourne, Australia, 29–31 May 2024; IEEE: Piscataway, NJ, USA, 2024; pp. 625–631. [Google Scholar]
- Eitel, F.; Soehler, E.; Bellmann-Strobl, J.; Brandt, A.U.; Ruprecht, K.; Giess, R.M.; Kuchling, J.; Asseyer, S.; Weygandt, M.; Haynes, J.D.; et al. Uncovering convolutional neural network decisions for diagnosing multiple sclerosis on conventional MRI using layer-wise relevance propagation. Neuroimage Clin. 2019, 24, 102003. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.W.; Heekeren, H.R.; Müller, K.R.; Samek, W. Analyzing neuroimaging data through recurrent deep learning models. Front. Neurosci. 2019, 13, 1321. [Google Scholar] [CrossRef] [PubMed]
- Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D. Attention gated networks: Learning to leverage salient regions in medical images. Med. Image Anal. 2019, 53, 197–207. [Google Scholar] [CrossRef]
- Katar, O.; Yildirim, O. An explainable vision transformer model based white blood cells classification and localization. Diagnostics 2023, 13, 2459. [Google Scholar] [CrossRef]
- Jetley, S.; Lord, N.A.; Lee, N.; Torr, P.H. Learn to pay attention. arXiv 2018, arXiv:1804.02391. [Google Scholar]
- Li, S.; Dong, M.; Du, G.; Mu, X. Attention dense-u-net for automatic breast mass segmentation in digital mammogram. IEEE Access 2019, 7, 59037–59047. [Google Scholar] [CrossRef]
- Yan, Y.; Kawahara, J.; Hamarneh, G. Melanoma recognition via visual attention. In Proceedings of the Information Processing in Medical Imaging: 26th International Conference, IPMI 2019, Hong Kong, China, 2–7 June 2019; Proceedings 26. Springer: Berlin/Heidelberg, Germany, 2019; pp. 793–804. [Google Scholar]
- Górriz, M.; Antony, J.; McGuinness, K.; Giró-i Nieto, X.; O’Connor, N.E. Assessing knee OA severity with CNN attention-based end-to-end architectures. In Proceedings of the International Conference on Medical Imaging with Deep Learning, PMLR, London, UK, 8–10 July 2019; pp. 197–214. [Google Scholar]
- Xu, X.; Li, C.; Fan, X.; Lan, X.; Lu, X.; Ye, X.; Wu, T. Attention Mask R-CNN with edge refinement algorithm for identifying circulating genetically abnormal cells. Cytom. Part A 2023, 103, 227–239. [Google Scholar] [CrossRef]
- Bramlage, L.; Cortese, A. Generalized attention-weighted reinforcement learning. Neural Netw. 2022, 145, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Shao, Q. Single image super-resolution based on trainable feature matching attention network. Pattern Recognit. 2024, 149, 110289. [Google Scholar] [CrossRef]
- Dubost, F.; Adams, H.; Yilmaz, P.; Bortsova, G.; van Tulder, G.; Ikram, M.A.; Niessen, W.; Vernooij, M.W.; de Bruijne, M. Weakly supervised object detection with 2D and 3D regression neural networks. Med. Image Anal. 2020, 65, 101767. [Google Scholar] [CrossRef] [PubMed]
- Lian, C.; Liu, M.; Wang, L.; Shen, D. End-to-end dementia status prediction from brain mri using multi-task weakly-supervised attention network. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part IV 22. Springer: Berlin/Heidelberg, Germany, 2019; pp. 158–167. [Google Scholar]
- Wang, H.; Feng, J.; Zhang, Z.; Su, H.; Cui, L.; He, H.; Liu, L. Breast mass classification via deeply integrating the contextual information from multi-view data. Pattern Recognit. 2018, 80, 42–52. [Google Scholar] [CrossRef]
- Li, L.; Xu, M.; Liu, H.; Li, Y.; Wang, X.; Jiang, L.; Wang, Z.; Fan, X.; Wang, N. A large-scale database and a CNN model for attention-based glaucoma detection. IEEE Trans. Med Imaging 2019, 39, 413–424. [Google Scholar] [CrossRef]
- Yang, H.; Kim, J.Y.; Kim, H.; Adhikari, S.P. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans. Med Imaging 2019, 39, 1306–1315. [Google Scholar] [CrossRef]
- Pesce, E.; Withey, S.J.; Ypsilantis, P.P.; Bakewell, R.; Goh, V.; Montana, G. Learning to detect chest radiographs containing pulmonary lesions using visual attention networks. Med. Image Anal. 2019, 53, 26–38. [Google Scholar] [CrossRef]
- Singla, S.; Gong, M.; Ravanbakhsh, S.; Sciurba, F.; Poczos, B.; Batmanghelich, K.N. Subject2Vec: Generative-discriminative approach from a set of image patches to a vector. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part I. Springer: Berlin/Heidelberg, Germany, 2018; pp. 502–510. [Google Scholar]
- Sun, J.; Darbehani, F.; Zaidi, M.; Wang, B. Saunet: Shape attentive u-net for interpretable medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, 4–8 October 2020; Proceedings, Part IV 23. Springer: Berlin/Heidelberg, Germany, 2020; pp. 797–806. [Google Scholar]
- Zhu, Z.; Ding, X.; Zhang, D.; Wang, L. Weakly-supervised balanced attention network for gastric pathology image localization and classification. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: Piscataway, NJ, USA, 2020; pp. 1–4. [Google Scholar]
- Barata, C.; Celebi, M.E.; Marques, J.S. Explainable skin lesion diagnosis using taxonomies. Pattern Recognit. 2021, 110, 107413. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Srivastava, A.; Chandra, M.; Saha, A.; Saluja, S.; Bhati, D. Current Advances in Locality-Based and Feature-Based Transformers: A Review. In Proceedings of the International Conference on Data & Information Sciences, Edinburgh, UK, 11–13 August 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 321–335. [Google Scholar]
- Wu, J.; Mi, Q.; Zhang, Y.; Wu, T. SVTNet: Automatic bone age assessment network based on TW3 method and vision transformer. Int. J. Imaging Syst. Technol. 2024, 34, e22990. [Google Scholar] [CrossRef]
- Park, S.; Kim, G.; Oh, Y.; Seo, J.B.; Lee, S.M.; Kim, J.H.; Moon, S.; Lim, J.K.; Park, C.M.; Ye, J.C. Self-evolving vision transformer for chest X-ray diagnosis through knowledge distillation. Nat. Commun. 2022, 13, 3848. [Google Scholar] [CrossRef]
- Chen, J.; Frey, E.C.; He, Y.; Segars, W.P.; Li, Y.; Du, Y. Transmorph: Transformer for unsupervised medical image registration. Med. Image Anal. 2022, 82, 102615. [Google Scholar] [CrossRef]
- Gupte, S.R.; Hou, C.; Wu, G.H.; Galaz-Montoya, J.G.; Chiu, W.; Yeung-Levy, S. CryoViT: Efficient Segmentation of Cryogenic Electron Tomograms with Vision Foundation Models. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Karimi, D.; Vasylechko, S.D.; Gholipour, A. Convolution-free medical image segmentation using transformers. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 78–88. [Google Scholar]
- Yun, B.; Wang, Y.; Chen, J.; Wang, H.; Shen, W.; Li, Q. Spectr: Spectral transformer for hyperspectral pathology image segmentation. arXiv 2021, arXiv:2103.03604. [Google Scholar] [CrossRef]
- Wenxuan, W.; Chen, C.; Meng, D.; Hong, Y.; Sen, Z.; Jiangyun, L. Transbts: Multimodal brain tumor segmentation using transformer. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Strasbourg, France, 27 September–1 October 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 109–119. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. Unetr: Transformers for 3d medical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 3–8 January 2022; pp. 574–584. [Google Scholar]
- Li, S.; Sui, X.; Luo, X.; Xu, X.; Liu, Y.; Goh, R. Medical image segmentation using squeeze-and-expansion transformers. arXiv 2021, arXiv:2105.09511. [Google Scholar]
- Zhang, Y.; Higashita, R.; Fu, H.; Xu, Y.; Zhang, Y.; Liu, H.; Zhang, J.; Liu, J. A multi-branch hybrid transformer network for corneal endothelial cell segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part I 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 99–108. [Google Scholar]
- Lin, A.; Chen, B.; Xu, J.; Zhang, Z.; Lu, G.; Zhang, D. Ds-transunet: Dual swin transformer u-net for medical image segmentation. IEEE Trans. Instrum. Meas. 2022, 71, 1–15. [Google Scholar] [CrossRef]
- Li, Y.; Cai, W.; Gao, Y.; Li, C.; Hu, X. More than encoder: Introducing transformer decoder to upsample. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 1597–1602. [Google Scholar]
- Xu, G.; Zhang, X.; He, X.; Wu, X. Levit-unet: Make faster encoders with transformer for medical image segmentation. In Proceedings of the Chinese Conference on Pattern Recognition and Computer Vision (PRCV), Xiamen, China, 13–15 October 2023; Springer: Berlin/Heidelberg, Germany, 2023; pp. 42–53. [Google Scholar]
- Chang, Y.; Menghan, H.; Guangtao, Z.; Xiao-Ping, Z. Transclaw u-net: Claw u-net with transformers for medical image segmentation. arXiv 2021, arXiv:2107.05188. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 205–218. [Google Scholar]
- Petit, O.; Thome, N.; Rambour, C.; Themyr, L.; Collins, T.; Soler, L. U-net transformer: Self and cross attention for medical image segmentation. In Proceedings of the Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, 27 September 2021; Proceedings 12. Springer: Berlin/Heidelberg, Germany, 2021; pp. 267–276. [Google Scholar]
- Xie, Y.; Zhang, J.; Shen, C.; Xia, Y. Cotr: Efficiently bridging cnn and transformer for 3d medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part III 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 171–180. [Google Scholar]
- Gao, Y.; Zhou, M.; Metaxas, D.N. UTNet: A hybrid transformer architecture for medical image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part III 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 61–71. [Google Scholar]
- Chen, B.; Liu, Y.; Zhang, Z.; Lu, G.; Kong, A.W.K. Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. IEEE Trans. Emerg. Top. Comput. Intell. 2023. [Google Scholar] [CrossRef]
- Dong, B.; Wang, W.; Fan, D.P.; Li, J.; Fu, H.; Shao, L. Polyp-pvt: Polyp segmentation with pyramid vision transformers. arXiv 2021, arXiv:2108.06932. [Google Scholar] [CrossRef]
- Shen, Z.; Yang, H.; Zhang, Z.; Zheng, S. Automated kidney tumor segmentation with convolution and transformer network. In International Challenge on Kidney and Kidney Tumor Segmentation; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–12. [Google Scholar]
- Deng, K.; Meng, Y.; Gao, D.; Bridge, J.; Shen, Y.; Lip, G.; Zhao, Y.; Zheng, Y. Transbridge: A lightweight transformer for left ventricle segmentation in echocardiography. In Proceedings of the Simplifying Medical Ultrasound: Second International Workshop, ASMUS 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, 27 September 2021; Proceedings 2. Springer: Berlin/Heidelberg, Germany, 2021; pp. 63–72. [Google Scholar]
- Jia, Q.; Shu, H. Bitr-unet: A cnn-transformer combined network for mri brain tumor segmentation. In Proceedings of the International MICCAI Brainlesion Workshop, Singapore, 27 September 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 3–14. [Google Scholar]
- Hatamizadeh, A.; Nath, V.; Tang, Y.; Yang, D.; Roth, H.R.; Xu, D. Swin unetr: Swin transformers for semantic segmentation of brain tumors in mri images. In Proceedings of the International MICCAI Brainlesion Workshop, Singapore, 27 September 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 272–284. [Google Scholar]
- Li, Y.; Wang, S.; Wang, J.; Zeng, G.; Liu, W.; Zhang, Q.; Jin, Q.; Wang, Y. Gt u-net: A u-net like group transformer network for tooth root segmentation. In Proceedings of the Machine Learning in Medical Imaging: 12th International Workshop, MLMI 2021, Held in Conjunction with MICCAI 2021, Strasbourg, France, 27 September 2021; Proceedings 12. Springer: Berlin/Heidelberg, Germany, 2021; pp. 386–395. [Google Scholar]
- Gheflati, B.; Rivaz, H. Vision transformers for classification of breast ultrasound images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Glasgow, UK, 11–15 July 2022; IEEE: Piscataway, NJ, USA, 2022; pp. 480–483. [Google Scholar]
- Zheng, Y.; Gindra, R.; Betke, M.; Beane, J.E.; Kolachalama, V.B. A deep learning based graph-transformer for whole slide image classification. medRxiv 2021. [Google Scholar] [CrossRef]
- Yu, S.; Ma, K.; Bi, Q.; Bian, C.; Ning, M.; He, N.; Li, Y.; Liu, H.; Zheng, Y. Mil-vt: Multiple instance learning enhanced vision transformer for fundus image classification. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part VIII 24. Springer: Berlin/Heidelberg, Germany, 2021; pp. 45–54. [Google Scholar]
- Sun, R.; Li, Y.; Zhang, T.; Mao, Z.; Wu, F.; Zhang, Y. Lesion-aware transformers for diabetic retinopathy grading. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 10938–10947. [Google Scholar]
- Perera, S.; Adhikari, S.; Yilmaz, A. Pocformer: A lightweight transformer architecture for detection of covid-19 using point of care ultrasound. In Proceedings of the 2021 IEEE International Conference on Image Processing (ICIP), Anchorage, AK, USA, 9–22 September 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 195–199. [Google Scholar]
- Park, S.; Kim, G.; Kim, J.; Kim, B.; Ye, J.C. Federated split vision transformer for COVID-19 CXR diagnosis using task-agnostic training. arXiv 2021, arXiv:2111.01338. [Google Scholar]
- Shome, D.; Kar, T.; Mohanty, S.N.; Tiwari, P.; Muhammad, K.; AlTameem, A.; Zhang, Y.; Saudagar, A.K.J. Covid-transformer: Interpretable COVID-19 detection using vision transformer for healthcare. Int. J. Environ. Res. Public Health 2021, 18, 11086. [Google Scholar] [CrossRef]
- Liu, C.; Yin, Q. Automatic diagnosis of covid-19 using a tailored transformer-like network. In Proceedings of the Journal of Physics: Conference Series; IOP Publishing: Bristol, UK, 2021; Volume 2010, p. 012175. [Google Scholar]
- Park, S.; Kim, G.; Oh, Y.; Seo, J.B.; Lee, S.M.; Kim, J.H.; Moon, S.; Lim, J.K.; Ye, J.C. Vision transformer for COVID-19 cxr diagnosis using chest X-ray feature corpus. arXiv 2021, arXiv:2103.07055. [Google Scholar]
- Gao, X.; Qian, Y.; Gao, A. COVID-VIT: Classification of COVID-19 from CT chest images based on vision transformer models. arXiv 2021, arXiv:2107.01682. [Google Scholar]
- Mondal, A.K.; Bhattacharjee, A.; Singla, P.; Prathosh, A. xViTCOS: Explainable vision transformer based COVID-19 screening using radiography. IEEE J. Transl. Eng. Health Med. 2021, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.C.; Chen, G.L.; Wu, M.H. Visual transformer with statistical test for covid-19 classification. arXiv 2021, arXiv:2107.05334. [Google Scholar]
- Zhang, L.; Wen, Y. A transformer-based framework for automatic COVID19 diagnosis in chest CTs. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 513–518. [Google Scholar]
- Ambita, A.A.E.; Boquio, E.N.V.; Naval, P.C., Jr. Covit-gan: Vision transformer forcovid-19 detection in ct scan imageswith self-attention gan forDataAugmentation. In Proceedings of the International Conference on Artificial Neural Networks, Bratislava, Slovakia, 14 September 2021; Springer: Berlin/Heidelberg, Germany, 2021; pp. 587–598. [Google Scholar]
- Zhang, Y.; Pan, X.; Li, C.; Wu, T. 3D liver and tumor segmentation with CNNs based on region and distance metrics. Appl. Sci. 2020, 10, 3794. [Google Scholar] [CrossRef]
- Azad, R.; Kazerouni, A.; Heidari, M.; Aghdam, E.K.; Molaei, A.; Jia, Y.; Jose, A.; Roy, R.; Merhof, D. Advances in medical image analysis with vision transformers: A comprehensive review. Med. Image Anal. 2023, 103000. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Li, Q.; Wang, P.; Guo, D.; Lu, L.; Jin, D.; Zhang, Y.; Hong, Q. Lvit: Language meets vision transformer in medical image segmentation. IEEE Trans. Med. Imaging 2023. [Google Scholar] [CrossRef]


| Attributes | Perturbation | Gradient | Decomposition | Trainable Attention Models |
|---|---|---|---|---|
| Model Dependency | Model-agnostic | Differentiable | Model-specific | Model-specific |
| Access to Model Parameters | No | Yes | Yes | Yes |
| Computational Efficiency | Slower | Faster | Varies | Varies |
| Domain | Task | Modality | Performance | Technique | Citation |
|---|---|---|---|---|---|
| Breast | Classification | MRI | N/A | IG | [45] |
| Eye | Classification | DR | Accuracy: 95.5% | IG | [46] |
| Multiple | Classification | DR | N/A | IG | [47] |
| Chest | Detection | X-ray | Accuracy: 94.9%, AUC: 97.4% | LIME | [48] |
| Gastrointestinal | Classification | Endoscopy | Accuracy: 97.9% | LIME | [49] |
| Brain | Segmentation, Detection | MRI | ICC: 93.0% | OS | [50] |
| Brain | Classification | MRI | Accuracy: 85.0% | OS | [51] |
| Breast | Detection, Classification | Histology | Accuracy: 55.0% | OS | [52] |
| Eye, Chest | Classification, Detection | OCT, X-ray | Eye Accuracy: 94.7%, Chest Accuracy: 92.8% | OS | [53] |
| Chest | Classification | X-ray | AUC: 82.0% | OS, IG, LIME | [54] |
| Domain-Task | Modality | Performance | Citation |
|---|---|---|---|
| Bladder Classification | Histology | Mean Accuracy: 69.9% | [77] |
| Brain Classification | MRI | Accuracy: 86.7% | [78,79] |
| Brain Detection | MRI, PET, CT | Accuracy: 90.2–95.3%, F1: 91.6–94.3% | [80,81] |
| Breast Classification | X-ray, Ultrasound, MRI | Accuracy: 83.0–89.0% | [82,83,84,85] |
| Breast Detection | X-ray, Ultrasound | Mean AUC: 81.0%, AUC: Mt-Net 98.0%, Sn-Net 92.8%, Accuracy: 92.5% | [86,87,88,89] |
| Chest Classification | X-ray, CT | Accuracy: 97.8%, Average AUC: 75.5–96.0% | [90,91,92,93,94,95,96,97] |
| Chest Segmentation | X-ray | Accuracy: 95.8% | [98] |
| Eye Classification | Fundus Photography, OCT, CT | F1: 95.0%, Precision: 93.0%, AUC: 88.0–99.0% | [99,100,101,102,103] |
| Eye Detection | Fundus Photography | Accuracy: 73.2–99.1%, AUC: 99.0% | [104,105,106,107,108] |
| Gastrointestinal (GI) Classification | Endoscopy | Mean Accuracy: 93.2% | [109,110,111,112] |
| Liver Classification, Segmentation | Histology | Mean Accuracy: 87.5% | [113,114] |
| Musculoskeletal Classification | MRI, X-ray | Accuracy: 86.0%, AUC: 85.3% | [115,116] |
| Skin Classification, Segmentation | Dermatoscopy | Accuracy: 83.6%, F1: 82.7% | [117,118] |
| Skull Classification | X-ray | AUC: 88.0–93.0% | [119] |
| Thyroid Classification | Ultrasound | Accuracy: 87.3%, AUC: 90.1% | [120] |
| Lymph Node Classification, Detection | Histology | Accuracy: 91.9%, AUC: 97.0% | [121] |
| Various Classification | CT, MRI, Ultrasound, X-ray, Fundoscopy | F1: 98.0%, Accuracy: 98.0% | [122,123] |
| Domain-Task | Modality | Performance | Citation |
|---|---|---|---|
| Brain Classification | MRI | 81.6–94.2% accuracy | [125,126,127,128,129,130] |
| Brain Detection | Ultrasound | 94.2% accuracy | [131] |
| Breast Classification | MRI | 91.0% AUC | [132] |
| Breast Segmentation | Histology | 95.6% accuracy | [133] |
| Cardiovascular | CT, X-ray, Ultrasound | 81.2–92.7% accuracy, AUC (81.0–96.3%) | [134,135,136,137] |
| Chest Classification | X-ray, CT, Histology | 72.0–99.9% accuracy, AUC (70.0–97.9%) | [138,139,140,141,142,143,144,145,146,147,148,149] |
| Dental Classification | X-ray | 85.4% accuracy, 92.5% AUC | [150] |
| Eye Classification | Fundus, OCT | 81–97.5% accuracy, AUC (48.1–99.2%) | [151,152,153,154,155] |
| Gastrointestinal (GI) Classification | CT, Endoscopy, Histology, MRI | 86.9–93.7% accuracy | [156,157,158,159,160] |
| Musculoskeletal | X-ray | 74.8–96.3% accuracy | [161,162,163,164] |
| Thyroid Classification | CT | 82.8% accuracy, 88.4% AUC | [165] |
| Whole-Body Scans | MRI | R2 value of 83.0% | [166] |
| Liver segmentation | CT scans | 96% accuracy LiTS | [167] |
| Brain Tumor Detection | MRI images | 98.52% accuracy | [168] |
| Breast Cancer | DISH and FISH images | 97% accuracy | [169] |
| Domain | Task | Modality | Performance | Citation |
|---|---|---|---|---|
| Brain | Detection | MRI | Accuracy: 76.5% | [186] |
| Brain | Detection, Classification | MRI | CC: 61.3–64.8%, RMSE: 1.503–5.701 | [187] |
| Breast | Classification | X-ray | Accuracy: 85.0%, AUC: 89.0% | [188] |
| Breast | Segmentation | Mammo | Accuracy: 78.4%, F1: 82.2% | [189] |
| Breast | Classification | Histology | Accuracy: 90.3, AUC: 98.4% | [190] |
| Chest | Detection | X-ray | Accuracy: 73.0–84.0% | [191] |
| Chest | Classification | CT | Accuracy: 87.6% | [192] |
| Chest | Segmentation | MRI | Accuracy: 91.3% | [193] |
| Eye | Detection | Fundus Photography | Accuracy: 96.2%, AUC: 98.3% | [180] |
| Gastrointestinal (GI) | Classification | Histology | Accuracy: 88.4% | [194] |
| Skin | Dermatoscopy | [195] | ||
| Skin | Classification | Dermatoscopy | Average Precision: 67.2%, AUC: 88.3% | [181] |
| Female Reproductive System, Stomach | Classification, Segmentation | CT, Fetal Ultrasounds | Ultrasound Classification: Accuracy: 97.7–98.0%, F1: 92.2–93.3%, CT Segmentation: Recall: 75.1–83.5% | [177] |
| Skeletal (Joint) | Classification | X-ray | Accuracy: 64.3% | [182] |
| Domain | Task | Modality | Performance | Citation |
|---|---|---|---|---|
| Stomach | segmentation | CT, MRI | Dice Score: 77.5%, Hausdorff distance: 31.7% | [202] |
| Brain, Pancreas, Hippocampus | segmentation | MRI, CT | Dice Scores: Brain: 87.9%, Pancreas: 83.6%, Hippocampus: 88.1% | [203] |
| Bile-duct | segmentation | Hyperspectral | Average Dice Score: 75.2% | [204] |
| Brain | segmentation | MRI | Dice Scores: Enhancing Tumor Region: 78.7%, Whole Tumor Region: 90.1%, Regions of Tumor Core: 81.7% | [205] |
| Brain, Spleen | segmentation | MRI, CT | Dice Score: 89.1% | [206] |
| Eye, Rectal, Brain | segmentation | Fundus, Colonoscopy, MRI | Average Dice Score: 91.7% | [207] |
| Eye | segmentation | Pathology | Dice Score: 78.6%, F1: 82.1% | [208] |
| Multi-organ | segmentation | Colonoscopy, Histology | Average Dice Score: 86.8% | [209] |
| Aorta, Gallbladder, Kidney, Liver, Pancreas, Spleen, Stomach | segmentation | MRI, CT | Average Dice Score: 78.1–80.4% | [210,211,212,213,214,215] |
| Heart | segmentation | MRI | Average Dice Score: 88.3% | [216] |
| Skin, Chest | segmentation | X-ray, CT | Average Dice Score: Skin: 90.7%, Chest: 86.6% | [217] |
| Rectal | segmentation | Colonoscopy, Histology | Average Dice Score: 91.7% | [218] |
| Kidney | segmentation | CT | Dice Score: 92.3% | [219] |
| Heart | segmentation | Echocardio- graphy | Dice Score: 91.4% | [220] |
| Brain | segmentation | MRI | Dice Score: 91.3–93.5% | [221,222] |
| Teeth | segmentation | X-ray | Dice Score: 92.5% | [223] |
| Breast | classification | Ultrasound | Accuracy: 86.7%, AUC 95.0% | [224] |
| Lung | classification | Microscopy | Accuracy: 97.5% | [225] |
| Eye | classification | Fundus | Accuracy: 95.9%, AUC: 96.3% | [226,227] |
| Chest | classification | Ultrasound | Accuracy: 93.9% | [228] |
| Chest | classification | X-ray | Average AUC: 93.1%, Accuracy: COVID: 98.0%, Pneumonia: 92.0% | [229,230,231,232] |
| Lung | classification | CT | F1: 76.0% | [233] |
| Chest | classification | X-ray, CT | Overall Accuracy: 87.2–98.1%, F1: 93.5% | [234,235,236,237] |
| Visualization Technique | Task | Body Parts | Modality | Accuracy | Evaluation Metric |
|---|---|---|---|---|---|
| CAM | Image classification and localization | Brain, chest, abdomen | X-ray, MRI, CT scans | 85.0–95.0% | Accuracy for classification; IoU for localization tasks |
| Grad-CAM | Image classification and localization | Brain, chest, abdomen | X-ray, MRI, CT scans | 85.0–95.0% | Accuracy for classification; IoU for localization tasks |
| LRP | Segmentation, classification | Brain, liver, lungs | MRI, CT scans | 90.0% | Dice coefficient for segmentation accuracy |
| IG | Image classification | Breast, lung, spine | X-ray, MRI | 80.0–92.0% | AUC-ROC for classification |
| Attention-based | Image classification, object detection | Brain, chest | X-ray, MRI | 5.0% to 10.0% | Accuracy for classification; mAP for object detection |
| LIME | Local explanations for model predictions | N/A | N/A | N/A | Task-specific metrics |
| Gradient-based | Visualize feature importance | N/A | N/A | N/A | Feature importance metrics, SHAP values, Grad-CAM++ |
| Vision Transformer | Dynamically attend to relevant features | Various body parts | Various modalities | N/A | Task-specific metrics |
| Task | Techniques | Application | Performance Metrics | Examples |
|---|---|---|---|---|
| Classification | CAM, Grad-CAM, Attention, ViTs | Disease diagnosis, organ identification | Accuracy, AUC-ROC, Precision, Recall | Disease Diagnosis: High AUC for cancer detection (e.g., mammograms); Organ Identification: CAM for liver segmentation or brain MRI; ViTs: High accuracy in lung and breast cancer classification |
| Segmentation | LRP, IG, ViTs | Tumor segmentation, anatomical structure delineation | Dice Similarity Coefficient (DSC), Intersection over Union (IoU) | Tumor Segmentation: Accurate tumor boundary delineation; Anatomical Structure: IG for cardiac structure in CT scans; ViTs: High DSC scores in brain and stomach segmentation |
| Detection | Saliency maps, CAM, Attention, ViTs | Lesion detection, nodule localization | Mean Average Precision (mAP), Sensitivity, Specificity | Lesion Detection: Saliency maps for skin cancer detection; Nodule Localization: CAM for lung nodule detection in CT scans; ViTs: Improved lesion detection in various modalities |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhati, D.; Neha, F.; Amiruzzaman, M. A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging. J. Imaging 2024, 10, 239. https://doi.org/10.3390/jimaging10100239
Bhati D, Neha F, Amiruzzaman M. A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging. Journal of Imaging. 2024; 10(10):239. https://doi.org/10.3390/jimaging10100239
Chicago/Turabian StyleBhati, Deepshikha, Fnu Neha, and Md Amiruzzaman. 2024. "A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging" Journal of Imaging 10, no. 10: 239. https://doi.org/10.3390/jimaging10100239
APA StyleBhati, D., Neha, F., & Amiruzzaman, M. (2024). A Survey on Explainable Artificial Intelligence (XAI) Techniques for Visualizing Deep Learning Models in Medical Imaging. Journal of Imaging, 10(10), 239. https://doi.org/10.3390/jimaging10100239








