The Challenge of Single-Photon Emission Computed Tomography Image Segmentation in the Internal Dosimetry of 177Lu Molecular Therapies
Abstract
1. Introduction
2. Background Information
2.1. 177Lu Characteristics
2.2. 177Lu Therapy of Neuroendocrine Tumours
2.3. Lu177 -PSMA in the Treatment of Castration-Resistant Prostate Cancer
2.4. Radiobiological Consideration
2.5. Absorbed Dose Calculation
2.6. Calibration of the SPECT System
Image Acquisition
2.7. SPECT Image Degradation Factors
2.7.1. Photon Attenuation
2.7.2. Compton Scattering
2.7.3. Collimator–Detector Response (CDR)
2.8. Partial Volume Effect (PVE)
2.9. SPECT Image Reconstruction
3. Standard Segmentation Approaches
3.1. Evaluation Metrics and Validation
3.1.1. Gold standard—Manual Delineation by a Skilled Operator
3.1.2. Dice Similarity Coefficient
DSC Is Also Known as the F1 Score
3.1.3. Boundary-Based Measures
3.1.4. Statistical Methods—Spearman and Pearson Coefficients
4. Threshold-Based Methods
4.1. Phantoms Used in Assessments
4.2. Fixed Thresholding Methods
4.3. Adaptive and Automated Thresholding
5. Region Growing Approach
6. Boundary-Based Surface Adaptation—Parametrically Deformable Shape Models
7. Stochastic and Learning-Based Methods
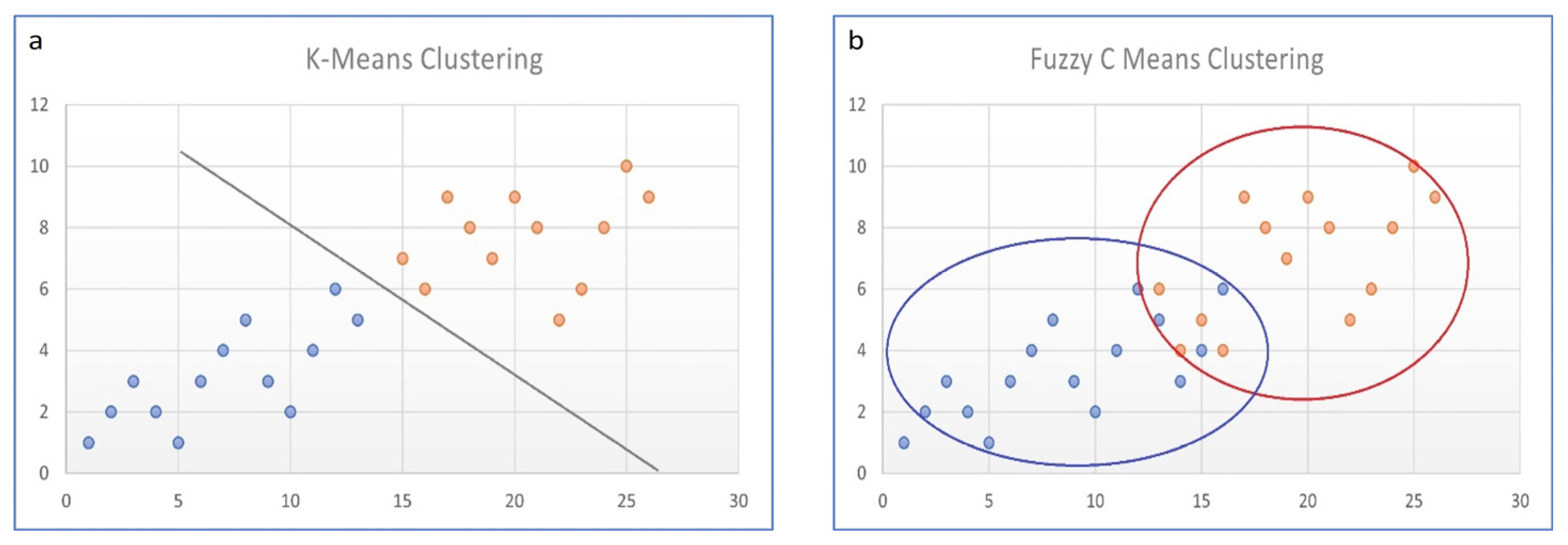
7.1. Fuzzy C-Means
7.2. AI Deep Learning Algorithms
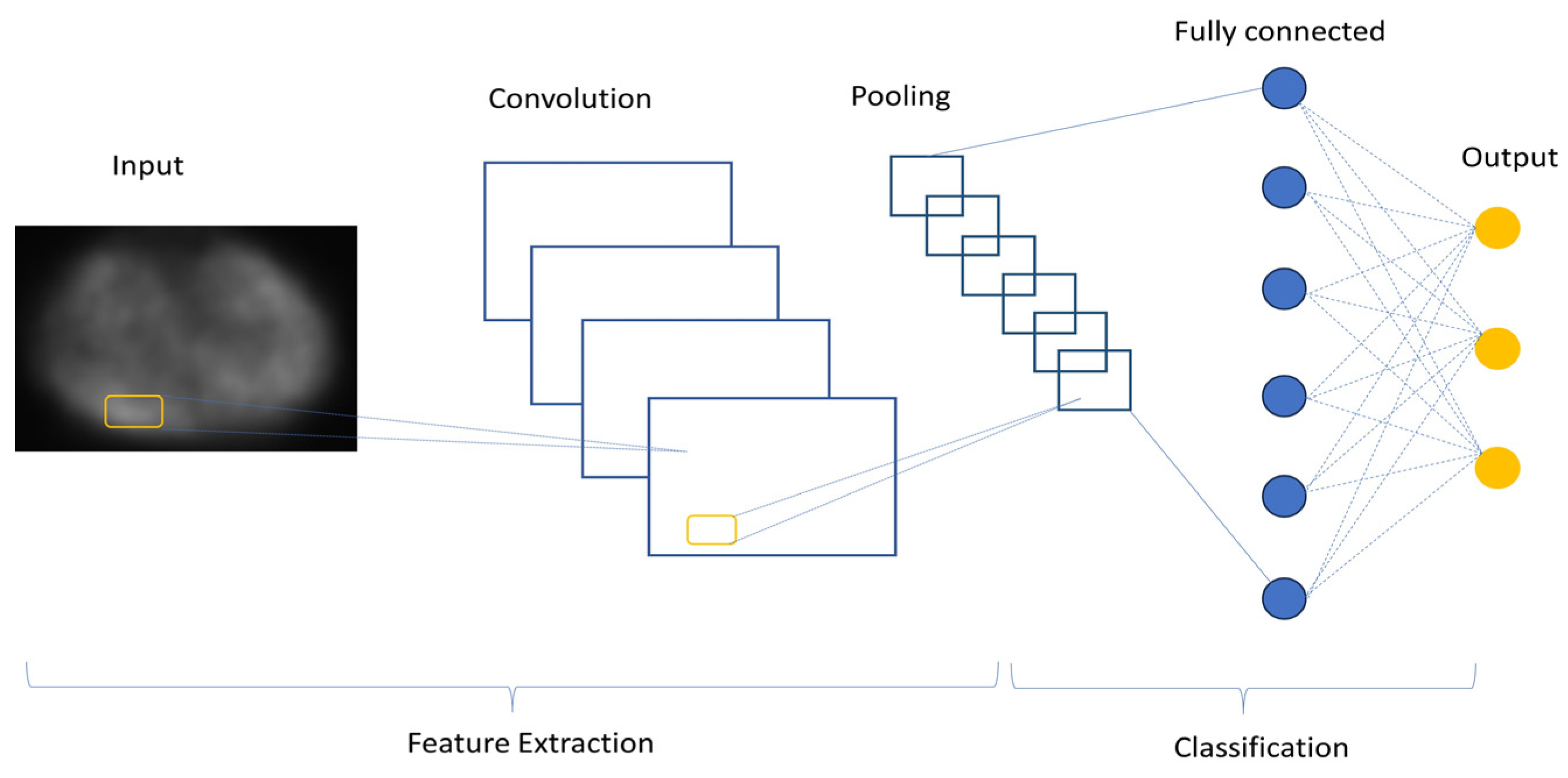
7.2.1. Convolutional Neural Networks (CNNs/ConvNet)
7.2.2. Generative Adversarial Networks (GANs)
8. Discussion
9. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dewaraja, Y.K.; Frey, E.C.; Sgouros, G.; Brill, A.B.; Roberson, P.; Zanzonico, P.B.; Ljungberg, M. MIRD Pamphlet No.23: Quantitative SPECT for Patient-Specific 3-Dimensional Dosimetry in Internal Radionuclide Therapy. J. Nucl. Med. 2012, 53, 1310–1325. [Google Scholar] [CrossRef] [PubMed]
- Baechler, S.; Hobbs, R.F.; Prideaux, A.R.; Wahl, R.L.; Sgouros, G. Extension of the biological effective dose to the MIRD schema and possible implications in radionuclide therapy dosimetry. Med. Phys. 2008, 35, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Sjögreen Gleisner, K.; Chouin, N.; Gabina, P.M.; Cicone, F.; Gnesin, S.; Stokke, C.; Konijnenberg, M.; Cremonesi, M.; Verburg, F.A.; Bernhardt, P.; et al. EANM dosimetry committee recommendations for dosimetry of 177Lu-labelled somatostatin-receptor- and PSMA-targeting ligands. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 1778–1809. [Google Scholar] [CrossRef]
- Banerjee, S.; Pillai, M.R.; Knapp, F.F. Lutetium-177 therapeutic radiopharmaceuticals: Linking chemistry, radiochemistry, and practical applications. Chem. Rev. 2015, 115, 2934–2974. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.J.; Hubbell, J.H.; Seltzer, S.M.; Chang, J.; Coursey, J.S.; Sukumar, R.; Zucker, D.S.; Olsen, K. XCOM: Photon Cross Section Database. NIST Standard Reference Database 8 (XGAM); National Institute of Standards and Technology: Gaithersburg, MD, USA, 2010. Available online: http://physics.nist.gov/xcom (accessed on 25 July 2023).
- Geenen, L.; Nonnekens, J.; Konijnenberg, M.; Baatout, S.; De Jong, M.; Aerts, A. Overcoming nephrotoxicity in peptide receptor radionuclide therapy using [177Lu]Lu-DOTA-TATE for the treatment of neuroendocrine tumours. Nucl. Med. Biol. 2021, 102–103, 1–11. [Google Scholar] [CrossRef]
- Oronsky, B.; Ma, P.C.; Morgensztern, D.; Carter, C.A. Nothing But NET: A Review of Neuroendocrine Tumors and Carcinomas. Neoplasia 2017, 19, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Scoville, S.D.; Cloyd, J.M.; Pawlik, T.M. New and emerging systemic therapy options for well-differentiated gastroenteropancreatic neuroendocrine tumors. Expert Opin. Pharmacother. 2020, 21, 183–191. [Google Scholar] [CrossRef]
- Strosberg, J.; El-Haddad, G.; Wolin, E.; Hendifar, A.; Yao, J.; Chasen, B.; Mittra, E.; Kunz, P.L.; Kulke, M.H.; Jacene, H.; et al. Phase 3 Trial of 177Lu-Dotatate for Midgut Neuroendocrine Tumors. N. Engl. J. Med. 2017, 376, 125–135. [Google Scholar] [CrossRef]
- EMA—Lutathera Product Information. Annex I Summary of Product Characteristics. Available online: https://www.ema.europa.eu/en/documents/product-information/lutathera-epar-product-information_en.pdf (accessed on 29 July 2023).
- Emmett, L.; Willowson, K.; Violet, J.; Shin, J.; Blanksby, A.; Lee, J. Lutetium 177PSMA radionuclide therapy for men with prostate cancer: A review of the current literature and discussion of practical aspects of therapy. J. Med. Radiat. Sci. 2017, 64, 52–60. [Google Scholar] [CrossRef]
- Kratochwil, C.; Fendler, W.P.; Eiber, M.; Hofman, M.S.; Emmett, L.; Calais, J.; Osborne, J.R.; Iravani, A.; Koo, P.; Lindenberg, L.; et al. Joint EANM/SNMMI procedure guideline for the use of 177Lu-labeled PSMA-targeted radioligand-therapy (177Lu-PSMA-RLT). Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 2830–2845. [Google Scholar] [CrossRef]
- Chang, S.S. Overview of prostate-specific membrane antigen. Rev. Urol. 2004, 6 (Suppl. S10), S13–S18. [Google Scholar] [PubMed]
- Bostwick, D.G.; Pacelli, A.; Blute, M.; Roche, P.; Murphy, G.P. Prostate specific membrane antigen expression in prostatic intraepithelial neoplasia and adenocarcinoma: A study of 184 cases. Cancer 1998, 82, 2256–2261. [Google Scholar] [CrossRef]
- Wright, G.L., Jr.; Grob, B.M.; Haley, C.; Grossman, K.; Newhall, K.; Petrylak, D.; Troyer, J.; Konchuba, A.; Schellhammer, P.F.; Moriarty, R. Upregulation of prostate-specific membrane antigen after androgen-deprivation therapy. Urology 1996, 48, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Pacilio, M.; Basile, C.; Shcherbinin, S.; Caselli, F.; Ventroni, G.; Aragno, D.; Mango, L.; Santini, E. An innovative iterative thresholding algorithm for tumour segmentation and volumetric quantification on SPECT images: Monte Carlo-based methodology and validation. Med. Phys. 2011, 38, 3050–3061. [Google Scholar] [CrossRef]
- Ross, J.S.; Sheehan, C.E.; Fisher, H.A.; Kaufman, R.P., Jr.; Kaur, P.; Gray, K.; Webb, I.; Gray, G.S.; Mosher, R.; Kallakury, B.V. Correlation of primary tumor prostate-specific membrane antigen expression with disease recurrence in prostate cancer. Clin. Cancer Res. 2003, 9, 6357–6362. [Google Scholar]
- Kozikowski, A.P.; Nan, F.; Conti, P.; Zhang, J.; Ramadan, E.; Bzdega, T.; Wroblewska, B.; Neale, J.H.; Pshenichkin, S.; Wroblewski, J.T. Design of remarkably simple, yet potent urea-based inhibitors of glutamate carboxypeptidase II (NAALADase). J. Med. Chem. 2001, 44, 298–301. [Google Scholar] [CrossRef]
- Eder, M.; Schäfer, M.; Bauder-Wüst, U.; Hull, W.E.; Wängler, C.; Mier, W.; Haberkorn, U.; Eisenhut, M. 68Ga-complex lipophilicity and the targeting property of a urea-based PSMA inhibitor for PET imaging. Bioconjug. Chem. 2012, 23, 688–697. [Google Scholar] [CrossRef]
- Benešová, M.; Schäfer, M.; Bauder-Wüst, U.; Afshar-Oromieh, A.; Kratochwil, C.; Mier, W.; Haberkorn, U.; Kopka, K.; Eder, M. Preclinical evaluation of a tailormade DOTA-conjugated PSMA inhibitor with optimized linker moiety for imaging and endoradiotherapy of PC. J. Nucl. Med. 2015, 56, 914–920. [Google Scholar] [CrossRef]
- Forrer, F.; Krenning, E.P.; Kooij, P.P.; Bernard, B.F.; Konijnenberg, M.; Bakker, W.H.; Teunissen, J.J.; de Jong, M.; van Lom, K.; de Herder, W.W.; et al. Bone marrow dosimetry in peptide receptor radionuclide therapy with [177Lu-DOTA0,Tyr3]octreotate. Eur. J. Nucl. Med. Mol. Imaging 2009, 36, 1138–1146. [Google Scholar] [CrossRef]
- Emami, B.; Lyman, J.; Brown, A.; Coia, L.; Goitein, M.; Munzenrider, J.E.; Shank, B.; Solin, L.J.; Wesson, M. Tolerance of normal tissue to therapeutic irradiation. Int. J. Radiat. Oncol. Biol. Phys. 1991, 21, 109–122. [Google Scholar] [CrossRef]
- Strigari, L.; Konijnenberg, M.; Chiesa, C.; Bardies, M.; Du, Y.; Gleisner, K.S.; Lassmann, M.; Flux, G. The evidence base for the use of internal dosimetry in the clinical practice of molecular radiotherapy. Eur. J. Nucl. Med. Mol. Imaging 2014, 41, 1976–1988. [Google Scholar] [CrossRef] [PubMed]
- Begum, N.J.; Thieme, A.; Eberhardt, N.; Tauber, R.; D’Alessandria, C.; Beer, A.J.; Glatting, G.; Eiber, M.; Kletting, P. The Effect of Total Tumor Volume on the Biologically Effective Dose to Tumor and Kidneys for 177Lu-Labeled PSMA Peptides. J. Nucl. Med. 2018, 59, 929–933. [Google Scholar] [CrossRef]
- Council of the European Union. Council Directive 2013/59/Euratom of 5 December 2013 laying down basic safety standards for protection against the dangers arising from exposure to ionising radiation, and repealing Directives 89/618/Euratom, 90/641/Euratom, 96/29/Euratom, 97/43/Euratom and 2003/122/Euratom. Off. J. Eur. Union 2014, 13, 1–73. [Google Scholar]
- Bolch, W.E.; Eckerman, K.F.; Sgouros, G.; Thomas, S.R. MIRD pamphlet No. 21: A generalized schema for radiopharmaceutical dosimetry--standardization of nomenclature. J. Nucl. Med. 2009, 50, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Gadd, R.; Baker, M.; Nijran, K.S.; Owens, S.; Thomas, W.; Woods, M.J.; Zananiri, F. Protocol for Establishing and Maintaining the Calibration of Medical Radionuclide Calibrators and Their Quality Control. Measurement Good Practice Guide. No. 93; NPL: Teddington, UK, 2006. [Google Scholar]
- Ljungberg, M.; Celler, A.; Konijnenberg, M.W.; Eckerman, K.F.; Dewaraja, Y.K.; Sjögreen Gleisner, K.; SNMMI MIRD Committee; Bolch, W.E.; Brill, A.B.; Fahey, F.; et al. MIRD Pamphlet No. 26: Joint EAMN/MIRD Guidelines for Quantitative 177Lu SPECT Applied for Dosimetry of Radiopharmaceutical Therapy. J. Nucl. Med. 2016, 57, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; El Fakhri, G. Quantitative Nuclear Medicine; Chapter 17; IAEA, Division of Human Health: Vienna, Austria, 2014; pp. 608–620. [Google Scholar]
- Ljungberg, M.; Strand, S.E. A Monte Carlo program for the simulation of scintillation camera characteristics. Comput. Methods Programs Biomed. 1989, 29, 257–272. [Google Scholar] [CrossRef]
- Zaidi, H.; Hasegawa, B.H. Overview of Nuclear Medical Imaging: Physics and Instrumentation. In Quantitative Analysis in Nuclear Medicine Imaging; Zaidi, H., Ed.; Springer: Boston, MA, USA, 2006; pp. 1–34. [Google Scholar] [CrossRef]
- Zaidi, H.; Koral, K.F. Scatter Correction Strategies in Emission Tomography. In Quantitative Analysis in Nuclear Medicine Imaging; Zaidi, H., Ed.; Springer: Boston, MA, USA, 2006; pp. 205–235. [Google Scholar]
- Zaidi, H.; Hasegawa, B.H. Attenuation Correction Strategies in Emission Tomography. In Quantitative Analysis in Nuclear Medicine Imaging; Zaidi, H., Ed.; Springer: Boston, MA, USA, 2006; pp. 167–204. [Google Scholar] [CrossRef]
- Ogawa, K.; Harata, Y.; Ichihara, T.; Kubo, A.; Hashimoto, S. A practical method for position-dependent Compton-scatter correction in single photon emission CT. IEEE Trans. Med. Imaging 1991, 10, 408–412. [Google Scholar] [CrossRef]
- Uribe, C.F.; Esquinas, P.L.; Tanguay, J.; Gonzalez, M.; Gaudin, E.; Beauregard, J.M.; Celler, A. Accuracy of 177Lu activity quantification in SPECT imaging: A phantom study. EJNMMI Phys. 2017, 4, 2. [Google Scholar] [CrossRef]
- Robinson, A.P.; Tipping, J.; Cullen, D.M.; Hamilton, D. The influence of triple energy window scatter correction on activity quantification for (1 7 7)Lu molecular radiotherapy. Phys. Med. Biol. 2016, 61, 5107–5127. [Google Scholar] [CrossRef]
- Karimi Ghodoosi, E.; D’Alessandria, C.; Li, Y.; Bartel, A.; Köhner, M.; Höllriegl, V.; Navab, N.; Eiber, M.; Li, W.B.; Frey, E.; et al. The effect of attenuation map, scatter energy window width, and volume of interest on the calibration factor calculation in quantitative 177Lu SPECT imaging: Simulation and phantom study. Phys. Med. 2018, 56, 74–80. [Google Scholar] [CrossRef]
- Tran-Gia, J.; Lassmann, M. Optimizing Image Quantification for 177Lu SPECT/CT Based on a 3D Printed 2-Compartment Kidney Phantom. J. Nucl. Med. 2018, 59, 616–624. [Google Scholar] [CrossRef] [PubMed]
- Frey, E.C.; Tsui, B.M.W. Collimator-Detector Response Compensation in SPECT. In Quantitative Analysis in Nuclear Medicine Imaging; Zaidi, H., Ed.; Springer: Boston, MA, USA, 2006; pp. 141–166. [Google Scholar] [CrossRef]
- Grings, A.; Jobic, C.; Kuwert, T.; Ritt, P. The magnitude of the partial volume effect in SPECT imaging of the kidney: A phantom study. EJNMMI Phys. 2022, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Mohy-ud-Din, H.; Boutagy, N.E.; Jiang, M.; Ren, S.; Stendahl, J.C.; Sinusas, A.J.; Liu, C. Fully automatic multi-atlas segmentation of CTA for partial volume correction in cardiac SPECT/CT. Phys. Med. Biol. 2017, 62, 3944. [Google Scholar] [CrossRef]
- Rundo, L.; Stefano, A.; Militello, C.; Russo, G.; Sabini, M.G.; D’Arrigo, C.; Marletta, F.; Ippolito, M.; Mauri, G.; Vitabile, S.; et al. A Fully Automatic Approach for Multimodal PET and MR Image Segmentation in Gamma Knife Treatment Planning. Comput. Methods Programs Biomed. 2017, 144, 77–96. [Google Scholar] [CrossRef]
- Foster, B.; Bagci, U.; Mansoor, A.; Xu, Z.; Mollura, D.J. A review on segmentation of positron emission tomography images. Comput. Biol. Med. 2014, 50, 76–96. [Google Scholar] [CrossRef] [PubMed]
- Warfield, S.K.; Zou, K.H.; Wells, W.M. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Trans. Med. Imaging 2004, 23, 903–921. [Google Scholar] [CrossRef] [PubMed]
- Moccia, S.; De Momi, E.; El Hadji, S.; Mattos, L.S. Blood vessel segmentation algorithms—Review of methods, datasets and evaluation metrics. Comput. Methods Programs Biomed. 2018, 158, 71–91. [Google Scholar] [CrossRef]
- Aydin, O.U.; Taha, A.A.; Hilbert, A.; Khalil, A.A.; Galinovic, I.; Fiebach, J.B.; Frey, D.; Madai, V.I. On the usage of average Hausdorff distance for segmentation performance assessment: Hidden error when used for ranking. Eur. Radiol. Exp. 2021, 5, 4, Erratum in Eur. Radiol. Exp. 2022, 6, 56. [Google Scholar] [CrossRef]
- Gear, J.I.; Cox, M.G.; Gustafsson, J.; Gleisner, K.S.; Murray, I.; Glatting, G.; Konijnenberg, M.; Flux, G.D. EANM practical guidance on uncertainty analysis for molecular radiotherapy absorbed dose calculations. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 2456–2474. [Google Scholar] [CrossRef]
- Gustafsson, J.; Sundlöv, A.; Sjögreen Gleisner, K. SPECT image segmentation for estimation of tumour volume and activity concentration in 177Lu-DOTATATE radionuclide therapy. EJNMMI Res. 2017, 7, 18. [Google Scholar] [CrossRef]
- Brolin, G.; Gustafsson, J.; Ljungberg, M.; Gleisner, K.S. Pharmacokinetic digital phantoms for accuracy assessment of image-based dosimetry in (177)Lu-DOTATATE peptide receptor radionuclide therapy. Phys. Med. Biol. 2015, 60, 6131–6149. [Google Scholar] [CrossRef] [PubMed]
- Materka, A.; Strumiłło, P. Wstęp do Komputerowej Analizy Obrazów; Politechnika Łódzka, Instytut Elektroniki: Łódź, Poland, 2009; pp. 108–133. [Google Scholar]
- Erdi, Y.E.; Wessels, B.W.; Loew, M.H.; Erdi, A.K. Threshold estimation in single photon emission computed tomography and planar imaging for clinical radioimmunotherapy. Cancer Res. 1995, 55 (Suppl. S23), 5823s–5826s. [Google Scholar]
- Brambilla, M.; Matheoud, R.; Secco, C.; Loi, G.; Krengli, M.; Inglese, E. Threshold segmentation for PET target volume delineation in radiation treatment planning: The role of target-to-background ratio and target size. Med. Phys. 2008, 35, 1207–1213. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.M.; Ocampo Ramos, J.C.; Kesner, A.L. Personalized dosimetry of177Lu-DOTATATE: A comparison of organ- and voxel-level approaches using open-access images. Biomed Phys Eng Express. 2021, 7, 057002. [Google Scholar] [CrossRef] [PubMed]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Kittler, J.; Illingworth, J. On threshold selection using clustering criteria. IEEE Trans. Syst. Man Cybern. 1985, SMC-15, 652–655. [Google Scholar] [CrossRef]
- Boudraa, A.O.; Zaidi, H. Image Segmentation Techniques in Nuclear Medicine Imaging. In Quantitative Analysis in Nuclear Medicine Imaging; Zaidi, H., Ed.; Springer: Boston, MA, USA, 2006; pp. 308–357. [Google Scholar] [CrossRef]
- Erdi, Y.E.; Mawlawi, O.; Larson, S.M.; Imbriaco, M.; Yeung, H.; Finn, R.; Humm, J.L. Segmentation of lung lesion volume by adaptive positron emission tomography image thresholding. Cancer 1997, 80 (Suppl. S12), 2505–2509. [Google Scholar] [CrossRef]
- Daisne, J.F.; Sibomana, M.; Bol, A.; Doumont, T.; Lonneux, M.; Grégoire, V. Tri-dimensional automatic segmentation of PET volumes based on measured source-to-background ratios: Influence of reconstruction algorithms. Radiother. Oncol. 2003, 69, 247–250. [Google Scholar] [CrossRef]
- Jentzen, W.; Freudenberg, L.; Eising, E.G.; Heinze, M.; Brandau, W.; Bockisch, A. Segmentation of PET volumes by iterative image thresholding. J. Nucl. Med. 2007, 48, 108–114. [Google Scholar]
- Grimes, J.; Celler, A.; Shcherbinin, S.; Piwowarska-Bilska, H.; Birkenfeld, B. The accuracy and reproducibility of SPECT target volumes and activities estimated using an iterative adaptive thresholding technique. Nucl. Med. Commun. 2012, 33, 1254–1266, Erratum in Nucl. Med. Commun. 2014, 35, 988. [Google Scholar] [CrossRef]
- Li, H.; Thorstad, W.L.; Biehl, K.J.; Laforest, R.; Su, Y.; Shoghi, K.I.; Donnelly, E.D.; Low, D.A.; Lu, W. A novel PET tumor delineation method based on adaptive region-growing and dual-front active contours. Med. Phys. 2008, 35, 3711–3721, Erratum in Med. Phys. 2008, 35, 5958. [Google Scholar] [CrossRef] [PubMed]
- Staib, L.H.; Duncan, J.S. Model-based deformable surface finding for medical images. IEEE Trans. Med. Imaging 1996, 15, 720–731. [Google Scholar] [CrossRef]
- Floreby, L.; Sornmo, L.; Sjogreen, K. Boundary finding using Fourier surfaces of increasing order [simulated medical images]. In Proceedings of the Fourteenth International Conference on Pattern Recognition (Cat. No.98EX170), Brisbane, QLD, Australia, 20 August 1998; Volume 1, pp. 465–467. [Google Scholar] [CrossRef]
- Floreby, L.; Sjogreen, K.; Sornmo, L.; Ljungberg, M. Deformable Fourier surfaces for volume segmentation in SPECT. In Proceedings of the Fourteenth International Conference on Pattern Recognition, (Cat. No.98EX170), Brisbane, QLD, Australia, 20 August 1998; Volume 1, pp. 358–360. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Luna, L.P.; Chung, H.W.; Rowe, S.P.; Du, Y.; Solnes, L.B.; Frey, E.C. Learning fuzzy clustering for SPECT/CT segmentation via convolutional neural networks. Med. Phys. 2021, 48, 3860–3877. [Google Scholar] [CrossRef]
- Belhassen, S.; Zaidi, H. A novel fuzzy C-means algorithm for unsupervised heterogeneous tumor quantification in PET. Med. Phys. 2010, 37, 1309–1324. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.L. Spatial Models for Fuzzy Clustering. Comput. Vis. Image Underst. 2001, 84, 285–297. [Google Scholar] [CrossRef]
- Bezdek, J.C.; Ehrlich, R.; Full, W. FCM: The fuzzy c-means clustering algorithm. Comput. Geosci. 1984, 10, 191–203. [Google Scholar] [CrossRef]
- Pham, D.L.; Jerry, L.; Prince, J.L. An adaptive fuzzy C-means algorithm for image segmentation in the presence of intensity inhomogeneities. Pattern Recognit. Lett. 1999, 20, 57–68. [Google Scholar] [CrossRef]
- Arabi, H.; AkhavanAllaf, A.; Sanaat, A.; Shiri, I.; Zaidi, H. The promise of artificial intelligence and deep learning in PET and SPECT imaging. Phys. Med. 2021, 83, 122–137. [Google Scholar] [CrossRef]
- Ramon, A.J.; Yang, Y.; Pretorius, P.H.; Johnson, K.L.; King, M.A.; Wernick, M.N. Improving Diagnostic Accuracy in Low-Dose SPECT Myocardial Perfusion Imaging with Convolutional Denoising Networks. IEEE Trans Med. Imaging 2020, 39, 2893–2903. [Google Scholar] [CrossRef]
- Dewaraja, Y.K.; Mirando, D.M.; Peterson, A.B.; Niedbala, J.; Millet, J.D.; Mikell, J.K.; Frey, K.A.; Wong, K.K.; Wilderman, S.J.; Nelson, A.S. A Pipeline for Automated Voxel Dosimetry: Application in Patients with Multi-SPECT/CT Imaging after 177Lu-Peptide Receptor Radionuclide Therapy. J. Nucl. Med. 2022, 63, 1665–1672. [Google Scholar] [CrossRef]
- Beauregard, J.M.; Hofman, M.S.; Pereira, J.M.; Eu, P.; Hicks, R.J. Quantitative (177)Lu SPECT (QSPECT) imaging using a commercially available SPECT/CT system. Cancer Imaging 2011, 11, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Hatt, M.; Lee, J.A.; Schmidtlein, C.R.; Naqa, I.E.; Caldwell, C.; De Bernardi, E.; Lu, W.; Das, S.; Geets, X.; Gregoire, V.; et al. Classification and evaluation strategies of auto-segmentation approaches for PET: Report of AAPM task group No. 211. Med. Phys. 2017, 44, e1–e42. [Google Scholar] [CrossRef] [PubMed]

| Lp | Reference | Radionuclide | Methods | Validation Phantom/Data Sets | Results |
|---|---|---|---|---|---|
| 1. | Dewaraja et al. [72] | 177Lu | Lesions: manual segmentation Organs: on CT with higher mAs, at a reference point, CNN algorithms | 20 patient data sets (77 lesion) | CNN defined kidneys with manual adjustment to manual segmentation— DSC (0.91–0.94) |
| 2. | Pacillo et al. [16] | 99mTc | Lesions: RIThM—upgraded ITM with RC (ITM as a comparator) | Jaszczak phantom + test objects Test images (hot spheres, Zubal head phantom simulated with SIMIND code) 3 brain metastasis + 2 gliomas | Accuracy level: 10% (for vol 20–110 mL); Difference between estimated and true volume: 15% (vol > 4 mL) for ITM and RIThM |
| 3. | Gustafsson et al. [48] | 177Lu | Lesions: Fixed threshold 42%—FT Otsu Method—OM Fourier Surfaces—FS | XCAT phantoms, MC simulated SPECT images in different time points | Volume and activity concentration root-mean-square error < 15% for tumours > 10 mL for OM and FS (FT worse); FS robust to noise, better for 366 h p.i. time point |
| 4. | Chen et al. [65] | 99mTc | Bone metastasis: DL, CNN with FCM and novel loss functions | Clinical datasets Simulated SPECT/CT images XCAT | DSC 0.75 and 0.74 for lesions and bone segmentation on SPECT images, respectively; DSC of 0.79 bone segmentation on CT scans |
| 5. | Grimes et al. [60] | 99mTc | Organs: (kidneys, liver, spleen, thyroid) and lesions: Iterative adaptive thresholding (ThV and ThA) Semiautomatic background selection | Phantoms with 20 different inserts 13 patient studies | Object volume and activity estimation separately |
| 6. | Uribe et al. [35] | 177Lu | FT of 40%; CT-based segmentation; Iterative Adaptive Dual Thresholding (IADT) | Phantoms—hot objects in warm water | Volumes > 34 mL—10% error (percent difference between experimental and true activities) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gawel, J.; Rogulski, Z. The Challenge of Single-Photon Emission Computed Tomography Image Segmentation in the Internal Dosimetry of 177Lu Molecular Therapies. J. Imaging 2024, 10, 27. https://doi.org/10.3390/jimaging10010027
Gawel J, Rogulski Z. The Challenge of Single-Photon Emission Computed Tomography Image Segmentation in the Internal Dosimetry of 177Lu Molecular Therapies. Journal of Imaging. 2024; 10(1):27. https://doi.org/10.3390/jimaging10010027
Chicago/Turabian StyleGawel, Joanna, and Zbigniew Rogulski. 2024. "The Challenge of Single-Photon Emission Computed Tomography Image Segmentation in the Internal Dosimetry of 177Lu Molecular Therapies" Journal of Imaging 10, no. 1: 27. https://doi.org/10.3390/jimaging10010027
APA StyleGawel, J., & Rogulski, Z. (2024). The Challenge of Single-Photon Emission Computed Tomography Image Segmentation in the Internal Dosimetry of 177Lu Molecular Therapies. Journal of Imaging, 10(1), 27. https://doi.org/10.3390/jimaging10010027






