Improved 19F{1H} Saturation Transfer Difference Experiments for Sensitive Detection to Fluorinated Compound Bound to Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Instrumentation and Chemicals
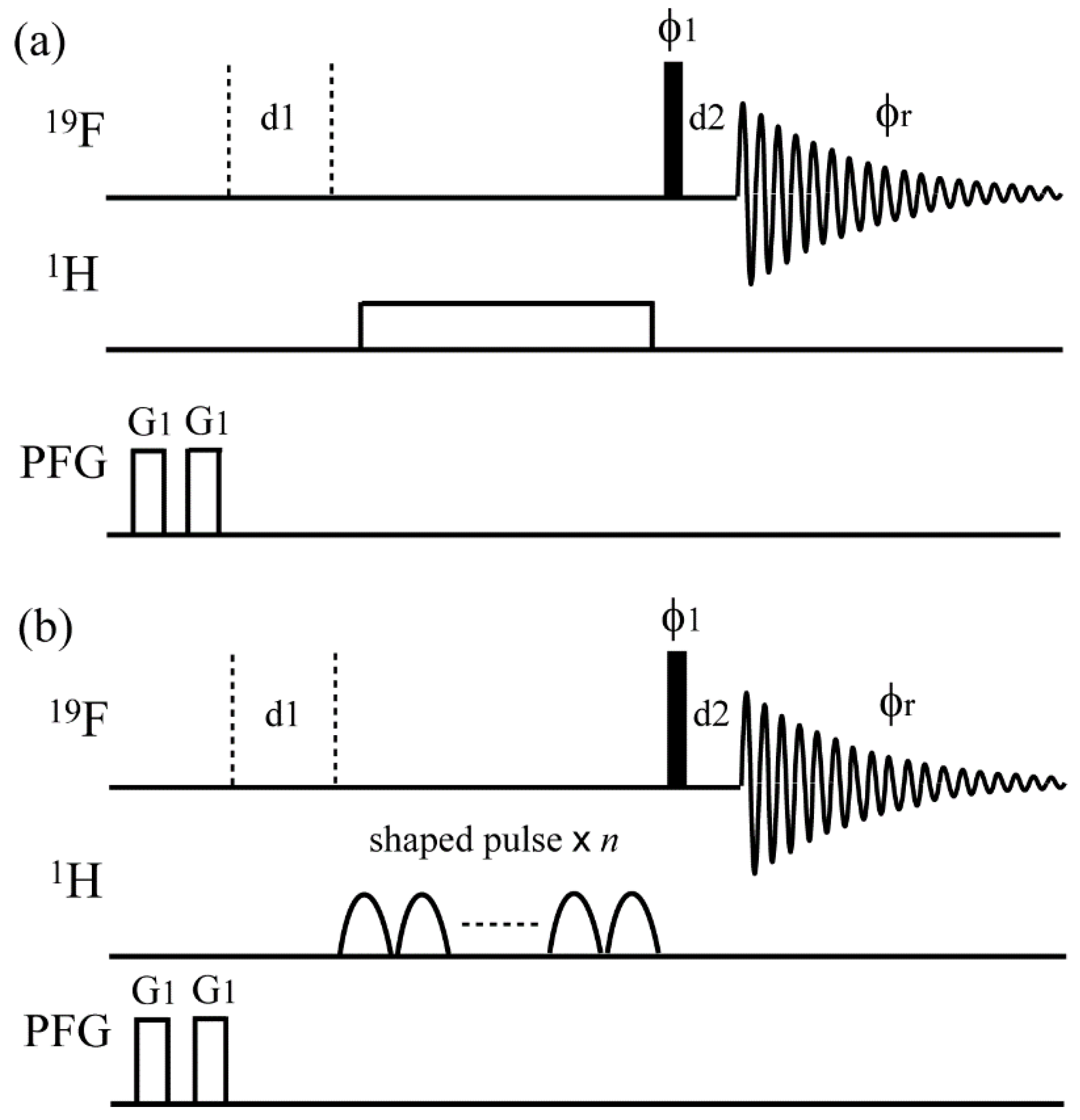
2.2. NMR Spectroscopy
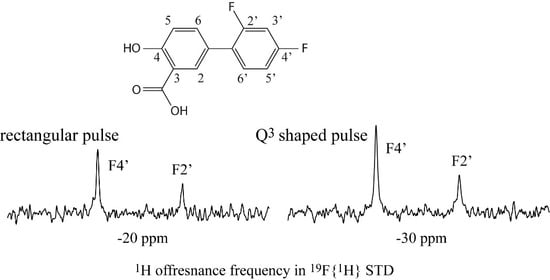
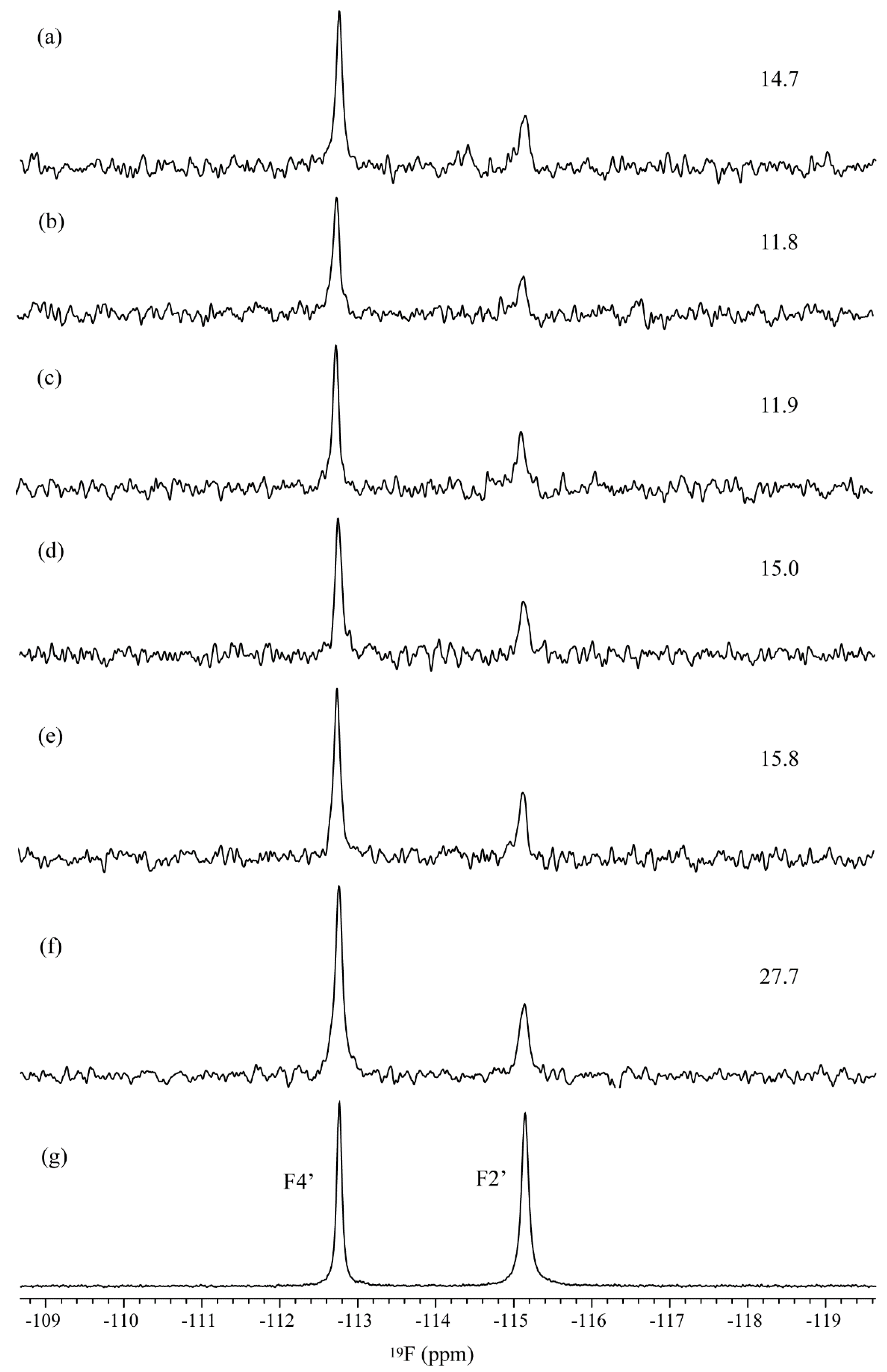
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, A.; Shapiro, M.J. NOE Pumping: A novel NMR technique for identification of compounds with binding affinity to macromolecules. J. Am. Chem. Soc. 1998, 120, 10258–10259. [Google Scholar] [CrossRef]
- Maye, M.; Meyer, B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar]
- Dalvit, C.; Pevarello, P.; Tatò, M.; Veronesi, M.; Vulpetti, A.; Sundström, M. Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J. Biomol. NMR 2000, 18, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Fogliatto, G.P.; Stewart, A.; Veronesi, M.; Stockman, B.J. WaterLOGSY as a method for primary NMR screening: Practical aspects and range of applicability. J. Biomol. NMR 2001, 21, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Shapiro, M.J. NOE Pumping. 2. A high-throughput method to determine compounds with binding affinity to macromolecules by NMR. J. Am. Chem. Soc. 2000, 122, 414–415. [Google Scholar] [CrossRef]
- Dalvit, C.; Fagerness, P.E.; Hadden, S.T.A.; Sarver, R.W.; Stockman, B.J. Fluorine-NMR experiments for high-throughput screening: Theoretical aspects, practical considerations, and range of applicability. J. Am. Chem. Soc. 2003, 125, 7696–7703. [Google Scholar] [CrossRef] [PubMed]
- Dalvit, C.; Flocco, M.; Stockman, B.J.; Veronesi, M. Competition binding experiments for rapidly ranking lead molecules for their binding affinity to human serum albumin. Comb. Chem. High Throughput Screen. 2002, 5, 645–650. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, C.; Kurita, J.; Furihata, K.; Tashiro, M. Achievement of 1H-19F heteronuclear experiments using the conventional spectrometer with a shared single high band amplifier. Magn. Reson. Chem. 2015, 53, 327–329. [Google Scholar] [CrossRef] [PubMed]
- Diercks, T.; Ribeiro, J.P.; Canada, F.J.; Andre, S.; Jimenez-Barbero, J.; Gabius, H. Fluorinated Carbohydrates as Lectin Ligands: Versatile Sensors in 19F-Detected Saturation Transfer Difference NMR Spectroscopy. Chem. Eur. J. 2009, 15, 5666–5668. [Google Scholar] [CrossRef] [PubMed]
- Park, B.K.; Kitteringham, N.R.; O’Neill, P.M. Metabolism of fluorine-containing drugs. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 443–470. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, D.; Harper, D.B. Fluorine-containing natural products. J. Fluorine Chem. 1999, 100, 127–133. [Google Scholar] [CrossRef]
- Furihata, K.; Usui, M.; Tashiro, M. Application of NMR screening methods with 19F detection to fluorinated compounds bound to proteins. Magnetochemistry 2018, 4, 3. [Google Scholar] [CrossRef]
- Geen, H.; Freeman, R. Band-selective radiofrequency pulses. J. Magn. Reson. 1991, 93, 93–141. [Google Scholar] [CrossRef]
- Emsley, L.; Bodenhausen, G. Optimization of shaped selective pulses for NMR using a quaternion description of their overall propagators. J. Magn. Reson. 1992, 97, 135–148. [Google Scholar] [CrossRef]
- Emsley, L.; Bodenhausen, G. Gaussian pulse cascades: New analytical functions for rectangular selective inversion and in-phase excitation in NMR. Chem. Phys. Lett. 1990, 165, 469–476. [Google Scholar] [CrossRef]
- Bauer, C.; Freeman, R.; Frenkiel, T.; Keeler, J.; Shaka, A.J. Gaussian pulses. J. Magn. Reson. 1984, 58, 442–457. [Google Scholar]
- Mayer, M.; James, T.L. NMR-based characterization of phenothiazines as a RNA binding scaffold. J. Am. Chem. Soc. 2004, 126, 4453–4460. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furihata, K.; Tashiro, M. Improved 19F{1H} Saturation Transfer Difference Experiments for Sensitive Detection to Fluorinated Compound Bound to Proteins. Magnetochemistry 2019, 5, 1. https://doi.org/10.3390/magnetochemistry5010001
Furihata K, Tashiro M. Improved 19F{1H} Saturation Transfer Difference Experiments for Sensitive Detection to Fluorinated Compound Bound to Proteins. Magnetochemistry. 2019; 5(1):1. https://doi.org/10.3390/magnetochemistry5010001
Chicago/Turabian StyleFurihata, Kazuo, and Mitsuru Tashiro. 2019. "Improved 19F{1H} Saturation Transfer Difference Experiments for Sensitive Detection to Fluorinated Compound Bound to Proteins" Magnetochemistry 5, no. 1: 1. https://doi.org/10.3390/magnetochemistry5010001
APA StyleFurihata, K., & Tashiro, M. (2019). Improved 19F{1H} Saturation Transfer Difference Experiments for Sensitive Detection to Fluorinated Compound Bound to Proteins. Magnetochemistry, 5(1), 1. https://doi.org/10.3390/magnetochemistry5010001