Comparative Transcriptome Analysis of High and Low Thujone-Producing Artemisia argyi Reveals Candidate Genes for Thujone Synthetic and Regulatory Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Headspace Solid-Phase Microextraction Gas Chromatography-Mass Spectrometry (HS-SPME-GC-MS)
2.3. RNA-Seq Analysis
3. Results
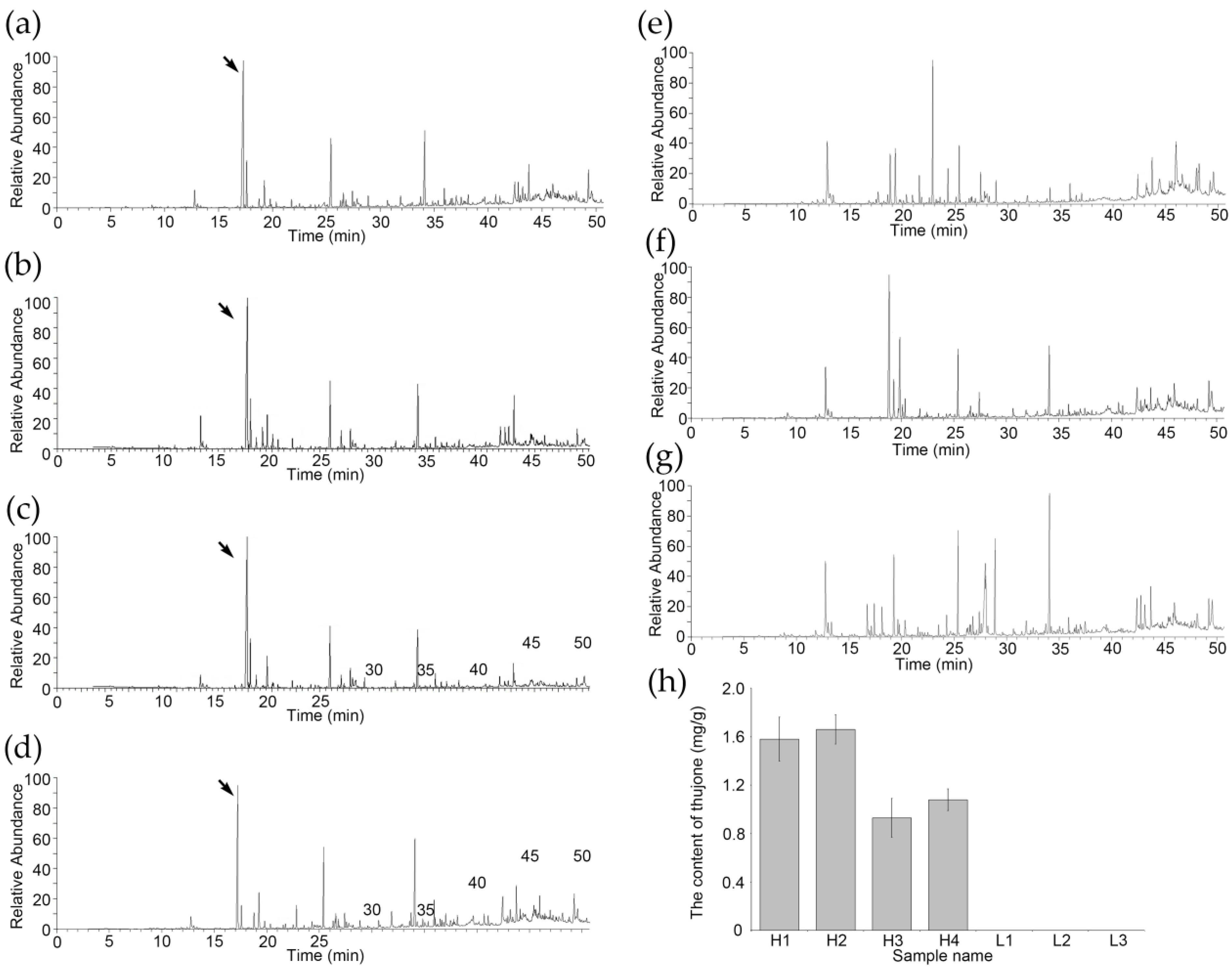
3.1. Thujone Content Has Rich Variation in A. argyi Varieties
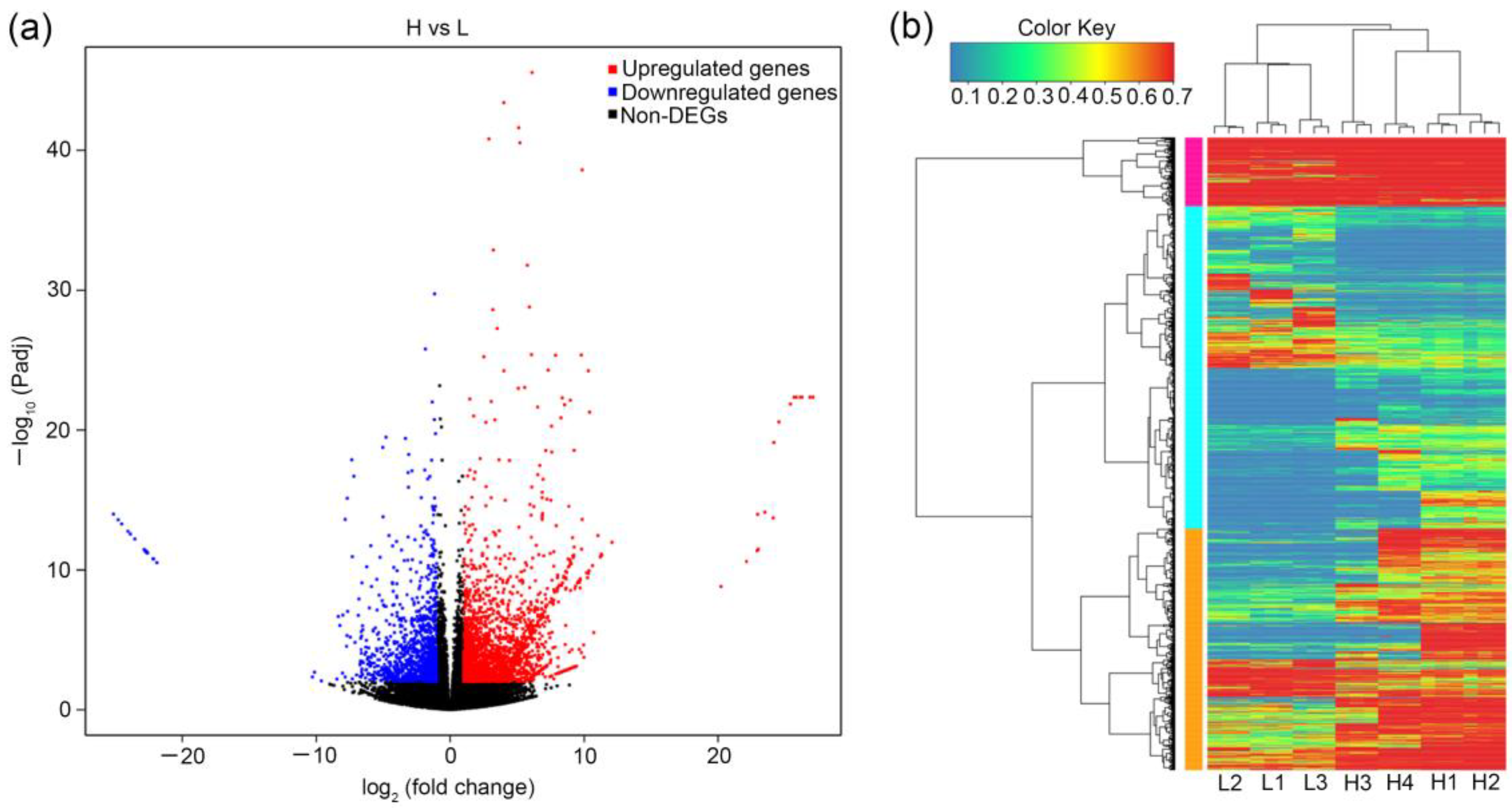
3.2. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Classification and Enrichment Analysis of Differentially Expressed Genes (DEGs) between Two Groups of Thujone Content High and Low Thujone-Producing Materials
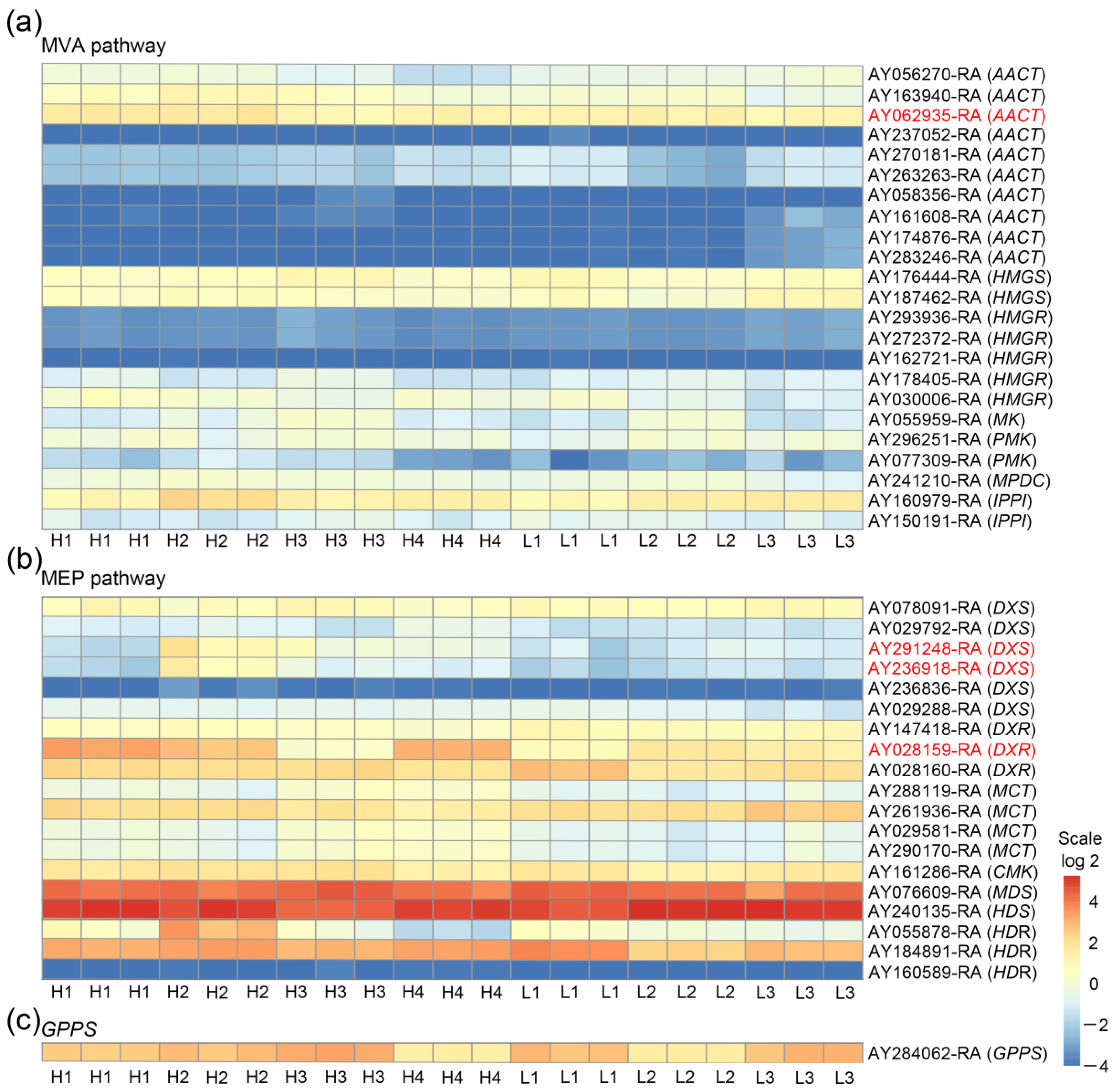
3.3. AACT, DXS, and DXR Are Three Main DEG Families of MVA and MEP Pathways in High and Low Thujone-Producing A. argyi Varieties with Different Thujone Content
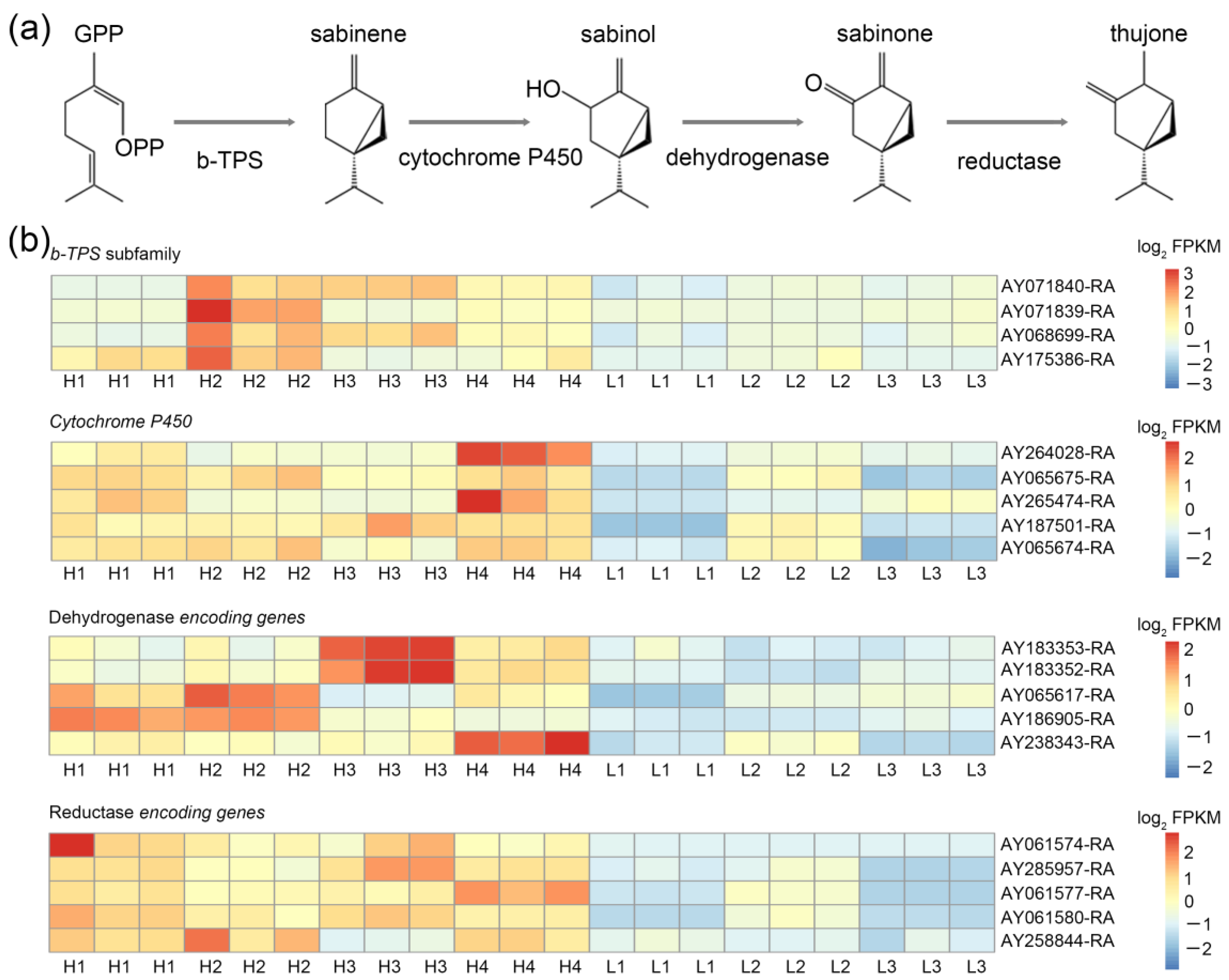
3.4. Identification of Candidate TPS, Cytochrome P450, Dehydrogenase, and Reductase Encoding Genes Involved in the Thujone Synthetic Pathway
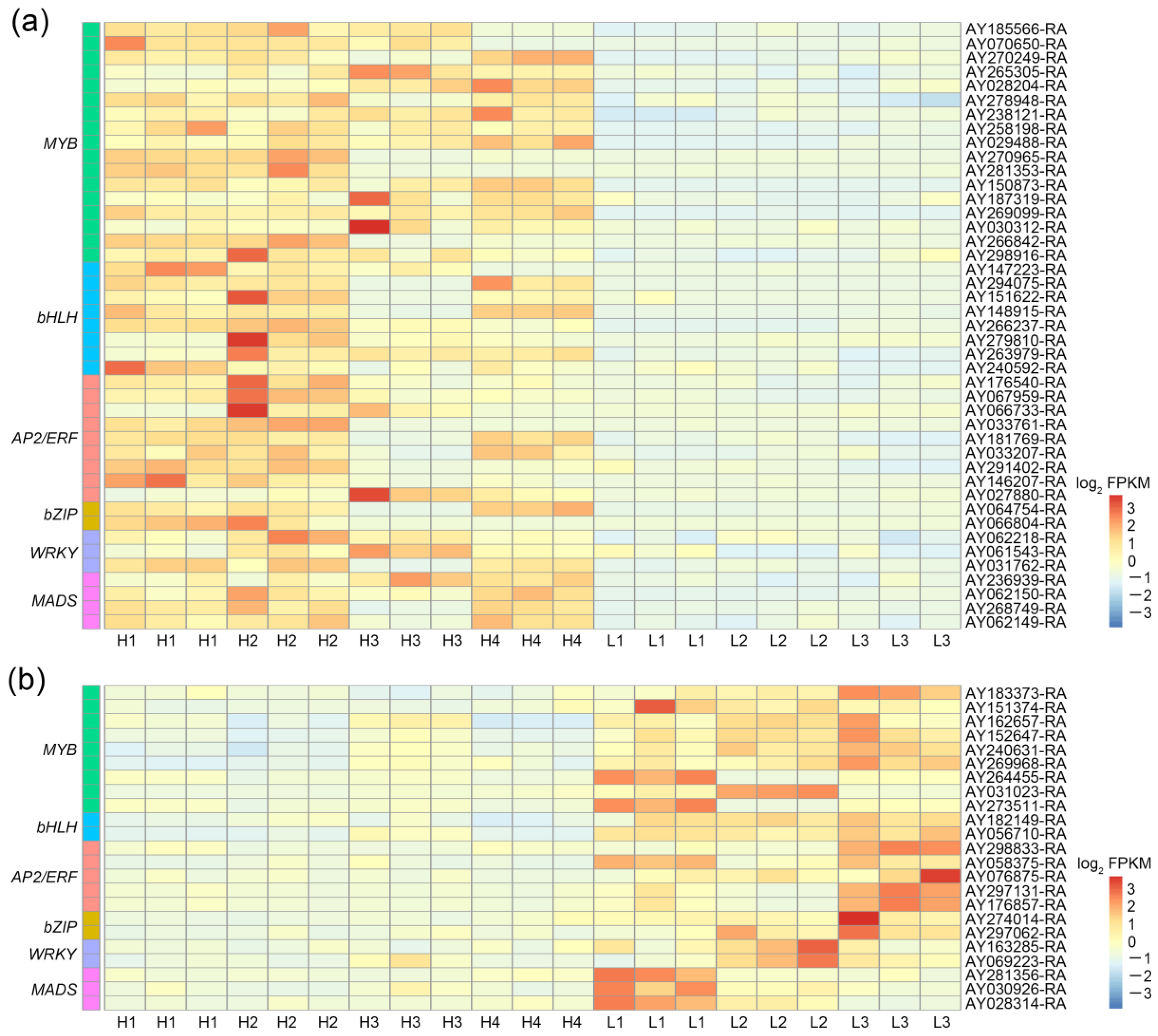
3.5. Identification of Positive and Negative Candidate Transcription Factors Related to Thujone Accumulation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, C.J.; Luo, D.D.; Miao, Y.H.; Kang, L.P.; Guo, L.P.; Liu, D.H.; Huang, L.Q. Analysis and evaluation of volatile oil content in leaves of different Artemisia argyi germplasm resources. Chung-Kuo Chung Yao Tsa Chih. 2021, 46, 3814–3823. [Google Scholar]
- Chen, C.J.; Luo, D.D.; Miao, Y.H.; Guo, L.P.; Liu, D.H. Diversity of Artemisia argyi germplasm resources based on agronomic and leaf phenotypic traits. Chung-Kuo Chung Yao Tsa Chih. 2021, 46, 2773–2782. [Google Scholar]
- Liang, H.; Lu, J.Q.; Dai, Y.; Li, X.S.; Guo, S.N. Comparative analysis of volatile components of Artemisiae argyi folium from different origins using HS-SPME-GC-MS combined with chemometrics analysis. Chin. J. Exp. Tradit. Med. Formulae 2014, 20, 85–90. [Google Scholar]
- Chen, L.L.; Zhang, H.J.; Chao, J.; Liu, J.F. Essential oil of Artemisia argyi suppresses inflammatory responses by inhibiting JAK/STATs activation. J. Ethnopharmacol. 2017, 204, 107–117. [Google Scholar]
- Hu, Q.; Liu, Z.; Guo, Y.; Lu, S.; Du, H.; Cao, Y. Antioxidant capacity of flavonoids from Folium Artemisiae Argyi and the molecular mechanism in Caenorhabditis elegans. J. Ethnopharmacol. 2021, 279, 114398. [Google Scholar]
- Xia, J.X.; Zhao, B.B.; Zan, J.F.; Wang, P.; Chen, L.L. Simultaneous determination of phenolic acids and flavonoids in Artemisiae Argyi Folium by HPLC-MS/MS and discovery of antioxidant ingredients based on relevance analysis. J. Pharm. Biomed. 2019, 175, 112734. [Google Scholar]
- Kim, K.O.; Lee, D.; Hiep, N.T.; Song, J.H.; Lee, H.J.; Kang, K.S. Protective effect of phenolic compounds isolated from mugwort (Artemisia argyi) against contrast-Induced apoptosis in kidney epithelium cell line LLC-PK1. Molecules 2019, 24, 195. [Google Scholar]
- Ge, Y.B.; Wang, Z.G.; Xiong, Y.; Huang, X.J.; Mei, Z.N.; Hong, Z.G. Anti-inflammatory and blood stasis activities of essential oil extracted from Artemisia argyi leaf in animals. J. Nat. Med. 2016, 70, 531–538. [Google Scholar]
- Guan, X.; Ge, D.; Li, S.; Huang, K.; Liu, J.; Li, F. Chemical composition and antimicrobial activities of Artemisia argyi Levl. et Vant essential oils extracted by simultaneous distillation-extraction, subcritical extraction and hydrodistillation. Molecules 2019, 24, 483. [Google Scholar]
- Liu, Y.; He, Y.; Wang, F.; Xu, R.; Yang, M.; Ci, Z.; Wu, Z.; Zhang, D.; Lin, J. From longevity grass to contemporary soft gold: Explore the chemical constituents, pharmacology, and toxicology of Artemisia argyi H. Lev. & vaniot essential oil. J. Ethnopharmacol. 2021, 279, 114404. [Google Scholar]
- Siveen, K.S.; Kuttan, G. Thujone inhibits lung metastasis induced by B16F-10 melanoma cells in C57BL/6 mice. Can. J. Physiol. Pharm. 2011, 89, 691–703. [Google Scholar]
- Alkhateeb, H.; Bonen, A. Thujone, a component of medicinal herbs, rescues palmitate-induced insulin resistance in skeletal muscle. Am. J. Physiol. Regul. 2010, 299, R804–R812. [Google Scholar]
- Nikolic, B.; Mitic-Culafic, D.; Vukovic-Gacic, B.; Knezevic-Vukcevic, J. Modulation of genotoxicity and DNA repair by plant monoterpenes camphor, eucalyptol and thujone in Escherichia coli and mammalian cells. Food Chem. Toxicol. 2011, 49, 2035–2045. [Google Scholar]
- Pu, X.; Dong, X.; Li, Q.; Chen, Z.; Liu, L. An update on the function and regulation of methylerythritol phosphate and mevalonate pathways and their evolutionary dynamics. J. Integr. Plant Biol. 2021, 63, 1211–1226. [Google Scholar]
- Vranova, E.; Coman, D.; Gruissem, W. Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 2013, 64, 665–700. [Google Scholar]
- Miziorko, H.M. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 2011, 505, 131–143. [Google Scholar]
- Simkin, A.J.; Guirimand, G.; Papon, N.; Courdavault, V.; Thabet, I.; Ginis, O.; Bouzid, S.; Giglioli-Guivarc’h, N.; Clastre, M. Peroxisomal localisation of the final steps of the mevalonic acid pathway in planta. Planta 2011, 234, 903–914. [Google Scholar]
- Miao, Y.H.; Luo, D.D.; Zhao, T.T.; Du, H.Z.; Liu, Z.H.; Xu, Z.P.; Guo, L.P.; Chen, C.J.; Peng, S.N.; Li, J.X.; et al. Genome sequencing reveals chromosome fusion and extensive expansion of genes related to secondary metabolism in Artemisia argyi. Plant Biotechnol. J. 2022, 20, 1902–1915. [Google Scholar]
- Frank, A.; Groll, M. The methylerythritol phosphate pathway to isoprenoids. Chem. Rev. 2017, 117, 5675–5703. [Google Scholar]
- Zhao, L.; Chang, W.C.; Xiao, Y.; Liu, H.W.; Liu, P. Methylerythritol phosphate pathway of isoprenoid biosynthesis. Annu. Rev. Bio Chem. 2013, 82, 497–530. [Google Scholar]
- Tholl, D.; Lee, S. Terpene Specialized Metabolism in Arabidopsis thaliana. Arab. Book 2011, 9, e0143. [Google Scholar]
- Karp, F.; Harris, J.L.; Croteau, R. Metabolism of monoterpenes: Demonstration of the hydroxylation of (+)-sabinene to (+)-cis-sabinol by an enzyme preparation from sage (Salvia officinalis) leaves. Arch. Biochem. Biophys. 1987, 256, 179–193. [Google Scholar]
- Dehal, S.S.; Croteau, R. Metabolism of monoterpenes: Specificity of the dehydrogenases responsible for the biosynthesis of camphor, 3-thujone, and 3-isothujone. Arch. Biochem. Biophys. 1987, 258, 287–291. [Google Scholar]
- Foster, A.J.; Hall, D.E.; Mortimer, L.; Abercromby, S.; Gries, R.; Gries, G.; Bohlmann, J.; Russell, J.; Mattsson, J. Identification of genes in Thuja plicata foliar terpenoid defenses. Plant Physiol. 2013, 161, 1993–2004. [Google Scholar]
- Liu, M.; Zhu, J.; Wu, S.; Wang, C.; Guo, X.; Wu, J.; Zhou, M. De novo assembly and analysis of the Artemisia argyi transcriptome and identification of genes involved in terpenoid biosynthesis. Sci. Rep. 2018, 8, 5824. [Google Scholar]
- Xu, R.; Ming, Y.; Li, Y.; Li, S.; Zhu, W.; Wang, H.; Guo, J.; Shi, Z.; Shu, S.; Xiong, C.; et al. Full-Length transcriptomic sequencing and temporal transcriptome expression profiling analyses offer insights into terpenoid biosynthesis in Artemisia argyi. Molecules 2022, 27, 5948. [Google Scholar]
- Zhang, K.; Wang, N.; Gao, X.; Ma, Q. Integrated metabolite profiling and transcriptome analysis reveals tissue-specific regulation of terpenoid biosynthesis in Artemisia argyi. Genomics 2022, 114, 110388. [Google Scholar]
- Yi, X.; Wang, X.; Wu, L.; Wang, M.; Yang, L.; Liu, X.; Chen, S.; Shi, Y. Integrated analysis of basic helix loop helix transcription factor family and targeted terpenoids reveals candidate AarbHLH genes involved in terpenoid biosynthesis in Artemisia argyi. Front. Plant Sci. 2022, 12, 811166. [Google Scholar]
- Cui, Y.; Gao, X.; Wang, J.; Shang, Z.; Zhang, Z.; Zhou, Z.; Zhang, K. Full-length transcriptome analysis reveals candidate genes involved in terpenoid biosynthesis in Artemisia argyi. Front. Genet. 2021, 12, 659962. [Google Scholar]
- Liu, Y.F.; Wang, B.; Shu, S.H.; Li, Z.; Song, C.; Liu, D.; Niu, Y.; Liu, J.; Zhang, J.; Liu, H.; et al. Analysis of the Coptis chinensis genome reveals the diversification of protoberberine-type alkaloids. Nat. Commun. 2021, 12, 3276. [Google Scholar]
- He, S.; Yang, L.; Ye, S.; Lin, Y.; Li, X.; Wang, Y.; Chen, G.; Liu, G.; Zhao, M.; Zhao, X.; et al. MPOD: Applications of integrated multi-omics database for medicinal plants. Plant Biotechnol. Rep. 2022, 20, 797–799. [Google Scholar]
- Su, P.; Guan, H.; Zhao, Y.; Tong, Y.; Xu, M.; Zhang, Y.; Hu, T.; Yang, J.; Cheng, Q.; Gao, L.; et al. Identification and functional characterization of diterpene synthases for triptolide biosynthesis from Tripterygium wilfordii. Plant J. 2018, 93, 50–65. [Google Scholar]
- Wang, R.; Ren, C.; Dong, S.; Chen, C.; Xian, B.; Wu, Q.; Wang, J.; Pei, J.; Chen, J. Integrated metabolomics and transcriptome analysis of flavonoid biosynthesis in Safflower (Carthamus tinctorius L.) with different colors. Front. Plant Sci. 2021, 12, 712038. [Google Scholar]
- Wang, M.; Chen, L.; Liang, Z.; He, X.; Liu, W.; Jiang, B.; Yan, J.; Sun, P.; Cao, Z.; Peng, Q.; et al. Metabolome and transcriptome analyses reveal chlorophyll and anthocyanin metabolism pathway associated with cucumber fruit skin color. BMC Plant Biol. 2020, 20, 386. [Google Scholar]
- Shoji, T.; Yuan, L. ERF gene clusters: Working together to regulate metabolism. Trends Plant Sci. 2021, 26, 23–32. [Google Scholar]
- Wang, M.; Qiu, X.; Pan, X.; Li, C. Transcriptional factor-mediated regulation of active component biosynthesis in medicinal plants. Curr. Pharm. Biotechnol. 2021, 22, 848–866. [Google Scholar]
- Okada, K. The biosynthesis of isoprenoids and the mechanisms regulating it in plants. Biosci. Biotechnol. Biochem. 2011, 75, 1219–1225. [Google Scholar]
- Patra, B.; Schluttenhofer, C.; Wu, Y.; Pattanaik, S.; Yuan, L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta 2013, 1829, 1236–1247. [Google Scholar]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar]
- Ma, D.; Pu, G.; Lei, C.; Ma, L.; Wang, H.; Guo, Y.; Chen, J.; Du, Z.; Li, G.; Ye, H.; et al. Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol. 2009, 50, 2146–2161. [Google Scholar]
- Yu, Z.X.; Li, J.X.; Yang, C.Q.; Hu, W.L.; Wang, L.J.; Chen, X.Y. The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 2012, 5, 353–365. [Google Scholar]
- Chuang, Y.C.; Hung, Y.C.; Tsai, W.C.; Chen, W.H.; Chen, H.H. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 2018, 69, 4363–4377. [Google Scholar]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar]
- Siberil, Y.; Benhamron, S.; Memelink, J.; Giglioli-Guivarc’h, N.; Thiersault, M.; Boisson, B.; Doireau, P.; Gantet, P. Catharanthus roseus G-box binding factors 1 and 2 act as repressors of strictosidine synthase gene expression in cell cultures. Plant Mol. Biol. 2001, 45, 477–488. [Google Scholar]
- Reddy, V.A.; Wang, Q.; Dhar, N.; Kumar, N.; Venkatesh, P.N.; Rajan, C.; Panicker, D.; Sridhar, V.; Mao, H.Z.; Sarojam, R. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 2017, 15, 1105–1119. [Google Scholar]
- Pichersky, E.; Raguso, R.A. Why do plants produce so many terpenoid compounds? New Phytol. 2018, 220, 692–702. [Google Scholar]
- Block, A.K.; Vaughan, M.M.; Schmelz, E.A.; Christensen, S.A. Biosynthesis and function of terpenoid defense compounds in maize (Zea mays). Planta 2019, 249, 21–30. [Google Scholar]
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. 2015, 148, 63–106. [Google Scholar]
- Zulak, K.G.; Bohlmann, J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J. Integr. Plant Biol. 2010, 52, 86–97. [Google Scholar]
- Zhong, J.; Guo, Y.H.; Shi, H.J.; Liang, Y.L.; Guo, Z.Y.; Li, D.W.; Wang, C.; Li, H.; Zhang, Q.X.; Sun, M. Volatiles mediated an eco-friendly aphid control strategy of Chrysanthemum genus. Ind. Crops Prod. 2022, 180, 114734. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, T.; Chen, C.; Li, J.; Luo, D.; Miao, Y.; Gui, C.; Liu, Q.; Liu, D. Comparative Transcriptome Analysis of High and Low Thujone-Producing Artemisia argyi Reveals Candidate Genes for Thujone Synthetic and Regulatory Pathway. Horticulturae 2023, 9, 232. https://doi.org/10.3390/horticulturae9020232
Zhao T, Chen C, Li J, Luo D, Miao Y, Gui C, Liu Q, Liu D. Comparative Transcriptome Analysis of High and Low Thujone-Producing Artemisia argyi Reveals Candidate Genes for Thujone Synthetic and Regulatory Pathway. Horticulturae. 2023; 9(2):232. https://doi.org/10.3390/horticulturae9020232
Chicago/Turabian StyleZhao, Tingting, Changjie Chen, Jinxin Li, Dandan Luo, Yuhuan Miao, Chun Gui, Qi Liu, and Dahui Liu. 2023. "Comparative Transcriptome Analysis of High and Low Thujone-Producing Artemisia argyi Reveals Candidate Genes for Thujone Synthetic and Regulatory Pathway" Horticulturae 9, no. 2: 232. https://doi.org/10.3390/horticulturae9020232
APA StyleZhao, T., Chen, C., Li, J., Luo, D., Miao, Y., Gui, C., Liu, Q., & Liu, D. (2023). Comparative Transcriptome Analysis of High and Low Thujone-Producing Artemisia argyi Reveals Candidate Genes for Thujone Synthetic and Regulatory Pathway. Horticulturae, 9(2), 232. https://doi.org/10.3390/horticulturae9020232




