Characterizations of MYB Transcription Factors in Camellia oleifera Reveal the Key Regulators Involved in Oil Biosynthesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Analysis of MYB Transcription Factors
2.2. Sequence Features Analysis of CoR2R3-MYBs
2.3. Chromosomal Location and Synteny Analysis
2.4. Phylogenetic Analysis
2.5. Analysis of CoR2R3-MYBs Expression Pattern at Five Different Stages of Seed Development
2.6. Weighted Correlation Network Analysis (WGCNA) and Screening of Hub Genes
3. Results
3.1. Identification of MYB Transcription Factors in C. oleifera
3.2. Protein Sequences Analysis of CoR2R3-MYBs
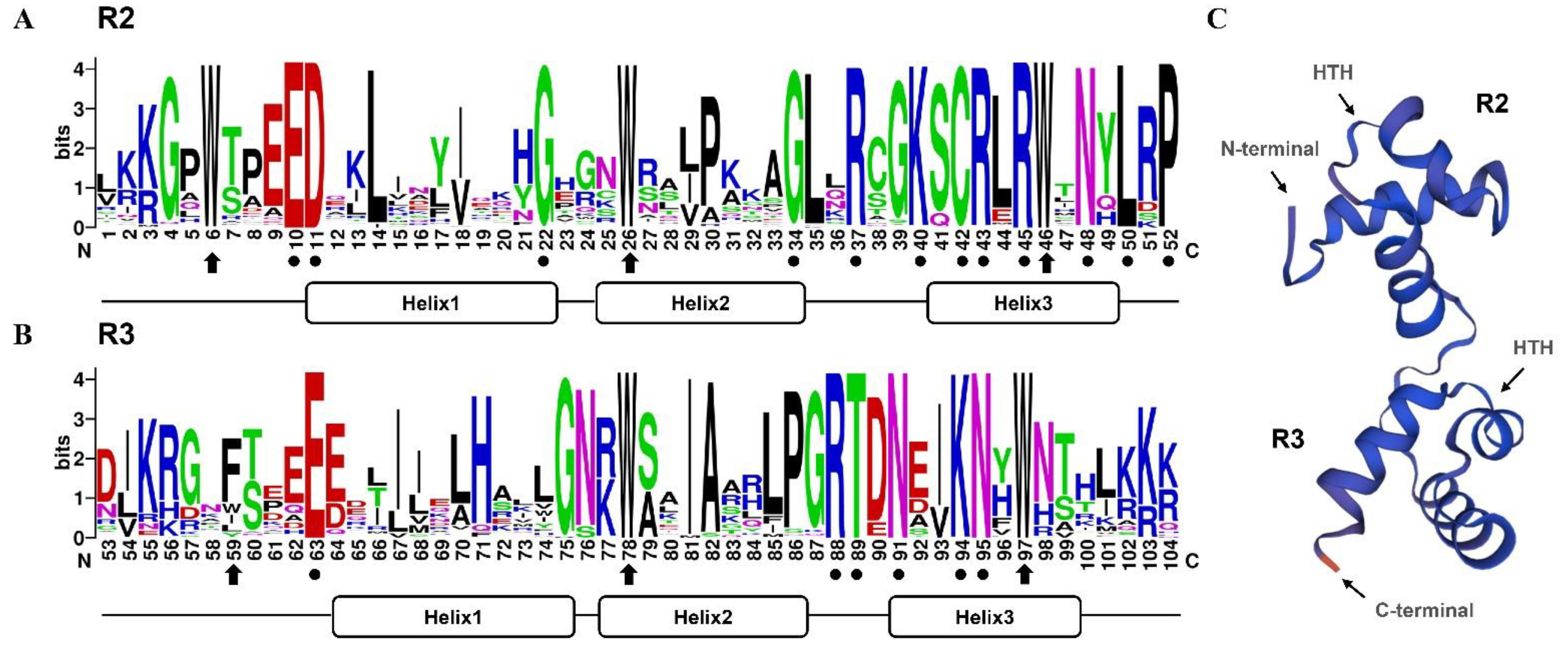
3.3. Sequence Conservation of the R2R3 Domain
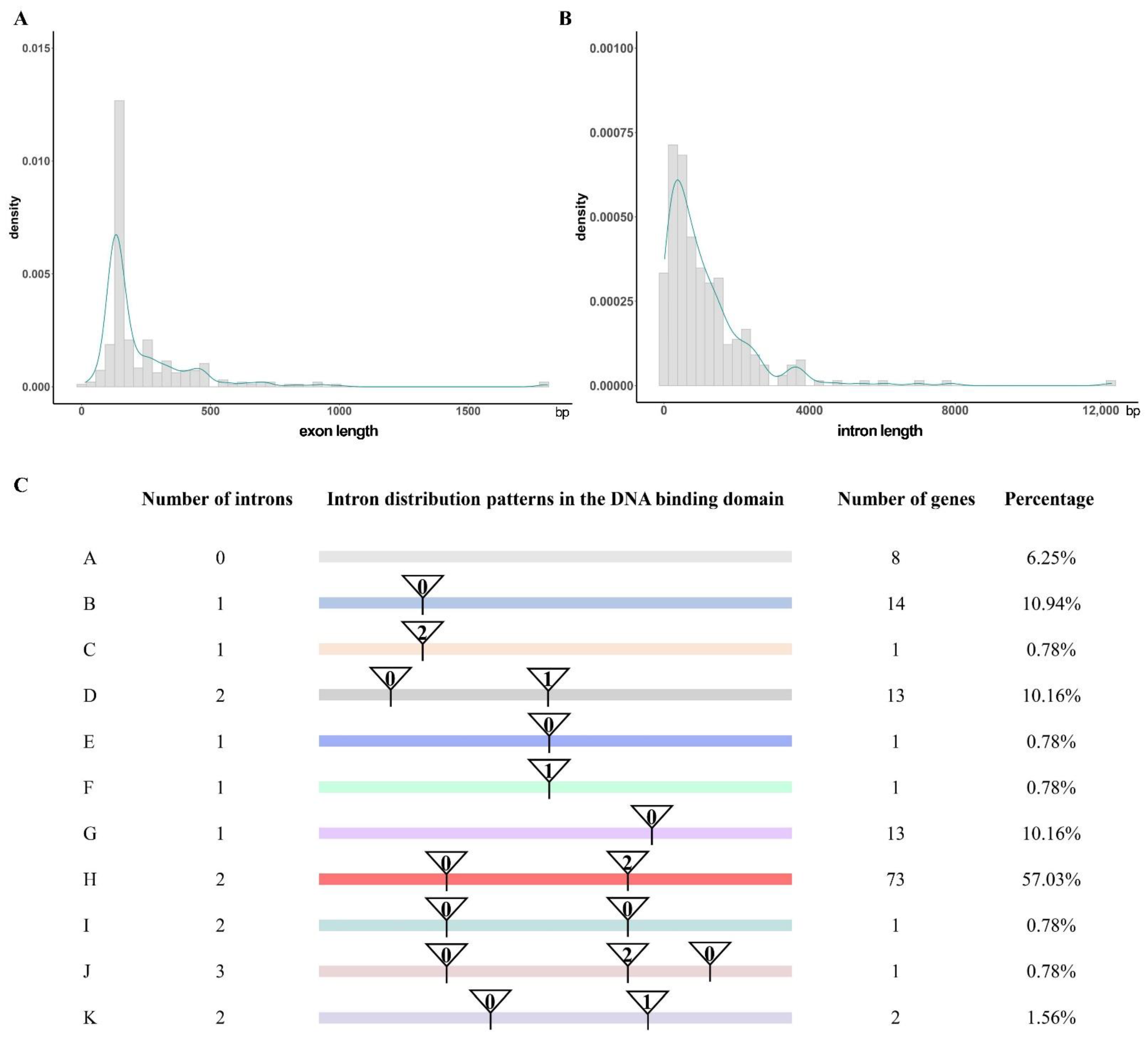
3.4. Gene Structure Analysis of CoR2R3-MYBs
3.5. Promoter Cis-Acting Elements Analysis of CoR2R3-MYBs
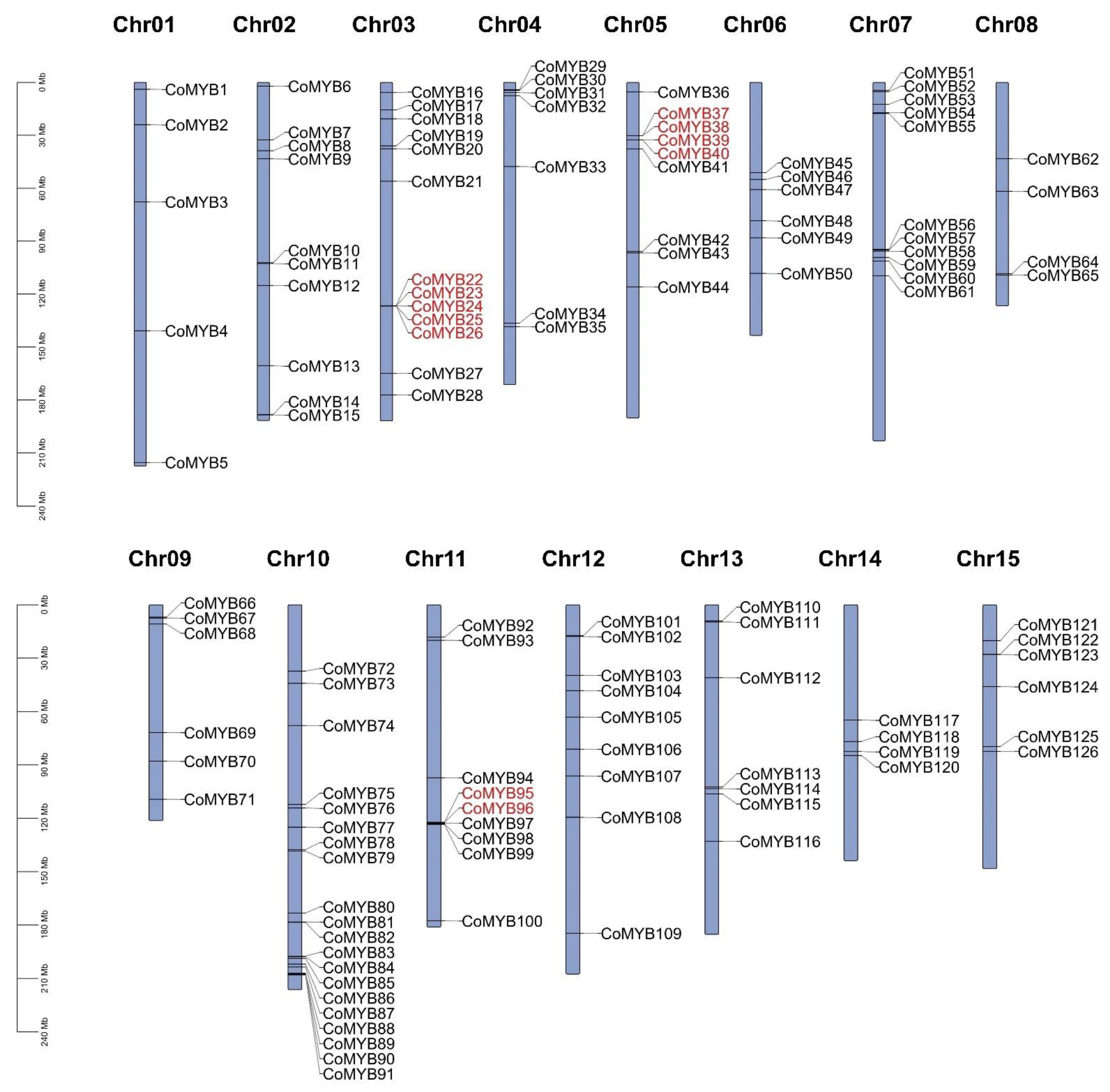
3.6. Chromosomal Location and Synteny Analysis
3.7. Phylogenetic Analysis
3.8. Expression Analysis of CoR2R3-MYBs Genes during Seed Development
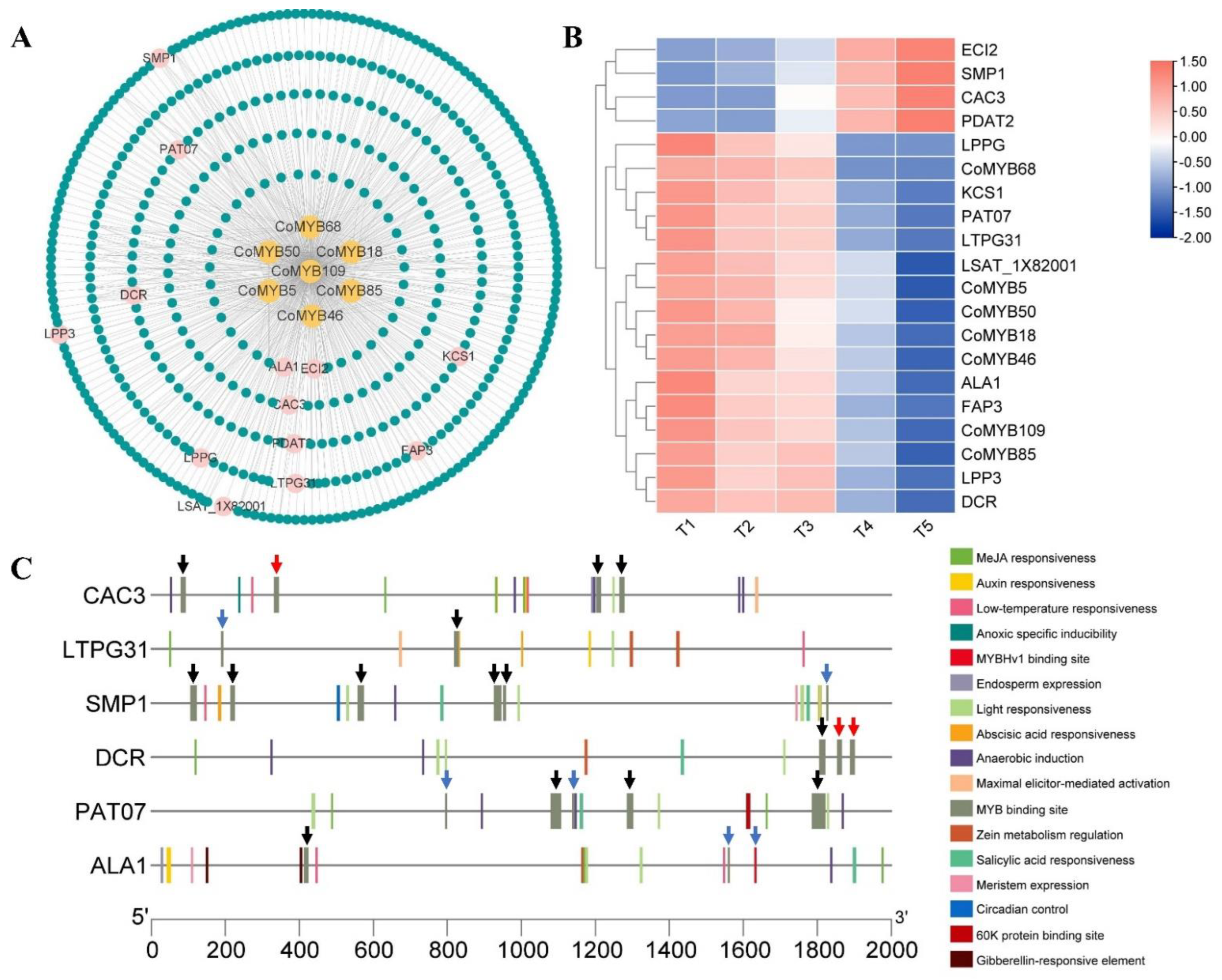
3.9. Gene Co-Expression Network Underlying the Seed Development
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CDD | Conserved domain database |
| GO | Gene ontology |
| MeJA | Methyl jasmonate |
| MEME | Multiple em for motif elicitation |
| Pfam | Database of protein families |
| PlantCARE | Plant cis-acting regulatory elements database |
| SMART | Simple modular architecture research tool |
References
- Hong, J.C. General Aspects of Plant Transcription Factor Families. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2016; pp. 35–56. [Google Scholar]
- Rombauts, S.; Florquin, K.; Lescot, M.; Marchal, K.; Rouze, P.; van de Peer, Y. Computational approaches to identify promoters and cis-regulatory elements in plant genomes. Plant Physiol. 2003, 132, 1162–1176. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xue, C.; Li, J.; Qiao, X.; Li, L.; Yu, L.; Huang, Y.; Wu, J. Genome-Wide Identification, Evolution and Functional Divergence of MYB Transcription Factors in Chinese White Pear (Pyrus bretschneideri). Plant Cell Physiol. 2016, 57, 824–847. [Google Scholar] [CrossRef] [PubMed]
- Kranz, H.D.; Denekamp, M.; Greco, R.; Jin, H.; Leyva, A.; Meissner, R.C.; Petroni, K.; Urzainqui, A.; Bevan, M.; Martin, C.; et al. Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 1998, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef]
- Yang, X.; Li, J.; Guo, T.; Guo, B.; Chen, Z.; An, X. Comprehensive analysis of the R2R3-MYB transcription factor gene family in Populus trichocarpa. Ind. Crops Prod. 2021, 168, 113614. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Haga, N.; Kato, K.; Murase, M.; Araki, S.; Kubo, M.; Demura, T.; Suzuki, K.; Muller, I.; Voss, U.; Jurgens, G.; et al. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development 2007, 134, 1101–1110. [Google Scholar] [CrossRef]
- Dubos, C.; Le Gourrierec, J.; Baudry, A.; Huep, G.; Lanet, E.; Debeaujon, I.; Routaboul, J.M.; Alboresi, A.; Weisshaar, B.; Lepiniec, L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 2008, 55, 940–953. [Google Scholar] [CrossRef]
- Hosoda, K.; Imamura, A.; Katoh, E.; Hatta, T.; Tachiki, M.; Yamada, H.; Mizuno, T.; Yamazaki, T. Molecular structure of the GARP family of plant Myb-related DNA binding motifs of the Arabidopsis response regulators. Plant Cell 2002, 14, 2015–2029. [Google Scholar] [CrossRef]
- Matsui, K.; Umemura, Y.; Ohme-Takagi, M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant J. 2008, 55, 954–967. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Jin, C.; Duan, S.; Zhu, Y.; Qi, S.; Liu, K.; Gao, C.; Ma, H.; Zhang, M.; Liao, Y.; et al. MYB89 Transcription Factor Represses Seed Oil Accumulation. Plant Physiol. 2017, 173, 1211–1225. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.H. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Reichelt, M.; Yoshida, K.; Gershenzon, J.; Constabel, C.P. Two R2R3-MYB proteins are broad repressors of flavonoid and phenylpropanoid metabolism in poplar. Plant J. 2018, 96, 949–965. [Google Scholar] [CrossRef]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef]
- Gu, Z.; Zhu, J.; Hao, Q.; Yuan, Y.W.; Duan, Y.W.; Men, S.; Wang, Q.; Hou, Q.; Liu, Z.A.; Shu, Q.; et al. A Novel R2R3-MYB Transcription Factor Contributes to Petal Blotch Formation by Regulating Organ-Specific Expression of PsCHS in Tree Peony (Paeonia suffruticosa). Plant Cell Physiol. 2019, 60, 599–611. [Google Scholar] [CrossRef]
- Schaart, J.G.; Dubos, C.; Romero De La Fuente, I.; van Houwelingen, A.; de Vos, R.C.H.; Jonker, H.H.; Xu, W.; Routaboul, J.M.; Lepiniec, L.; Bovy, A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria x ananassa) fruits. New Phytol. 2013, 197, 454–467. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Raffaele, S.; Vailleau, F.; Leger, A.; Joubes, J.; Miersch, O.; Huard, C.; Blee, E.; Mongrand, S.; Domergue, F.; Roby, D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 2008, 20, 752–767. [Google Scholar] [CrossRef]
- To, A.; Joubes, J.; Thueux, J.; Kazaz, S.; Lepiniec, L.; Baud, S. AtMYB92 enhances fatty acid synthesis and suberin deposition in leaves of Nicotiana benthamiana. Plant J. 2020, 103, 660–676. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Jin, C.; Li, D.; Gao, C.; Qi, S.; Liu, K.; Hai, J.; Ma, H.; Chen, M. MYB76 Inhibits Seed Fatty Acid Accumulation in Arabidopsis. Front. Plant Sci. 2017, 8, 226. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Z.; Zhu, Y.; Li, Z.; Hussain, N.; Xuan, L.; Guo, W.; Zhang, G.; Jiang, L. The effect of transparent TESTA2 on seed fatty acid biosynthesis and tolerance to environmental stresses during young seedling establishment in Arabidopsis. Plant Physiol. 2012, 160, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Barthole, G.; To, A.; Marchive, C.; Brunaud, V.; Soubigou-Taconnat, L.; Berger, N.; Dubreucq, B.; Lepiniec, L.; Baud, S. MYB118 represses endosperm maturation in seeds of Arabidopsis. Plant Cell 2014, 26, 3519–3537. [Google Scholar] [CrossRef] [PubMed]
- Luan, F.; Zeng, J.; Yang, Y.; He, X.; Wang, B.; Gao, Y.; Zeng, N. Recent advances in Camellia oleifera Abel: A review of nutritional constituents, biofunctional properties, and potential industrial applications. J. Funct. Foods 2020, 75, 104242. [Google Scholar] [CrossRef]
- Zhao, Y.; Su, R.; Zhang, W.; Yao, G.-L.; Chen, J. Antibacterial activity of tea saponin from Camellia oleifera shell by novel extraction method. Ind. Crops Prod. 2020, 153, 112604. [Google Scholar] [CrossRef]
- Lin, P.; Wang, K.; Zhou, C.; Xie, Y.; Yao, X.; Yin, H. Seed Transcriptomics Analysis in Camellia oleifera Uncovers Genes Associated with Oil Content and Fatty Acid Composition. Int. J. Mol. Sci. 2018, 19, 118. [Google Scholar] [CrossRef]
- Gong, W.; Song, Q.; Ji, K.; Gong, S.; Wang, L.; Chen, L.; Zhang, J.; Yuan, D. Full-Length Transcriptome from Camellia oleifera Seed Provides Insight into the Transcript Variants Involved in Oil Biosynthesis. J. Agric. Food Chem. 2020, 68, 14670–14683. [Google Scholar] [CrossRef]
- Su, M.H.; Shih, M.C.; Lin, K.H. Chemical composition of seed oils in native Taiwanese Camellia species. Food Chem. 2014, 156, 369–373. [Google Scholar] [CrossRef]
- Lin, P.; Wang, K.; Wang, Y.; Hu, Z.; Yan, C.; Huang, H.; Ma, X.; Cao, Y.; Long, W.; Liu, W.; et al. The genome of oil-Camellia and population genomics analysis provide insights into seed oil domestication. Genome Biol. 2022, 23, 14. [Google Scholar] [CrossRef]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Gómez-Rubio, V. ggplot2—Elegant Graphics for Data Analysis (2nd Edition). J. Stat. Softw. 2017, 77, 1–3. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Li, K.B. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 2003, 19, 1585–1586. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Boeckmann, B.; Bairoch, A.; Apweiler, R.; Blatter, M.C.; Estreicher, A.; Gasteiger, E.; Martin, M.J.; Michoud, K.; O’Donovan, C.; Phan, I.; et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003, 31, 365–370. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Litwack, G. Nucleic Acids and Molecular Genetics. In Human Biochemistry; Academic Press: Cambridge, MA, USA, 2018; pp. 257–317. [Google Scholar]
- Lee, H.G.; Park, B.-Y.; Kim, H.U.; Seo, P.J. MYB96 stimulates C18 fatty acid elongation in Arabidopsis seeds. Plant Biotechnol. Rep. 2015, 9, 161–166. [Google Scholar] [CrossRef]
- Steiner-Lange, S.; Unte, U.S.; Eckstein, L.; Yang, C.; Wilson, Z.A.; Schmelzer, E.; Dekker, K.; Saedler, H. Disruption of Arabidopsis thaliana MYB26 results in male sterility due to non-dehiscent anthers. Plant J. 2003, 34, 519–528. [Google Scholar] [CrossRef]
- Gonzalez, A.; Mendenhall, J.; Huo, Y.; Lloyd, A. TTG1 complex MYBs, MYB5 and TT2, control outer seed coat differentiation. Dev. Biol. 2009, 325, 412–421. [Google Scholar] [CrossRef]
- Li, S.F.; Milliken, O.N.; Pham, H.; Seyit, R.; Napoli, R.; Preston, J.; Koltunow, A.M.; Parish, R.W. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 2009, 21, 72–89. [Google Scholar] [CrossRef]
- Newman, L.J.; Perazza, D.E.; Juda, L.; Campbell, M.M. Involvement of the R2R3-MYB, AtMYB61, in the ectopic lignification and dark-photomorphogenic components of the det3 mutant phenotype. Plant J. 2004, 37, 239–250. [Google Scholar] [CrossRef]
- Penfield, S.; Meissner, R.C.; Shoue, D.A.; Carpita, N.C.; Bevan, M.W. MYB61 is required for mucilage deposition and extrusion in the Arabidopsis seed coat. Plant Cell 2001, 13, 2777–2791. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Burma, P.K. Regulation of tapetum-specific A9 promoter by transcription factors AtMYB80, AtMYB1 and AtMYB4 in Arabidopsis thaliana and Nicotiana tabacum. Plant J. 2017, 92, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.A.; Gubler, F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 2005, 17, 705–721. [Google Scholar] [CrossRef]
- Agarwal, P.; Mitra, M.; Banerjee, S.; Roy, S. MYB4 transcription factor, a member of R2R3-subfamily of MYB domain protein, regulates cadmium tolerance via enhanced protection against oxidative damage and increases expression of PCS1 and MT1C in Arabidopsis. Plant Sci. 2020, 297, 110501. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Madhuvanthi, R.; Karthikeyan, A.S.; Raghothama, K.G. Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2009, 2, 43–58. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Han, S.W.; Koo, Y.J.; Kim, C.H.; Song, S.I.; Nahm, B.H.; Choi, Y.D.; Cheong, J.J. Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Nguyen, N.H.; Jeong, C.Y.; Nguyen, N.T.; Hong, S.W.; Lee, H. Loss of the R2R3 MYB, AtMyb73, causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 2013, 170, 1461–1465. [Google Scholar] [CrossRef]
- Keereetaweep, J.; Liu, H.; Zhai, Z.; Shanklin, J. Biotin Attachment Domain-Containing Proteins Irreversibly Inhibit Acetyl CoA Carboxylase. Plant Physiol. 2018, 177, 208–215. [Google Scholar] [CrossRef]
- Toll-Riera, M.; Rado-Trilla, N.; Martys, F.; Alba, M.M. Role of low-complexity sequences in the formation of novel protein coding sequences. Mol. Biol. Evol. 2012, 29, 883–886. [Google Scholar] [CrossRef]
- Larkin, J.C.; Oppenheimer, D.G.; Pollock, S.; Marks, M.D. Arabidopsis GLABROUS7 Gene Requires Downstream Sequences for Function. Plant Cell 1993, 5, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Holtgräwe, D.; Schneider, J.; Pucker, B.; Sörensen, T.R.; Weisshaar, B. Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 2014, 14, 249. [Google Scholar] [CrossRef] [PubMed]



| Species | R2R3 | 3R | 1R and MYB-Related | “Unusual” MYB Genes with Two or More Repeats | Total | Reference |
|---|---|---|---|---|---|---|
| A. thaliana | 126 | 5 | 64 | 2 | 197 | [5] |
| P. trichocarpa | 196 | 5 | 152 | 1 | 354 | [6] |
| C. oleifera | 128 | 5 | 44 | 9 | 186 | This study. |
| Subgroup | Conserved Motif |
|---|---|
| Subgroup1 (S1) | YASS |
| Subgroup2 (S2) | MxFW//SFW |
| Subgroup3 (S3) | WFKHLESELGLEExDNQQQ |
| Subgroup4 (S4) | LNL[E/D]L |
| Subgroup5 (S5) | TKAxRC |
| Subgroup6 (S6) | PRPRxF |
| Subgroup7 (S7) | Sx(14)GRT |
| Subgroup8 (S8) | LRKMGIDPLTHKPL |
| Subgroup9 (S9) | AQWESARxxAExRLxR |
| Subgroup10 (S10) | QxxAAAxxN//KxQLxHxMxQ//DDxxSDSxWK |
| Subgroup11 (S11) | PRxDLLD |
| Subgroup12 (S12) | [L/F]LN[K/R]VA |
| Subgroup13 (S13) | GIDPxTHK[P/L]L[S/I]xx[E/G] |
| Subgroup14 (S14) | R2R3: [W]-x(20)-[W]-x(19)-[W]-x(12)-[F]-x(18)-[W]-x(18)-[W] |
| Subgroup15 (S15) | WVxxDxFELSxL |
| Subgroup16 (S16) | PxLxFxEW |
| Subgroup17 (S17) | QQ[F/E]QQ |
| Subgroup18 (S18) | GLPxYP |
| Subgroup19 (S19) | PxLxFSEW |
| Subgroup20 (S20) | WxPRL |
| Subgroup21 (S21) | FxDFL |
| Subgroup22 (S22) | QEMIxxEVRSYM |
| Subgroup23 (S23) | RVxRxxxF//PxxGxxGC |
| Subgroup24 (S24) | QxGxDPxTH |
| Subgroup25 (S25) | LxxYIxxxN |
| CoR2R3-MYBs | Subgroup | Representative within a Subgroup or Most Homologous R2R3-MYB Genes of Arabidopsis Thaliana | Function or Biological Process of AtMYBs | Reference |
|---|---|---|---|---|
| CoMYB46; CoMYB47 | S21 | AtMYB89; AtMYB110 | Inhibit seed FA accumulation by regulating WRI1, BCCP1 | [13] |
| CoMYB67 | S5 | AtMYB123 | Inhibit seed FA biosynthesis | [24] |
| CoMYB85; CoMYB45; CoMYB60; CoMYB70 | S1 | AtMYB30; AtMYB96 | Regulate VLCFAs Biosynthesis | [21,49] |
| CoMYB89; CoMYB90 | S25 | AtMYB118; AtMYB119 | Negatively regulate FA biosynthesis in the endosperm | [25] |
| CoMYB68 | None | AtMYB26 | Male sterility | [50] |
| CoMYB5; CoMYB50 | None | AtMYB5 | Control outer seed coat differentiation | [51,52] |
| CoMYB109; CoMYB3 | S13 | AtMYB61 | Mucilage deposition; lignin biosynthesis | [53,54] |
| CoMYB116 | S23 | AtMYB1 | Upregulate Tapetum-specific promoter A9 activity | [55] |
| CoMYB74; CoMYB118 | None | AtMYB99 | Unknown | - |
| CoMYB106; CoMYB119 | S18 | AtMYB33; AtMYB65 | Stamen development; leaf development | [56] |
| CoMYB41 | S17 | AtMYB71; AtMYB79 | Unknown | - |
| CoMYB103; CoMYB54 | S4 | AtMYB4 | Antioxidant defense | [57] |
| CoMYB61 | S20 | AtMYB116; AtMYB62 | Phosphate starvation responses | [58] |
| CoMYB18; CoMYB2; CoMYB81; CoMYB82; CoMYB17 | S22 | AtMYB44; AtMYB73 | Abiotic stress response | [59,60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Huang, H.; Ma, X.; Hu, Z.; Li, J.; Yin, H. Characterizations of MYB Transcription Factors in Camellia oleifera Reveal the Key Regulators Involved in Oil Biosynthesis. Horticulturae 2022, 8, 742. https://doi.org/10.3390/horticulturae8080742
Li S, Huang H, Ma X, Hu Z, Li J, Yin H. Characterizations of MYB Transcription Factors in Camellia oleifera Reveal the Key Regulators Involved in Oil Biosynthesis. Horticulturae. 2022; 8(8):742. https://doi.org/10.3390/horticulturae8080742
Chicago/Turabian StyleLi, Sijia, Hu Huang, Xianjin Ma, Zhikang Hu, Jiyuan Li, and Hengfu Yin. 2022. "Characterizations of MYB Transcription Factors in Camellia oleifera Reveal the Key Regulators Involved in Oil Biosynthesis" Horticulturae 8, no. 8: 742. https://doi.org/10.3390/horticulturae8080742
APA StyleLi, S., Huang, H., Ma, X., Hu, Z., Li, J., & Yin, H. (2022). Characterizations of MYB Transcription Factors in Camellia oleifera Reveal the Key Regulators Involved in Oil Biosynthesis. Horticulturae, 8(8), 742. https://doi.org/10.3390/horticulturae8080742






