Abstract
Fusarium oxysporum f. sp. conglutinans (FOC) is the dominant pathogen of vascular wilt disease on cabbage and other crucifers. Foc-Nto1 was confirmed to be the homologous protein of Nto1, a subunit of the NuA3 (nucleosomal acetyltransferase of histone H3) complex in Saccharomyces cerevisiae. FOC contains two races, race 1 and race 2. The functions of Nto1 in both races were investigated through functional genetics analyses. The Nto1-deleted mutants were decreased in conidium production and displayed increased sensitivity to hydrogen peroxide. These mutants also had reduced virulence on cabbage. The study provided evidence that Nto1 is a potential metabolic- and pathogenic-related factor in F. oxysporum.
1. Introduction
The soilborne, asexual fungus Fusarium oxysporum f. sp. conglutinans (FOC) is the dominant pathogen of Fusarium wilt on cabbage (Brassica oleracea) and other Cruciferae [1]. FOC contains two races, R1 and R2 [2,3]. R2 infects quickly and causes more severe disease symptoms. In the past two decades, many studies have been published in the field that investigated the roles of the infection process, genomics, transcriptomics, proteomics, and avirulence/effector proteins [1,4,5]. These efforts provided a better understanding of this fungus and its interaction with hosts. A number of metabolism-, development-, and pathogenicity-related genes in Fusarium species have been identified by genetic engineering approaches. The genes related to the infection process in vascular tissues, signal transduction, and cell wall degradation, were well studied during successful infection by F. oxysporum [6]. Some of them encode transcriptional regulators (TFs). For example, TFs REN1 and FoSTUA were essential for conidiation in F. oxysporum [7,8]. FoHTF1 was conserved in Fusarium species and required for phialide development and conidiogenesis [9]. Snt2, a BAH/PHD-containing TF, was related to pathogenicity and morphology in F. oxysporum [10]. The SIX effectors contribute to virulence during infection of host [11].
The fundamental repeating unit of chromatin is scherm the nucleosome which is formed by DNA association with histone. Chromatin-modifying activities are one of the basic ways to reinforce or alleviate transcription. Histone modifications are believed to contribute to a mechanism that can alter chromatin structure and regulate chromatin function [12]. Several types of enzymatic activities participate in histone modifications and among those the histone acetyltransferases (HATs) are the best studied [13,14]. Histone acetylation plays an important role in the interaction between pathogens and their host plants [15]. For example, the Fusarium graminearum histone acetyltransferases are important for morphogenesis, deoxynivalenol (DON) biosynthesis, and pathogenicity [16]. HAT enzymes catalyze an acetyl group from acetyl-CoA transfer to the lysine ε-amino groups on the N-terminal tails of histones [17]. There are five families of the histone acetyltransferases, the Gcn5-related acetyltransferases (GNATs), the MYST (MOZ, Ybf2/Sas3, Sas2 and Tip60)-related HATs, p300/CBP HATs, the general transcription factor HATs, and the nuclear hormone-related HATs SRC1 and ACTR (SRC3) [18]. Studies suggest that the MYST-type HAT NuA3 is primarily responsible for the H3K14 acetylation events in vivo which strongly correlates with transcriptional activity [17,19]. The conserved yeast histone acetyltransferase complex, NuA3, is generally thought to contain at least five subunits, including Eaf6p (13 kDa), Taf14p (27 kDa), Nto1p (86 kDa), Sas3p (98 kDa), and Yng1p (25 kDa). The NuA3 complex, which contains Sas3p as its histone acetyltransferase domain, acetylates Lys-14 of histone H3 and activates the transcription of downstream genes [20]. Histone acetylation is very important for the normal functioning of cells and the development of organisms. Sas3 is required for both the HAT activity and the integrity of the NuA3 complex in S. cerevisiae [21]. Sas3 is also required for development and pathogenicity in Magnaporthe oryzae. The deletion mutant affected asexual reproduction, germination, and appressorium formation so that the pathogenicity was completely lost [22]. The deletion mutant ΔSas3 also showed reduced growth, increased sensitivity to oxidative and osmotic stresses, reduced asexual sporulation and perithecium formation, and lack of DON production in F. graminearum [16]. The PHD finger of Yng1 binds to H3K4me3, whereas its N-terminal region is critical for the integrity of NuA3 HAT [23]. Deletion of FNG1 (the homolog of Yng1 in F. graminearum) led to severe defects on growth, conidiation, sexual reproduction, DON production, and virulence [24]. Fng1 is associated with the NuA4 complex in F. graminearum. Fng3 is part of the NuA3 HAT complex [25]. The Nto1 PHD finger weakly binds to H3K36me3 in vitro [26]. In F. graminearum, the growth rate was slightly reduced in the deletion mutant ΔNto1. Nto1 mediated the FgSas3-Fng3 interaction in the NuA3 HAT complex [25].
In the current study, we searched the amino acid sequence of Nto1 through the genomic databases of F. oxysporum f. sp. conglutinans by BLASTP search. As expected, its homologous protein which had three protein domains was identified in FOC. We conducted a functional analysis of Nto1 in FOC. Deletion of Nto1 impaired virulence, conidium production, and the colony color of FOC in both R1 and R2. In addition, the mutants were more sensitive to hydrogen peroxide (H2O2) than the wild type. Our results suggest that Nto1 is a potential factor related to metabolism and pathogenicity in F. oxysporum.
2. Materials and Methods
2.1. Fungal Strains and Media
F. oxysporum f. sp. conglutinans wild type strains R1 (52557-TM) and R2 (58385-TM), obtained from the American Type Culture Collection (ATCC), were used as the parental strains for the transformation experiments and were maintained as a stock culture in our lab. Potato dextrose agar (PDA) was used as routine culture medium for wild types of two races. The deletion mutants were cultured on PDA supplemented with 60 μg/mL hygromycin B. The cultures of the complemented strains were supplemented with an additional 200 μg/mL neomycin (Amresco, Solon, OH, USA) [27]. Potato dextrose broth (PDB) was used for fungal mycelia and conidium production or DNA and RNA extraction. PDA containing 0.06% H2O2 was used for oxidative stress assays. All cultures, purified from single spores, were grown at 28 °C and stored in 30% (v/v) glycerol at −80 °C.
2.2. Identification and Phylogenetic Analysis of Nto1
Nto1 (EXL85392.1) was originally obtained by the BLASTP program for homology search of the F. oxysporum f. sp. conglutinans genome sequence (GCA_000260215.2) using the Nto1 sequences of S. cerevisiae as query (DAA11457.1). Other Nto1 sequences were downloaded from the National Center for Biotechnology Information (NCBI). The strain names and accession numbers are provided in Table S1. The amino acid sequences of Nto1 were aligned by ClustalW, and a maximum likelihood phylogeny was constructed by MEGA X software using 1000 bootstrap replicates [28]. Conserved domains of Nto1 in FOC were analyzed based on Pfam analysis [29].
2.3. Construction of Nto1Deleted Mutants
The deletion cassettes were generated by split-marker strategy based on fusion PCR [30,31]. All primers used in this study are listed in Table S2. First, a 1.3 kb upstream flanking sequence fragment and a 1.4 kb downstream flanking sequence fragment were amplified from genomic DNA of F. oxysporum with primer pairs UpF/UpR and DpF/DpR. The first two-thirds sequence fragment (1.1 kb) and last two-thirds sequence fragment (0.7 kb) of the hygromycin resistance gene were amplified from plasmid pKOV21 using primer pairs HuF/HuR and HdF/HdR, respectively. Next, the upstream flanking sequence fragment and first two-thirds sequence fragment of the hygromycin resistance gene were joined together by overlap PCR with primer pairs UpF/HuR. Similarly, the downstream flanking sequence fragment and last two-thirds sequence fragment of the hygromycin resistance gene were joined together with primer pairs HdF/DpR. Lastly, the amplified 2.4 kb and 2.1 kb fusion productions of PCR were purified with an EasyPure Quick Gel Extraction Kit (TransGen Biotech, Beijing, China) for subsequent experiments.
Nto1-deleted mutants were obtained by using PEG-mediated protoplast transformation [32]. The putative transformants were purified from single spores and cultured on PDA plates containing 60 μg/mL hygromycin B. The extraction of genomic DNA of wild types and transformants was performed following a CTAB-based protocol [33]. The transformants were screened for hygromycin B resistance to identify the gene-deleted mutants. The primer pairs HuF/HdR and PeF/PeR were used to identify the hygromycin B resistance gene and Nto1 gene, respectively (Table S2). The primer pairs CupF/CupR and CdpF/CdpR were used to amplify the fragment of the connecting area to confirm that the hygromycin B resistance gene as a selectable marker replaced the gene Nto1 on the correct location in the genome. The deletion mutants were further confirmed by Southern blot analysis on EcoRV-digested genomic DNA using a PrF/PrR PCR-amplified 568 bp fragment as a probe (Table S2). The genetically stable gene deletion transformants for FOC wild type strains R1 were named 1-ΔFonto1-1 and 1-ΔFonto1-2. The mutants of R2 were named 2-ΔFonto1-1 and 2-ΔFonto1-2.
2.4. Complementation of Nto1-Deleted Mutants
To confirm that the phenotypic changes of deleted mutants were due to deletion of the gene Nto1, the Nto1-deleted mutants (1-ΔFonto1-1 and 2-ΔFonto1-1) were complemented by transformation with a modified plasmid carrying an intact copy of the Foc-Nto1 fragment, including 1300 bp of the promotor region, 330 bp of the terminator region, and neomycin resistance marker. The Nto1 gene was amplified as a 5.3 kb fragment using primers CpF/CpR (Table S2). The primers created SpeI and ApaLI sites at the ends of the fragment. Then the amplicon was cloned into SpeI/ApaLI-digested pKN which carries a selectable marker for neomycin resistance. As described above, the protoplast transformation of 1-ΔFonto1-1 and 2-ΔFonto1-1 with pKN-Nto1 were performed, and neomycin at 200 μg/mL was used as a screening marker. The genetically stable gene complementation transformants for 1-ΔFonto1-1 and 2-ΔFonto1-1 were named 1-ΔFonto1-C and 2-ΔFonto1-C, respectively.
2.5. Conidiation
When the colonies of the wild type strains and mutants reached 3–4 cm in diameter, three mycelial plugs with 9 mm diameter from the colony were transferred into 100 mL of PDB at 28 °C for shake culture at 150 rpm for 96 h. Then, the number of spores was calculated with a hemocytometer. In addition, the spores were also observed microscopically to detect changes in conidium morphology. Each strain had three replicates, and each experiment was repeated three times.
2.6. Oxidant Stressor Treatment
Mycelial plugs with 9 mm diameter, which were taken from the edge of the fungal colony, were grown on PDA plates amended with 0.06% of H2O2 at 28 °C in the dark for 4 days. The diameters of the colonies were measured daily. Tolerance to H2O2 was assessed by mycelial radial growth inhibition (RGI) as follows:
where C is the colony diameter of the control and N is that of a treatment. Three technical replicates were conducted for each strain and each experiment was repeated three times independently [34].
RGI = [(C − N)/(C − 9)] ×100%,
2.7. Plant Material and Inoculation
Cabbage cultivar ZHONG GAN 21 (China Vegetable Seed Technology Co., Ltd., Beijing, China) was used in inoculation tests for its susceptibility to FOC. Cabbage seedlings with 2–3 true leaves were inoculated by the root-dipping method. The seedling roots were dipped in conidial suspensions with the concentration of 1 × 106 conidia/mL for 20 min, followed by transferring to soil medium and maintaining in a growth chamber at 28 °C with 16 h light and 8 h darkness. The disease severity of each plant was evaluated depending on disease index (DI) over a two-week period after inoculation. The scoring standard was evaluated from 0 to 5 based on leaf yellowing and wilting as follows: 0, no symptoms; 1, yellowing of one lower leaf; 2, yellowing and wilting of two lower leaves; 3, yellowing and wilting of all leaves except heart; 4, yellowing and wilting of all leaves; 5, wilting of all leaves or plant death. The DI was calculated using the formula:
where Xi is the scoring standard of wilting symptoms (Xi = 1, 2, 3, 4 and 5), n is the number of seedlings in each symptom class for each strain, and N is the total number of cabbage seedlings for each strain. For each strain, three independent biological experiments were conducted. At least 30 seedlings were inoculated in each replicate [32].
DI = Σ (Xi × n)/(5 × N) × 100,
3. Results
3.1. Identification of Nto1 in F. oxysporum
A total of 14 homologous proteins from 9 Fusarium species, 2 Zygosaccharomyces species, and 3 Saccharomyces species were used to construct the phylogenetic tree. Nto1 from Fusarium oxysporum was the most related scherm to each other (Figure 1a). Nto1 (EXL85392.1) was identified from NCBI through homology searches. The full Nto1 gene in FOC is 3579 bp long, containing a 102 bp intron and encoding a protein of 1158 amino acids. It contains three kinds of conserved protein domains, enhancer of polycomb-like (EPL1), PHD-finger, and PHD-zinc-finger-like domain based on Pfam analysis (Figure 1b).
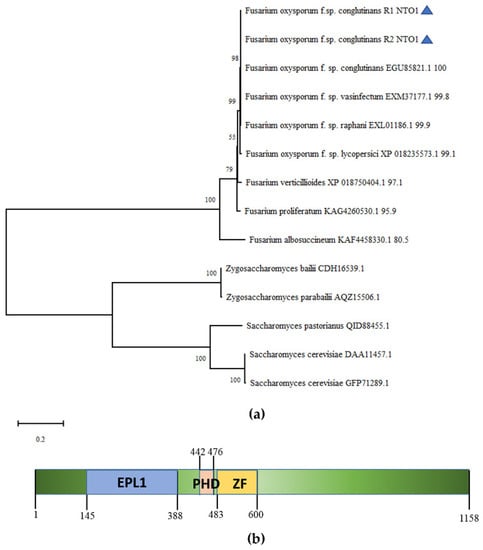
Figure 1.
Phylogenetic relationship and structural domain analysis of Nto1. (a) Phylogenetic relationship of the Nto1 homologs from different fungal species. Races R1 and R2 are marked with blue triangles; (b) conserved domains of Nto1 in F. oxysporum f. sp. conglutinans based on Pfam analysis.
3.2. Deletion of Nto1 in R1 and R2
To investigate the functions of Nto1 in race 1 and race 2 of FOC, fusion PCR-based deletion methods were used for homologous gene knockout. The flanking region sequences of the targeted gene and the selectable marker were amplified with primer pairs PeF/PeR, HL/HR respectively (Figure 2a). After PCR screening (Figure 2b), two stable mutants of each race were used for Southern analysis. Genomic DNA of the mutants and the wild-type strain were digested with EcoRV. A 523-bp fragment located upstream amplified with primer pair PrF/PrR was used as probe in Southern blot analysis. When hybridized with the probe, the wild-type strain showed a 7016 bp band, whereas the deletion mutants instead had a 4813 bp band (Figure 2c). All mutants showed that the hyg gene replaced the Nto1 gene at the correct position in the genome of FOC.
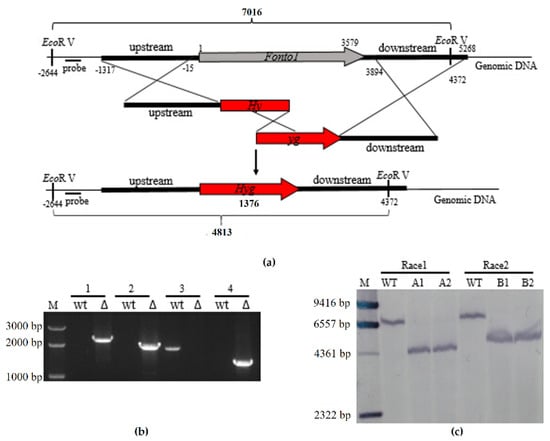
Figure 2.
Deletion of Nto1 in F. oxysporum f. sp. conglutinans. (a) Schematic diagram of gene-disruption and Southern blot analysis verification strategy; (b) PCR strategy to screen ΔNto1 transformants. M: marker; wt: wild type; Δ: Nto1-deleted mutant. 1 = PCR performed with primer pair CupF/CupR; a 2.3-kb amplified fragment indicates ΔNto1 integration at the left junction. 2 = PCR performed with primer pair CdpF/CdpR; a 2.2-kb amplified fragment indicates ΔNto1 integration at the right junction. 3 = PCR performed with primer pair PeF/PeR; the result indicates that Nto1 was deleted. 4 = PCR performed with primer pair HL/HR; a 1.4-kb amplified fragment indicates that Hyg was inserted to the F. oxysporum f. sp. conglutinans genome; (c) Southern blot analysis confirms the gene deletion mutants. The genetically stable gene deletion transformants of FOC wild type strains race 1 were named 1-ΔFonto1-1 and 1-ΔFonto1-2. The mutants of race 2 were named 2-ΔFonto1-1 and 2-ΔFonto1-2. M: Marker; WT: wild type; A1: 1-ΔFonto1-1; A2: 1-ΔFonto1-2; B1: 2-ΔFonto1-1; B2: 2-ΔFonto1-2.
3.3. Complementation of Nto1-Deleted Mutants
In order to confirm that the changes of phenotype in the Nto1-deleted mutants were caused by the gene deletion, the Nto1-deleted mutants (1-ΔFonto-1 and 2-ΔFonto-1) were complemented by transformation with a modified plasmid carrying the Nto1 sequence (Figure 3a). PCR (Figure 3b) and RT-PCR (Figure 3c) were conducted to screen transformants which expressed the target gene. Then, one complementation transformant of each race was used for Southern analysis. In order to determine the copies of Nto1 in complementation transformants, Southern blot analysis, using a 800 bp Nto1 fragment as probe, was performed on the wild-type strain and complementation transformants genomic DNA digested by EcoRV, HindIII, and EcoRI separately. Results showed that each complementation transformant only had one insert copy (Figure 3d).
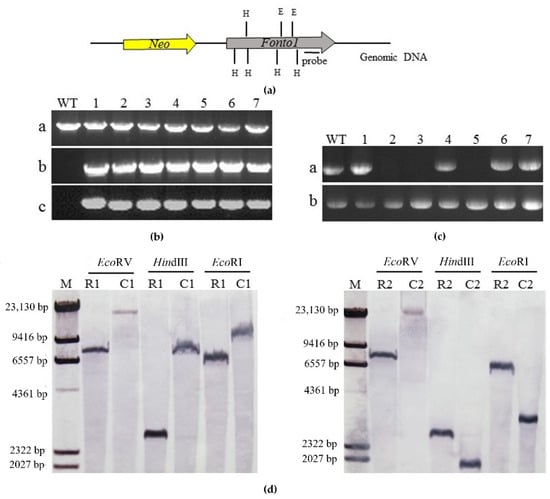
Figure 3.
Complementation of the Nto1 deletion mutants. (a) Schematic representation of the Nto1 complementation strategy; Nto1 and Neo (selectable marker gene) fragments are denoted by large gray and yellow arrows, respectively. H = HindIII digest site; E = EcoRI digest site; (b) PCR strategy to screen complemented transformants. WT: wild type; 1–7 = candidate complemented strains. a: PCR performed with primer pair PeF/PeR; a 1.7-kb fragment amplified from Nto1. b: PCR performed with primer pair HL/HR; a 1.4-kb fragment amplified from Hyg. c: PCR performed with primer pair NF/NR; a 1.8-kb fragment amplified from Neo; (c) RT-PCR screen complemented transformants; WT: wild type; 1–7 = candidate complemented strains. a: PCR performed with primer pair PeF/PeR with genomic cDNA as templates; a 1.7-kb fragment amplified from Nto1. b: PCR performed with primer pair IrF/IrR; a 144 bp fragment amplified from elongation factor 2 gene as reference; (d) Results of Southern blot hybridization analyses. The genetically stable gene complementation transformants for 1-ΔFonto1-1 and 2-ΔFonto1-1 were named 1-ΔFonto1-C and 2-ΔFonto1-C, respectively. M: marker; R1: race1; C1: 1- ΔFonto1-C; R2: race2; C2: 2-ΔFonto1-C; A 485-bp fragment amplified by primer pair PcF/Pcr from Nto1 was used as a probe. Genomic DNA preparations from the wild-type strain (R1 and R2) and the complemented strain (C1 and C2) were digested with EcoRV, HindIII and EcoRI.
3.4. Nto1 Disruption Resulted in Reduction in Microconidium Formation and Abnormal Conidium Color
In order to analyze the effects of Nto1 on the conidiation in F. oxysporum, wild types, Nto1-deleted mutant strains and complementation strains were grown in PDB, and their microconidia were observed. There was no difference in the morphology of microconidia among wild strains, Nto1-deleted mutants, and complementation strains. However, the number of microconidia produced by the deletion mutants was less than that of the wild types (Figure 4a). The conidium numbers of the two races and their corresponding complemented mutants showed no obvious difference. When collected by centrifuging, microconidia of all the mutants exhibited a pink color while wild-type strains and complementation strains exhibited white pigmentation (Figure 4b).
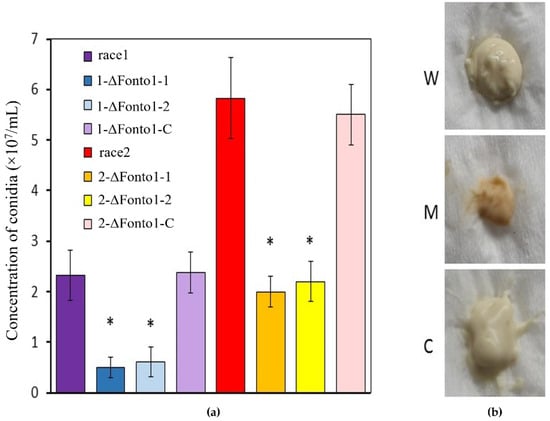
Figure 4.
Number and color of conidia of all F. oxysporum strains. (a) Conidium numbers of race1 and race2 and two each of their corresponding Nto1 gene deletion mutants (1-ΔFonto1-1, 1-ΔFonto1-2, 2-ΔFonto1-1 and 2-ΔFonto1-2) and one each of their corresponding complemented mutants (1-ΔFonto1-C and 2-ΔFonto1-C). * represents the significant differences at p < 0.05; (b) Conidium color of wild types, deletion mutants and complemented mutants. Microconidia of all the mutants exhibited a pink color while wild-type strains and complemented mutants exhibited white pigmentation. W: wild type; M: Nto1 deletion mutants; C: Nto1 complementation mutants.
3.5. Nto1 Disruption Leads to Hypersensitivity to Oxidative Stress
To further analyze whether Nto1 plays a role in the oxidative stress response, the wild type, deletion mutants and complemented strains were cultured on PDA plates containing 0.06% H2O2 (Figure 5a). The mycelial growth of Nto1-deleted mutants of R1 was significantly affected and its growth inhibition was 32.7% greater than that of R1 (Figure 5b). However, the mycelial growth of Nto1-deleted mutants of R2 showed no difference compared to the wild type.
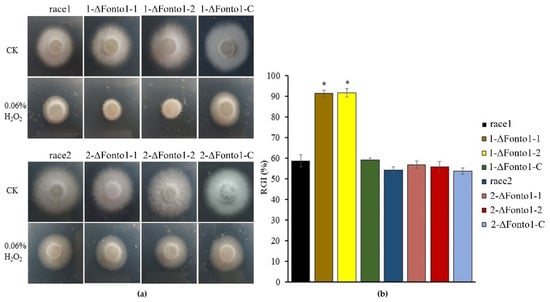
Figure 5.
Sensitivity to oxidative stress. (a) Colony morphology of race 1, race 2 and their corresponding Nto1 deletion mutants (1-ΔFonto1-1, 1-ΔFonto1-2, 2-ΔFonto1-1 and 2-ΔFonto1-2) and complementation mutants (1-ΔFonto1-C and 2-ΔFonto1-C) after 4 days of inoculation on PDA amended with 0.06% H2O2. Three replicates were performed for each race; (b) mycelial radial growth inhibition (RGI) of race 1, race 2 and their corresponding Nto1 deletion mutants and complementation mutants. * represents the significant differences at p < 0.05.
3.6. Nto1 Is Essential for Full Virulence of F. oxysporum
To investigate the virulence of the Nto1-deleted mutants, plant infection assays were performed. Cabbage seedlings inoculated with wild types or mutants all showed characteristic wilt symptoms (Figure 6). However, the disease index of plants inoculated with mutants was significantly lower than that inoculated with wild types. Results demonstrated that Nto1 was essential for pathogenicity in F. oxysporum.
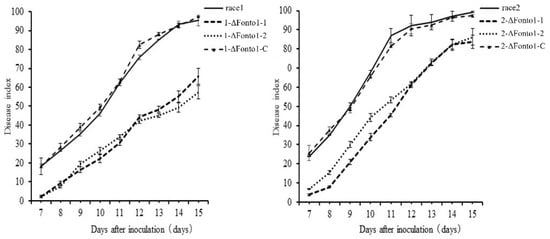
Figure 6.
Disease indexes of race 1, race 2 and their corresponding Nto1 deletion mutants (1-ΔFonto1-1, 1-ΔFonto1-2, 2-ΔFonto1-1 and 2-ΔFonto1-2) and complementation mutants (1-ΔFonto1-C and 2-ΔFonto1-C) from 7 to 15 days after inoculation. Means and standard errors were calculated from three independent experiments.
4. Discussion
In this study, we investigated the possible involvement of Nto1 in microconidium development and virulence in both races of F. oxysporum f. sp. conglutinans. Relative to oxidative stress, Nto1 is essential for resistance to oxidative damage in R1 but not in R2. In plants, there are multiple efficient defense systems against the invasion of pathogenic microbes. When the pathogen infects the plants, reactive oxygen species (ROS), primarily superoxide and H2O2, are considered to be defense molecules [35]. Low concentrations of ROS act as signal molecules in plants to induce the expression of various defense-related genes against pathogen attack, which is necessary for pathogen-associated molecular patterns (PAMP) to trigger immunity (PTI) responses in plant-microbe interactions [36,37]. ROS can prevent pathogens from invading the plants through the synthesis of lignin and other phenolic polymers by apoplastic peroxidases [38]. ROS also directly kill the pathogen depending on its toxicity [39]. Concurrently, plant pathogens have also developed many strategies to recognize and inhibit host immune response for invading successfully, such as ROS detoxification and signal transduction disruption [40,41]. The relative sensitivity of fungal pathogens to ROS can, to a certain extent, reflect its ability to infect the host [37]. In our study, the wild type R2 was more able to tolerate oxidative stress than R1. When Nto1 was deleted, the oxidative tolerance of R2 did not change while that of R1 was significantly decreased. Hence, R2 appears to have more regulation pathways for oxidative stress. Additionally, microconidium production and the mycelial growth rate of R2 were higher than R1 [5]. The combined effect of all these factors on invasion into the host may explain why R2 is more virulent than R1.
Nto1 is a subunit of the histone acetyltransferase NuA3 complex and its specific function is currently unknown. More and more studies indicate that histone acetylation is linked not only to transcription activation, but also to regulation of diverse cellular processes, such as gene silencing, DNA repair, and cell-cycle progression [17]. The Nto1 protein of FOC carries three conserved domains: EPL1, PHD, and Zn-finger. It shares two of the same domains with Snt2. Snt2 was identified and characterized as transcriptional regulator involved in hyphal growth, conidiation and pathogenicity in F. oxysporum f. sp. melonis [10,42]. Moreover, it was reported that Snt2 physically associates with Ecm5 and Rpd3 deacetylases and regulates transcription related to oxidative stress [43]. Snt2 was also characterized as histone E3 ligase and plays a crucial part in regulating histone protein levels in S. cerevisiae [44]. Interestingly, several proteins related to histone modification were reported to be involved in the pathogenesis in Fusarium specie. Cti6, associated with the Rpd3-Sin3 histone deacetylase, is a PHD finger-containing protein, whose homologous protein FoCti6 is required for full pathogenicity in F. oxysporum f. sp. lycopersici [45,46]. Spt3 and spt8 are important components of the histone acetyltransferase SAGA complex [47,48]. Their homologous genes FgSpt3 and FgSpt8 play important roles in conidiation, mycelial growth, and pathogenicity in F. graminearum [34]. Seeing that Nto1 participates in histone modification, especially histone acetylation, the character changes may be a result of the histone modification which could cause epigenetic changes.
Fungi are known to synthesize a variety of pigments as secondary metabolites, such as carotenoids, melanins, azaphilones, flavins, phenazines, quinones, monascin, violacein, indigo, among others [49]. Many species of Fusarium have been reported for their capability to produce pigments that vary in color [49]. Studies have reported pigments such as naphthoquinones, anthraquinones compounds, and polyketide pigment (bikaverin) from F. oxysporum [50]. It was also reported that the biosynthesis of pigments in Fusarium species may be considered a fungal response to stress [49]. In this research, the Nto1 gene deletion mutants exhibited a different microconidium color compared to the wild type. The pigmented secondary metabolites and their encoding gene or gene cluster regulated by Nto1 are the focus of our next research.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8060540/s1, Table S1. The names and accession numbers of other strains used for comparison. Table S2: All primers used in the study.
Author Contributions
Conceptualization, B.X. and Z.M.; methodology, E.L., J.X. and Y.Y.; validation, E.L., J.X. and Y.Y.; formal analysis, E.L. and Y.Y.; data curation, E.L. and J.X.; writing—original draft preparation, E.L. and J.X.; writing—review and editing, B.X. and Z.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Sciences Foundation of China, grant number “31801682”.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data sets supporting the results of this article will be freely available upon request to the first author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Correll, J.C. The relationship between formae speciales, races, and vegetative compatibility groups in Fusarium oxysporum. Phytopathology 1991, 81, 1061–1064. [Google Scholar]
- Liu, X.; Ling, J.; Xiao, Z.; Xie, B.; Fang, Z.; Yang, L.; Zhang, Y.; Lv, H.; Yang, Y. Characterization of emerging populations of Fusarium oxysporum f. sp. conglutinans causing cabbage wilt in China. J. Phytopathol. 2017, 165, 813–821. [Google Scholar] [CrossRef]
- Ma, L.J.; van der Does, H.C.; Borkovich, K.A.; Coleman, J.J.; Daboussi, M.J.; Di Pietro, A.; Dufresne, M.; Freitag, M.; Grabherr, M.; Henrissat, B.; et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 2010, 464, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Ling, J.; Wang, G.; Xiao, J.; Yang, Y.; Mao, Z.; Wang, X.; Xie, B. Comparative proteomics analyses of two races of Fusarium oxysporum f. sp. conglutinans that differ in pathogenicity. Sci. Rep. 2015, 5, 13663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietro, A.D.; Madrid, M.P.; Caracuel, Z.; Delgado-Jarana, J.; Roncero, M.J.G. Fusarium oxysporum: Exploring the molecular arsenal of a vascular wilt fungus. Mol. Plant Pathol. 2003, 4, 315–325. [Google Scholar] [CrossRef]
- Ohara, T.; Inoue, I.; Namiki, F.; Kunoh, H.; Tsuge, T. REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum. Genetics 2004, 166, 113–124. [Google Scholar] [CrossRef] [Green Version]
- Ohara, T.; Tsuge, T. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryot. Cell 2004, 3, 1412–1422. [Google Scholar] [CrossRef] [Green Version]
- Wenhui, Z.; Xu, Z.; Qiurong, X.; Qingping, H.; Chengkang, Z.; Huanchen, Z.; Liping, X.; Guodong, L.; Won-Bo, S.; Zonghua, W. A conserved homeobox transcription factor Htf1 is required for phialide development and conidiogenesis in Fusarium species. PLoS ONE 2012, 7, e45432. [Google Scholar]
- Denisov, Y.; Freeman, S.; Yarden, O. Inactivation of Snt2, a BAH/PHD-containing transcription factor, impairs pathogenicity and increases autophagosome abundance in Fusarium oxysporum. Mol. Plant Pathol. 2011, 12, 449–461. [Google Scholar] [CrossRef]
- Bart, L.; Houterman, P.M.; Martijn, R. Effector gene screening allows unambiguous identification of Fusarium oxysporum f. sp. lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 2009, 300, 201–215. [Google Scholar]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef]
- Gómez-Diaz, E.; Jordà, M.; Peinado, M.A.; Rivero, A. Epigenetics of host-pathogen interactions: The road ahead and the road behind. PLoS Pathog. 2012, 8, e1003007. [Google Scholar] [CrossRef] [Green Version]
- Xiangjiu, K.; Van, D.A.D.; Van, D.L.T.A.J.; Cees, W.; Jingsheng, X.; Jin, X.; Hao, Z.; Wanquan, C.; Jie, F. The Fusarium graminearum histone acetyltransferases are important for morphogenesis, DON biosynthesis, and pathogenicity. Front. Microbiol. 2018, 9, 654. [Google Scholar]
- Carrozza, M.J.; Utley, R.T.; Workman, J.L.; Cote, J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003, 19, 321–329. [Google Scholar] [CrossRef]
- Joseph, T.; Christopher, G.; Michael, G.R. Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 1998, 10, 373–383. [Google Scholar]
- Vicente-Muoz, S.; Romero, P.; Magraner-Pardo, L.; Martinez-Jimenez, C.P.; Tordera, V.; Pamblanco, M. Comprehensive analysis of interacting proteins and genome-wide location studies of the Sas3-dependent NuA3 histone acetyltransferase complex. FEBS Open Bio 2014, 4, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, T.M.; Mcdaniel, S.L.; Byrum, S.D.; Cades, J.A.; Dancy, B.C.R.; Wade, H.; Tackett, A.J.; Strahl, B.D.; Taverna, S.D. A PWWP domain-containing protein targets the NuA3 acetyltransferase complex via histone H3 lysine 36 trimethylation to coordinate transcriptional elongation at coding regions. Mol. Cell Proteom. 2014, 13, 2883–2895. [Google Scholar] [CrossRef] [Green Version]
- John, S.; Howe, L.A.; Tafrov, S.T.; Grant, P.A.; Sternglanz, R.; Workman, J.L. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 2000, 14, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Lee, J.; Kwon, S.; Lee, Y.; Jeon, J. A MYST family histone acetyltransferase, MoSAS3, is required for development and pathogenicity in the rice blast fungus. Mol. Plant Pathol. 2019, 20, 1491–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Yoon, C.Y.; Jun, Y.; Lee, B.B.; Lee, J.E.; Ha, S.D.; Woo, H.; Choi, A.; Lee, S.; Jeong, W.; et al. NuA3 HAT antagonizes the Rpd3S and Rpd3L HDACs to optimize mRNA and lncRNA expression dynamics. Nucleic Acids Res. 2020, 48, 10753–10767. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Xia, A.; Ye, M.; Ren, J.; Jiang, C. Opposing functions of Fng1 and the Rpd3 HDAC complex in H4 acetylation in Fusarium graminearum. PLoS Genet. 2020, 16, e1009185. [Google Scholar] [CrossRef]
- Xu, H.; Ye, M.; Xia, A.; Jiang, H.; Huang, P.; Liu, H.; Hou, R.; Wang, Q.; Li, D.; Xu, J.R.; et al. The Fng3 ING protein regulates H3 acetylation and H4 deacetylation by interacting with two distinct histone modifying complexes. New Phytol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Kachirskaia, L.; Walter, K.L.; Kuo, J.H.A.; Lake, A.; Davrazou, F.; Chan, S.M.; Martin, D.G.E.; Fingerman, I.M.; Briggs, S.D.; et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007, 282, 2450–2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, L.; Li, J.; Cheng, L.; Ling, J.; Luo, Z.; Bai, M.; Xie, B. A high efficiency gene disruption strategy using a positive-negative split selection marker and electroporation for Fusarium oxysporum. Microbiol. Res. 2014, 169, 835–843. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Wang, G.; Liu, Z.; Lin, R.; Li, E.; Mao, Z.; Ling, J.; Yang, Y.; Yin, W.B.; Xie, B. Biosynthesis of antibiotic leucinostatins in bio-control fungus Purpureocillium lilacinum and their inhibition on Phytophthora revealed by genome mining. PLoS Pathog. 2016, 12, e1005685. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Zhang, H.; Wang, Y.; Liu, F.; Li, E.; Ma, J.; Yang, B.; Zhang, C.; Li, L.; Liu, Y. Requirement of LaeA, VeA, and VelB on asexual development, ochratoxin A biosynthesis, and fungal virulence in Aspergillus ochraceus. Front. Microbiol. 2019, 10, 2759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, E.; Wang, G.; Xiao, J.; Ling, J.; Yang, Y.; Xie, B. A SIX1 homolog in Fusarium oxysporum f. sp. conglutinans is required for full virulence on cabbage. PLoS ONE 2016, 11, e0152273. [Google Scholar]
- Nicholson, P.; Parry, D.W. Development and use of a PCR assay to detect Rhizoctonia cerealis, the cause of sharp eyespot in wheat. Plant Pathol. 1996, 45, 872–883. [Google Scholar] [CrossRef]
- Gao, T.; Zheng, Z.; Hou, Y.; Zhou, M. Transcription factors spt3 and spt8 are associated with conidiation, mycelium growth, and pathogenicity in Fusarium graminearum. FEMS Microbiol. Lett. 2014, 351, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doke, N.; Miura, Y.; Sanchez, L.; Park, H.; Noritake, T.; Yoshioka, H.; Kawakita, K. The oxidative burst protects plants against pathogen attack: Mechanism and role as an emergency signal for plant bio-defence-a review. Gene 1996, 179, 45–51. [Google Scholar] [CrossRef]
- Gan, Y.; Zhang, L.; Zhang, Z.; Dong, S.; Li, J.; Wang, Y.; Zheng, X. The LCB2 subunit of the sphingolip biosynthesis enzyme serine palmitoyltransferase can function as an attenuator of the hypersensitive response and Bax-induced cell death. New Phytol. 2009, 181, 127–146. [Google Scholar] [CrossRef]
- Guo, M.; Chen, Y.; Du, Y.; Dong, Y.; Guo, W.; Zhai, S.; Zhang, H.; Dong, S.; Zhang, Z.; Wang, Y.; et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011, 7, e1001302. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Schopfer, P. Hydroxyl-radical production in physiological reactions: A novel function of peroxidase. Eur. J. Biochem. 1999, 260, 726–735. [Google Scholar] [CrossRef]
- Averyanov, A. Oxidative burst and plant disease resistance. Front. Biosci. 2009, 1, 142–152. [Google Scholar]
- Mayer, A.M.; Staples, R.C.; Gil-ad, N.L. Mechanisms of survival of necrotrophic fungal plant pathogens in hosts expressing the hypersensitive response. Phytochemistry 2001, 58, 33–41. [Google Scholar] [CrossRef]
- Moye-Rowley, W.S. Regulation of the transcriptional response to oxidative stress in fungi: Similarities and differences. Eukaryot. Cell 2003, 2, 381–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denisov, Y.; Yarden, O.; Freeman, S. The transcription factor SNT2 is involved in fungal respiration and reactive oxidative stress in Fusarium oxysporum and Neurospora crassa. Physiol. Mol. Plant Pathol. 2011, 76, 137–143. [Google Scholar] [CrossRef]
- Baker, L.A.; Ueberheide, B.M.; Dewell, S.; Chait, B.T.; Zheng, D.; Allis, C.D. The yeast Snt2 protein coordinates the transcriptional response to hydrogen peroxide-mediated oxidative stress. Mol. Cell. Biol. 2013, 33, 3735–3748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.K.; Gonzalez, M.; Kabbaj, M.H.M.; Gunjan, A. Novel E3 ubiquitin ligases that regulate histone protein levels in the budding yeast Saccharomyces cerevisiae. PLoS ONE 2012, 7, e36295. [Google Scholar] [CrossRef] [Green Version]
- Papamichos-Chronakis, M.; Petrakis, T.; Ktistaki, E.; Topalidou, I.; Tzamarias, D. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 2002, 9, 1297–1305. [Google Scholar] [CrossRef]
- Puig, S.; Lau, M.; Thiele, D.J. Cti6 is an Rpd3-Sin3 histone deacetylase-associated protein required for growth under iron-limiting conditions in Saccharomyces cerevisiae. J. Biol. Chem. 2004, 279, 30298–30306. [Google Scholar] [CrossRef] [Green Version]
- Bian, C.; Xu, C.; Ruan, J.; Lee, K.K.; Burke, T.L.; Tempel, W.; Barsyte, D.; Li, J.; Wu, M.; Zhou, B.; et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 2011, 30, 2829–2842. [Google Scholar] [CrossRef] [Green Version]
- Pray-Grant, M.G.; Daniel, J.A.; Schieltz, D.; Yates, J.R.; Grant, P.A. Chd1 chromodomain links histone H3 methylation with SAGA-and SLIK-dependent acetylation. Nature 2005, 433, 434–438. [Google Scholar] [CrossRef]
- Medentsev, A.G.; Arinbasarova, A.Y.; Akimenko, V.K. Microbiology, biosynthesis of naphthoquinone pigments by fungi of the genus Fusarium. Appl. Biochem. Microbiol. 2005, 41, 503–507. [Google Scholar] [CrossRef]
- Lagashetti, A.C.; Dufossé, L.; Singh, S.K.; Singh, P.N. Fungal pigments and their prospects in different industries. Microorganisms 2019, 7, 604. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).