Molecular Characterization of Prunus Cultivars from Romania by Microsatellite Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. SSR Analysis
| Locus | Primer Sequence (5′→ 3′) | Repeat Motif | References | Size-Range (bp) | Peach | Apricot | Nectarine | No. of Unique Alleles | PIC |
|---|---|---|---|---|---|---|---|---|---|
| BPPCT-001 | * AAT TCC CAA AGG ATG TGT ATG AG CAG GTG AAT GAG CCA AAG C | (GA)27 | [22] | 133–176 | 4 | 2 | 3 | 6 | 0.8076 |
| BPPCT-002 | * TCG ACA GCT TGA TCT TGA CC CAA TGC CTA CGG AGA TAA AAG AC | (AG)25 | 196–221 | 3 | 9 | 2 | 12 | 0.9102 | |
| BPPCT-004 | CTG AGT GAT CCA TTT GCA GG AGG GCA TCT AGA CCT CAT TGT T | (CT)22 | 175–230 | 1 | 3 | 1 | 4 | 0.7005 | |
| BPPCT-010 | AAA GCA CAG CCC ATA ATG C GTA CTG TTA CTG CTG GGA ATG C | AG)4 GG(AG)10 | 125–150 | 1 | 2 | 1 | 3 | 0.5907 | |
| BPPCT-014 | * TTG TCT GCC TCT CAT CTT AAC C CAT CGC AGA GAA CTG AGA GC | (AG)23 | 204–233 | 2 | 1 | 1 | 3 | 0.5916 | |
| BPPCT-025 | * TCC TGC GTA GAA GAA GGT AGC CGA CAT AAA GTC CAA ATG GC | (GA)29 | 168–215 | 5 | 4 | 4 | 9 | 0.8769 | |
| CPPCT-022 | CAATTAGCTAGAGAGAATTATTGGACAAGAAGCAAGTAGTTTG | (CT)28CAA (CT)20 | [20] | 250–800 | 2 | 2 | 1 | 4 | 0.6363 |
| CPPCT-029 | CCAAATTCCAAATCTCCTAACATGATCAACTTTGAGATTTGTTGAA | (CT)24 | 80–200 | 2 | 1 | 1 | 3 | 0.5513 | |
| CPPCT-030 | TGAATATTGTTCCTCAATTCCTCTAGGCAAGAGATGAGA | (CT)30 | 175–220 | 2 | 2 | 2 | 3 | 0.5897 | |
| Pchgms-001 | GGG TAA ATA TGC CCA TTG TGC AAT C GGA TCA TTG AAC TAC GTC AAT CCT C | (AC)12(AT)6 | [18] | 160–200 | 1 | 2 | 1 | 3 | 0.5898 |
| Pchgms-003 | GGA TCA TTG AAC TAC GTC AAT CCT C CAA CCT GTG ATT GCT CCT ATT AAA C | (CT)14 | 200–220 | 2 | 2 | 1 | 3 | 0.592 | |
| Pchgms-004 | ATC TTC ACA ACC CTA ATG TC GTT GAG GCA AAA GAC TTC AAT | (CT)21 | 150–200 | 2 | 3 | 2 | 4 | 0.7002 | |
| Pchgms-010 | GGTCACGCATCCTTTCATTT GACACCTCCATTTGTATCAAAGC | T19A10 | [48] | 180–200 | 1 | 2 | 1 | 2 | 0.3743 |
| Pchgms-011 | AAGCAATAAAACCAGCAGCAA TCAATCAATTGGCATGTTCG TTGAGGCCCACTTATTAGCC CCCCCATTATTCAAACTTCTG | (TA)11 | 250–300 | 1 | 1 | 1 | 3 | 0.5907 | |
| Pchgms-012 | CGACACTTAGCTAGAAGTTGCCTTA TCAAGCTCAAGGTACCAGCA | (CT)9(TC)20(CA)9 | 200–450 | 2 | 3 | 2 | 7 | 0.8245 | |
| Pchgms-020 | * AATTGCATCACAGCAAGAGC GGGGGTTTGGTTAAGATCG CCCTTACCCCCTTACCACTT | (TA)15(TC)11 | 265–280 | 2 | 4 | 2 | 6 | 0.8101 | |
| Pchgms-021 | * ACCACCATTTTGGCTCTCTG ACCACCACAACCAAACCATT | (TA)14 | 289–306 | 3 | 1 | 3 | 4 | 0.703 | |
| Pchgms-022 | ATAATCCGGCAGGGGTCTTA TTGGGGTTTGTCAGTATTTTACA | (GA)14(AT)9 | 100–500 | 1 | 1 | 1 | 2 | 0.2392 | |
| Pchgms-023 | CTGCCGAAAGCATTTTGAAT GAGCTCATGGCAACACAGAA | (TTC)5 | 300–500 | 1 | 2 | 1 | 3 | 0.5769 | |
| UDP96-001 | * AGTTTGATTTTCTGATGCATCC TGCCATAAGGACCGGTATGT | (CA)17 | [19] | 123–146 | 3 | 1 | 3 | 6 | 0.8096 |
| UDP97-401 | TAAGAGGATCATTTTTGCCTTG CCCTGGAGGACTGAGGGT | (GA)19 | 100–150 | 1 | 2 | 1 | 2 | 0.3648 | |
| UDP97-402 | TCCCATAACCAAAAAAAACACC TGGAGAAGGGTGGGTACTTG | (AG)17 | 125–170 | 1 | 1 | 2 | 3 | 0.5874 | |
| UDP97-403 | CTGGCTTACAACTCGCAAGC CGTCGACCAACTGAGACTCA | (AG)22 | 100–110 | 1 | 1 | 1 | 2 | 0.3744 | |
| UDP98-405 | ACGTGATGAACTGACACCCA GAGTCTTTGCTCTGCCATCC | (AG)9 | 100–150 | 2 | 3 | 2 | 4 | 0.6975 | |
| Total | 46 | 55 | 40 | 101 | |||||
| Average | 1.9 | 2.3 | 1.7 | 4.2 |
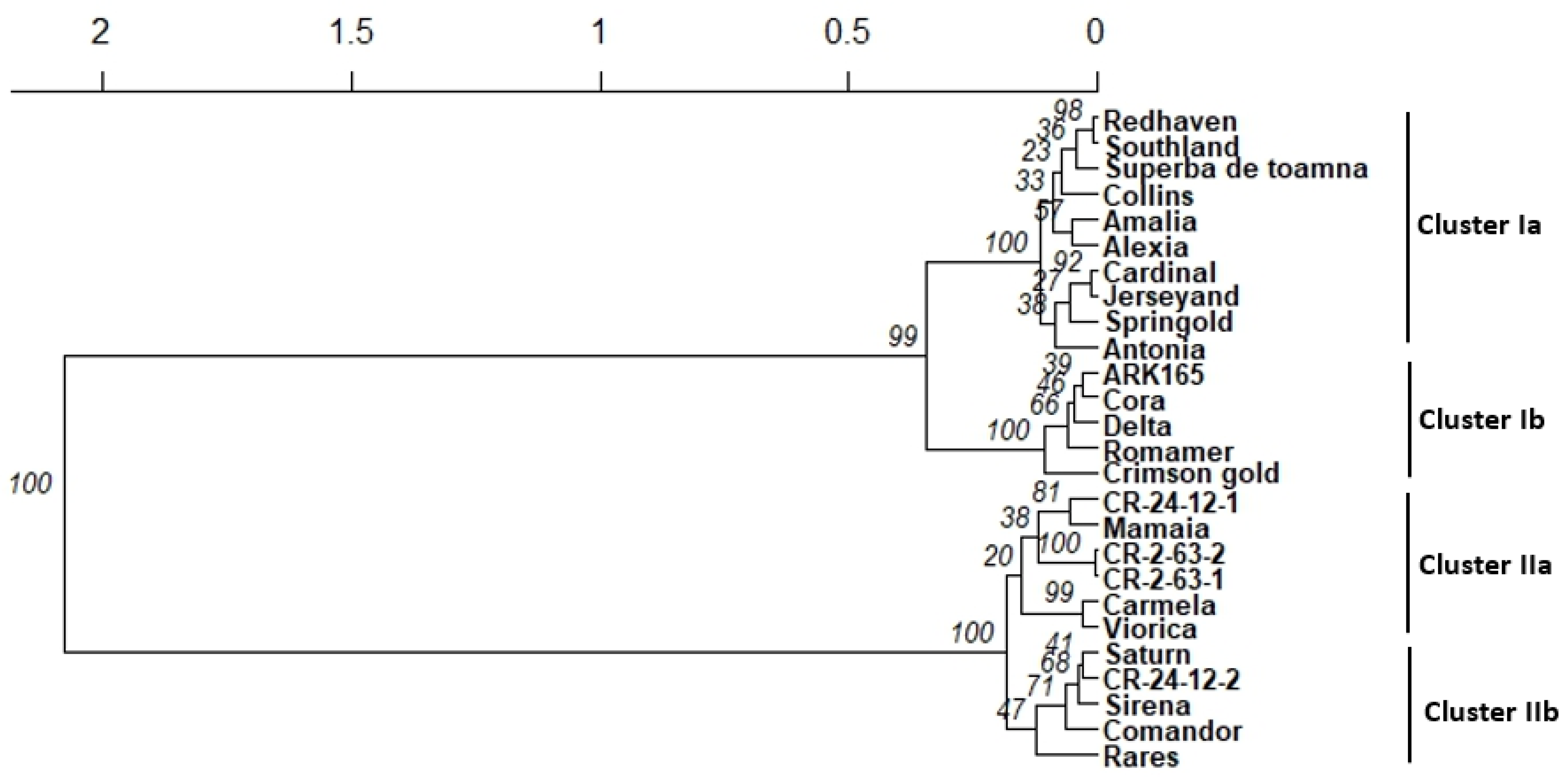
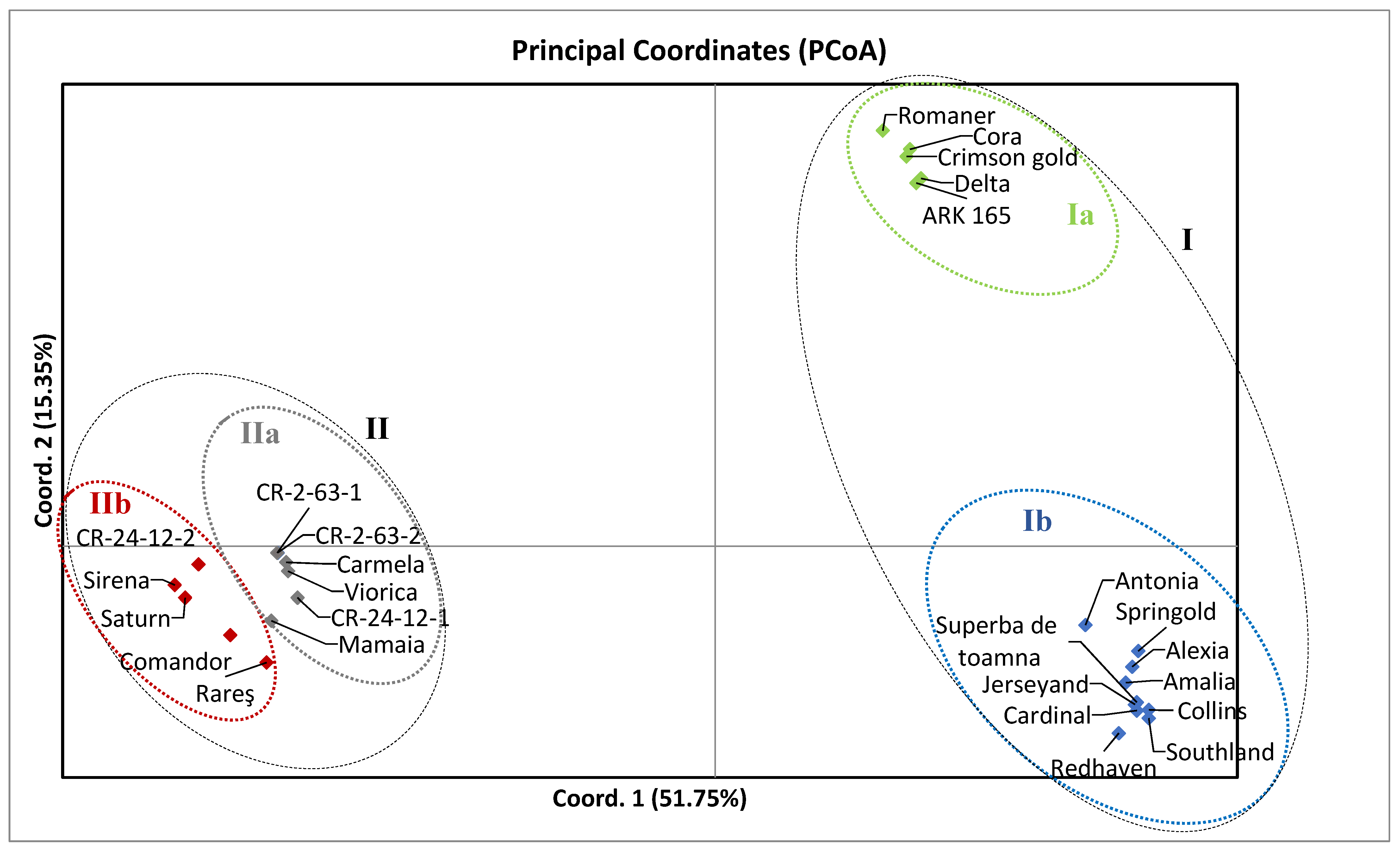
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Vavilov, N.I. Phytogeographical basis of plant breeding. In: Chester KS (transl.), The origin, variation, immunity and breeding of cultivated plants. Chron. Bot. 1951, 13, 13–54. [Google Scholar]
- Rostova, N.S.; Sokolova, E.A. Variability of anatomical and morphological leaf characters in apricot (Armeniaca scop.) species and varieties. Bull. Appl. Bot. Genet. Plant Breed. 1992, 146, 74–86. (In Russian) [Google Scholar]
- Kostina, K.F. The use of varietal resources of apricots for breeding. Trud. Nikit. Bot. Sad. 1969, 40, 45–63. [Google Scholar]
- Martín, C.; Herrero, M.; Hormaza, J.I. Molecular Characterization of Apricot Germplasm from an Old Stone Collection. PLoS ONE 2011, 6, e23979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verde, I.; Abbott, A.G.; Scalabrin, S.; Jung, S.; Shu, S.; Marroni, F.; Zhebentyayeva, T.; Dettori, M.T.; Grimwood, J.; Cattonaro, F.; et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat. Genet. 2013, 45, 487–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Kang, M.; Liu, H.; Gao, J.; Zhang, Z.; Li, Y.; Wu, R.; Pang, X. High-level genetic diversity and complex population structure of siberian apricot (Prunus sibirica L.) in China as revealed by nuclear SSR Markers. PLoS ONE 2014, 9, e87381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akpınar, A.E.; Koçal, H.; Ergül, A.; Kazan, K.; Şelli, M.E.; Bakır, M.; Aslantaş, S.; Kaymak, S.; Sarıbaş, R. SSR-based molecular analysis of economically important Turkish apricot cultivars. Genet. Mol. Res. 2010, 9, 324–332. [Google Scholar] [CrossRef]
- Khadivi-Khub, A.; Yarahmadi, M.; Jannatizadeh, A.; Ebrahimi, A. Genetic relationships and diversity of common apricot (Prunus armeniaca L.) based on simple sequence repeat (SSR) markers. Biochem. Syst. Ecol. 2015, 61, 366–371. [Google Scholar] [CrossRef]
- Scorza, R.; Sherman, W.B.; Lightner, G.W. Inbreeding and co-ancestry of low chill short fruit development period freestone peaches and nectarines produced by the University of Florida breeding program. Fruit Var. J. 1988, 43, 79–85. [Google Scholar]
- Llácer, G.; Alonso, M.; Rubio, M.J.; Batlle, I.; Iglesias, I.; Vargas, F.J.; García-Brunton, J.; Badenes, M.L. Situación del material vegetal del melocotonero utilizado en España. ITEA 2009, 105, 67–83. [Google Scholar]
- Aranzana, M.J.; Carbó, J.; Arús, P. Microsatellite variability in peach [Prunus persica (L.) Batsch] cultivar identification, marker mutation, pedigree inferences and population structure. Theor. Appl. Genet. 2003, 106, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Aranzana, M.J.; Abbassi, E.-K.; Howad, W.; Arus, P. Genetic variation, population structure and linkage disequilibrium in peach commercial varieties. BMC Genet. 2010, 11, 69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, K.; Zheng, Z.; Wang, L.; Liu, X.; Zhu, G.; Fang, W.; Cheng, S.; Zeng, P.; Chen, C.; Wang, X.; et al. Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 2014, 15, 415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abidi, W.; Moreno, M.A.; Gogorcena, Y. Genetic diversity among nectarine [Prunus persica (L.) Batsch] seedlings in agronomical and biochemical fruit quality traits Journal of new sciences. Agric. Biotechnol. 2017, 48, 2888–2896. [Google Scholar]
- Llácer, G.; Alonso, J.M.; Rubio-Cabetas, M.J.; Battle, I.; Iglesias, I.; Vargas, F.J.; García-Brunton, J.; Badenes, M.L. Peach industry in Spain. J. Am. Pomol. Soc. 2009, 63, 128–133. [Google Scholar]
- Morgante, M.; Olivieri, A. PCR-amplified microsatellites as markers in plant genetics. Plant J. 1993, 3, 175–182. [Google Scholar] [CrossRef]
- Dettori, M.T.; Micali, S.; Giovinazzi, J.; Scalabrin, S.; Verde, I.; Cipriani, G. Mining microsatellites in the peach genome: Development of new long-core SSR markers for genetic analyses in five Prunus species. SpringerPlus 2015, 4, 337. [Google Scholar] [CrossRef] [Green Version]
- Sosinski, B.; Gannavarapu, M.; Hager, L.D.; Beck, E.; King, G.J.; Ryder, C.D.; Rajapakse, S.; Baird, W.V.; Ballard, R.E.; Abbott, A.G. Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. Theor. Appl. Genet. 2000, 101, 421–428. [Google Scholar] [CrossRef]
- Testolin, R.; Marrazo, T.; Cipriani, G.; Quarta, R.; Verde, I.; Dettori, T.; Pancaldi, M.; Sansavini, S. Microsatellite DNA in peach [Prunus persica (L.) Batsch] and its use in fingerprinting and testing the genetic origin of cultivars. Genome 2000, 43, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Aranzana, M.J.; Garcia-Mas, J.; Carbó, J.; Arús, P. Development and variability of microsatellite markers in peach. Plant Breed. 2002, 121, 87–92. [Google Scholar] [CrossRef]
- Aranzana, M.J.; Pineda, A.; Cosson, P.; Dirlewanger, E.; Ascasibar, J.; Cipriani, G.; Ryder, C.D.; Testolin, R.; Abbott, A.; King, G.J.; et al. A set of simple-sequence repeat (SSR) markers covering the Prunus genome. Theor. Appl. Genet. 2003, 106, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Dirlewanger, E.; Cosson, P.; Tavaud, M.; Aranzana, M.J.; Poizat, C.; Zanetto, A.; Arús, P.; Laigret, F. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor. Appl. Genet. 2002, 105, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Potter, D.; Southwick, S.M. Genotyping of peach and nectarine cultivars with SSR and SRAP molecular markers. J. Am. Soc. Hortic. Sci. 2004, 129, 204–210. [Google Scholar] [CrossRef] [Green Version]
- Wünsch, A.; Carrera, M.; Hormaza, J.I. Molecular characterization of local Spanish peach [Prunus persica (L.) Batsch] germplasm. Genet. Resour. Crop Evol. 2006, 53, 925–932. [Google Scholar] [CrossRef]
- Messina, R.; Lain, O.; Marrazzo, M.T.; Cipriani, G.; Testolin, R. New set of microsatellite loci isolated in apricot. Mol. Ecol. Notes 2004, 4, 432–434. [Google Scholar] [CrossRef]
- Maghuly, F.; Fernandez, E.B.; Ruthner, S.; Pedryc, A.; Laimer, M. Microsatellite variability in apricots (Prunus armeniaca L.) reflects their geographic origin and breeding history. Tree Genet. Genomes 2005, 1, 151–165. [Google Scholar] [CrossRef]
- Anderson, N.A. Diversity of Low Chill Peaches (Prunus persica) from Asia, Brazil, Europe and the USA. Master’s Thesis, Office of Graduate Studies of Texas A&M University, College Station, TX, USA, 2010. [Google Scholar]
- Fernández, I.; Martí, A.; Font i Forcada, C.; Kamali, K.; Rubio-Cabetas, M.J.; Wirthensohn, M.; Socias, I.; Company, R. Molecular analyses of evolution and population structure in a worldwide almond [Prunus dulcis (Mill.) D.A. Webb syn. P. amygdalus Batsch] pool assessed by microsatellite markers. Genet. Resour. Crop Evol. 2015, 62, 205–219. [Google Scholar] [CrossRef]
- Urrestarazu, J.; Errea, P.; Miranda, C.; Santesteban, L.G.; Pina, A. Genetic diversity of Spanish Prunus domestica L. germplasm reveals a complex genetic structure underlying. PLoS ONE 2018, 13, e0195591. [Google Scholar] [CrossRef] [Green Version]
- Comlekcioglu, S.; Kuden, A.B. Evaluation of peach and nectarine hybrids using SSR marker assisted selection approach. Acta Hortic. 2015, 1084, 105–111. [Google Scholar] [CrossRef]
- Minas, I.S.; Font i Forcada, C.; Dangl, G.S.; Gradziel, T.; Dandekar, A.M.; Crisosto, C.H. Discovery of non-climacteric and suppressed-climacteric bud sport mutations originating from a climacteric Japanese plum cultivar (Prunus salicina lindl.). Front. Plant Sci. 2015, 6, 316. [Google Scholar] [CrossRef] [Green Version]
- Forcada, C.; Velascom, L.; Socias, I.; Company, R.; Fernández i Martí, Á. Association mapping for kernel phytosterol content in almond. Front. Plant Sci. 2015, 6, 530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández Mora, J.R.; Micheletti, D.; Bink, M.; Van de Weg, E.; Cantín, C.; Nazzicari, N.; Caprera, A.; Dettori, M.T.; Micali, S.; Banchi, E.; et al. Integrated QTL detection for key breeding traits in multiple peach progenies. BMC Genom. 2017, 18, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Quero-García, J.; Barreneche, T.; Dirlewanger, E.; Saski, C.; Iezzoni, A. A fruit firmness QTL identified on linkage group 4 in sweet cherry (Prunus avium L.) is associated with domesticated and bred germplasm. Sci. Rep. 2019, 9, 5008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cociu, V. 50 years of apricot varieties breeding in Romania. Acta Hortic. 2001, 701, 355–358. [Google Scholar] [CrossRef]
- Botu, M. Status of the Prunus collections in Romania. In Report of a Working Group on Prunus; Maggioni, L., Lipman, E., Eds.; Biodiversity International: Rome, Italy, 2006; pp. 87–88. [Google Scholar]
- Balan, V.; Tudor, V.; Topor, E. Apricot genetics and breeding in Romania. Acta Hortic. 2007, 760, 483–489. [Google Scholar] [CrossRef]
- Catalogue of Cultivated Plants from Romania, București. 2003; pp. 69–75. (In Romanian)
- Germplasm colletions of fruit tree and strawberry from Romania. I.C.D.P. Pitești-Mărăcineni. 2006; pp. 163–205. (In Romanian)
- Doyle, J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Salin, F. Autobin. 2010. Available online: https://hal.inrae.fr/hal-02824452 (accessed on 21 March 2021).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5:genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
- Miks, S.; Bińkowski, J. Gene Calc. Available online: https://gene-calc.pl/pic (accessed on 22 March 2022).
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Des Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Rohlf, F.J. NTSyS-p.c. Numerical Taxonomy and Multivariate Analysis System (Version 2.0); Exeter Software Publishers Ltd.: Setauket, NY, USA, 1998. [Google Scholar]
- Wang, Y.; Georgi, L.L.; Zhebentyayeva, T.N.; Reighard, G.L.; Scorza, R.; Abbott, A.G. High-throughput targeted SSR marker development in peach [Prunus persica (L.) Batsch]. Genome 2002, 45, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Wünsch, A. Cross-transferable polymorphic SSR loci in Prunus species. Sci. Hortic. 2009, 120, 348–352. [Google Scholar] [CrossRef]
- Rojas, G.; Méndezm, M.A.; Muñoz, C.; Lemus, G.; Hinrichsen, P. Identification of a minimal microsatellite marker panel for the fingerprinting of peach and nectarine cultivars. Electron. J. Biotechnol. 2008, 11, 4–5. [Google Scholar] [CrossRef]
- Zhang, Q.P.; Liu, D.C.; Liu, S.; Liu, N.; Wei, X.; Zhang, A.M.; Liu, W.S. Genetic diversity and relationships of common apricot (Prunus armeniaca L.) in China based on simple sequence repeat (SSR) markers. Genet. Resour. Crop Evol. 2014, 61, 357–368. [Google Scholar] [CrossRef]
- Romero, C.; Pedryc, A.; Muñoz, V.; Llácer, G.; Badenes, M.L. Genetic diversity of different apricot geographical groups determined by SSR markers. Genome 2003, 46, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pérez, R.; Ruiz, D.; Dicenta, F.; Egea, J.; Martinez-Gomez, P. Application of simple sequence repeat (SSR) markers in apricot breeding: Molecular characterisation, protection, and genetic relationships. Sci. Hortic. 2005, 103, 305–315. [Google Scholar] [CrossRef]
- Arismendi, M.J.; Almada, R.; Pimentel, P.; Bastias, A.; Salvatierra, A.; Rojas, P.; Hinrichsen, P.; Pinto, M.; Di Genova, A.; Travisany, D.; et al. Transcriptome sequencing of Prunus sp. rootstocks roots to identify candidate genes involved in the response to root hypoxia. Tree Genet. Genomes 2015, 11, 11. [Google Scholar] [CrossRef]
- Dondini, L.; Lain, O.; Geuna, F.; Banfi, R.; Gaiotti, F.; Tartarini, S.; Bassi, D.; Testolin, R. Development of a new SSR-based linkage map in apricot and analysis of synteny with existing Prunus maps. Tree Genet. Genomes 2007, 3, 239–249. [Google Scholar] [CrossRef]
- Köse, M.A.; Çetinsağ, N.; Gürcan, K. De novo transcriptome assembly and SSR marker development in apricot (Prunus armeniaca). Turk. J. Agric. For. 2017, 41, 305–315. [Google Scholar] [CrossRef]
- Balan, V.; Oprea, M.; Drosu, S.; Chireceanu, C.; Tudor, V.; Petrisor, C. Maintenance of biodiversity of apricot tree phenotypes in Romania. Acta Hortic. 2001, 701, 199–206. [Google Scholar] [CrossRef]
- Balan, V.; Tudor, V.; Dumitrescu, E.C.; Topor, E. Genetic particularities for the biology of early apricot phenotypes created in Romania. Acta Hortic. 2010, 862, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Krška, B. Genetic Apricot Resources and their Utilisation in Breeding. In Breeding and Health Benefits of Fruit and Nut Crops; Soneji, J.R., Nageswara-Rao, M., Eds.; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar]
| Genotype | Country | Vigor of the Tree | Blooming Age | Harvest Maturity | Size of the Fruit | Shape of the Fruit | Color of the Fruit Skin | Extension Color Coverage | Color of the Pulp | Firmness of the Pulp | Taste of the Pulp | Adhesion of the Kernel to the Pulp | Origin | Compatibility | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
| Peach cultivars | |||||||||||||||
| P1 | Alexia | RO | 5 | 3 | 3 | 8 | 2 | 4 | 3 | 2 | 5 | 9 | 2 | Cross Flacara x Marygold SCDP Baneasa | SC |
| P2 | Collins | USA | 5 | 5 | 4 | 6 | 2 | 4 | 6 | 5 | 6 | 6 | 1 | Jerseyland x NJ188 (=Raritan Rose x NJ125). NJ125 = NJ66325 op (=J.H. Halle x Goldfinch) | SC |
| P3 | Amalia | RO | 5 | 3 | 5 | 7 | 3 | 3 | 3 | 3 | 5 | 8 | 1 | Roubidoux x Flacara | SC |
| P4 | Jerseyland | USA | 7 | 5 | 6 | 7 | 3 | 5 | 8 | 5 | 7 | 7 | 1 | NJ104325 op [= J.H. Halle x NJ41 (=Slappey x Dewey)] | SC |
| P5 | Springold | USA | 5 | 4 | 2 | 4 | 2 | 4 | 9 | 5 | 7 | 7 | 3 | Cross [(Fireglow x Hiley) x Fireglow] x Springtime | SC |
| P6 | Cardinal | USA | 7 | 4 | 4 | 6 | 1 | 5 | 7 | 5 | 7 | 6 | 3 | Self-fertilization of the Redhaven variety | SC |
| P7 | Antonia | RO | 4 | 3 | 4 | 7 | 3 | 3 | 8 | 3 | 5 | 8 | 2 | - | SC |
| P8 | Southland | USA | 5 | 3 | 6 | 7 | 3 | 4 | 8 | 3 | 5 | 8 | 1 | Self-fertilization of the Halehaven variety | SC |
| P9 | Redhaven | USA | 5 | 5 | 6 | 6 | 1 | 5 | 7 | 6 | 5 | 6 | 1 | Cross Halehaven x Kalehaven | SC |
| P10 | Superbă de toamnă | RO | 5 | 4 | 7 | 7 | 3 | 3 | 9 | 2 | 5 | 8 | 1 | Elberta x Mayflower | SC |
| Apricot cultivars | |||||||||||||||
| C1 | Rareş | RO | 3 | 3 | 1 | 6 | 3 | 5 | 5 | 5 | 5 | 7 | 1 | Cross B12/6 x NJA13 | SC |
| C2 | Mamaia | RO | 5 | 8 | 7 | 6 | 3 | 6 | 5 | 6 | 5 | 7 | 1 | Complex hybridization between Marculesti1 (Ananas x Ananas) x Marculesti 5 (Targii de Bucuresti x Ananas) | SC |
| C3 | Comandor | RO | 6 | 7 | 6 | 8 | 3 | 4 | 5 | 5 | 5 | 7 | 1 | Cross B 17/52 x Mr 43/1 | SC |
| C4 | CR-2-63-1 (Cream Ridge 2–63) | USA | American selection | SI | |||||||||||
| C5 | CR-2-63-2 (Cream Ridge 2–63) | USA | 6 | 4 | 4 | 7 | 2 | 6 | 3 | 7 | 3 | 7 | 1 | American selection | SI |
| C6 | CR-24-12-1 | USA | 7 | 2 | 2 | 8 | 1 | 5 | 5 | 7 | 7 | 9 | 1 | American selection | |
| C7 | CR-24-12-2 | USA | American selection | ||||||||||||
| C8 | Saturn | RO | 5 | 6 | 5 | 6 | 2 | 4 | 5 | 7 | 9 | 5 | 2 | Hybridization of Marculesti selection 40 | SC |
| C9 | Viorica | RO | 5 | 5 | 3 | 8 | 1 | 6 | - | 7 | 7 | 7 | 1 | Cross B 3/9 (P1) x NJA20 | SC |
| C10 | Sirena | RO | 5 | 7 | 8 | 7 | 4 | 5 | 4 | 6 | 6 | 7 | 1 | Cross Mr 37/1 x Mr 21/50 | SC |
| C11 | Carmela | RO | 5 | 6 | 5 | 8 | 3 | 5 | 7 | 8 | 6 | 7 | 1 | Cross Farmigdale x NJA20 | SC |
| Nectarine cultivars | |||||||||||||||
| N1 | Crimsongold | USA | 7 | 3 | 4 | 5 | 3 | 6 | 9 | 5 | 5 | 7 | 2 | Nectarine selection x July Elberta | SC |
| N2 | Romamer | RO | 7 | 5 | 2 | 5 | 4 | 6 | 3 | 4 | 5 | 5 | 1 | Cross 624029148 x RR 48–153 | SC |
| N3 | Delta | RO | 5 | 5 | 4 | 5 | 3 | 5 | 7 | 5 | 5 | 8 | 2 | Romania, SCDP-Constanta | SC |
| N4 | Cora | RO | 5 | 5 | 6 | 7 | 3 | 5 | 7 | 5 | 6 | 7 | 2 | Romania, SCDP-Constanta | SC |
| N5 | ARK 165 | USA | 3 | 1 | 6 | 6 | 6 | - | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Butiuc-Keul, A.; Coste, A.; Postolache, D.; Laslo, V.; Halmagyi, A.; Cristea, V.; Farkas, A. Molecular Characterization of Prunus Cultivars from Romania by Microsatellite Markers. Horticulturae 2022, 8, 291. https://doi.org/10.3390/horticulturae8040291
Butiuc-Keul A, Coste A, Postolache D, Laslo V, Halmagyi A, Cristea V, Farkas A. Molecular Characterization of Prunus Cultivars from Romania by Microsatellite Markers. Horticulturae. 2022; 8(4):291. https://doi.org/10.3390/horticulturae8040291
Chicago/Turabian StyleButiuc-Keul, Anca, Ana Coste, Dragoș Postolache, Vasile Laslo, Adela Halmagyi, Victoria Cristea, and Anca Farkas. 2022. "Molecular Characterization of Prunus Cultivars from Romania by Microsatellite Markers" Horticulturae 8, no. 4: 291. https://doi.org/10.3390/horticulturae8040291
APA StyleButiuc-Keul, A., Coste, A., Postolache, D., Laslo, V., Halmagyi, A., Cristea, V., & Farkas, A. (2022). Molecular Characterization of Prunus Cultivars from Romania by Microsatellite Markers. Horticulturae, 8(4), 291. https://doi.org/10.3390/horticulturae8040291






