Abstract
Chinese cherry (Cerasus pseudocerasus (Lindl.) G.Don) is an economically important tetraploid fruiting cherry species native to China. Simple sequence repeats (SSRs)—due to their codominance, polymorphism, and stability—have been widely applied in genetic identification and trait-association analysis. In this study, using comparative genomics strategy and the data of one high-quality whole genome and seven preliminarily assembled genome sequences, we constructed a database containing 25,779 polymorphic SSR loci to efficiently develop novel markers. Sixty-four SSR loci covering eight linkage groups were selected to design primer pairs. Sixty (93.75%) primer pairs yielded specific bands and 32 (50.00%) exhibited moderate-to-high levels of informativeness (PIC ranging from 0.264 to 0.728) in 94 Chinese cherry accessions. A total of 38 primer pairs exhibited high transferability across 13 Cerasus taxa. The marker SAUCps203 was species-specific in C. pseudocerasus by checking with 114 accessions from Cerasus and 16 relatives, suggesting its potential application in accurate identification of Chinese cherry or its interspecific hybrid. Moreover, 1081 out of 1122 individuals from three cross F1 populations of Chinese cherry were identified as true hybrid offspring by using only five SSR markers. Trait association analysis suggested that 20 SSR loci were significantly associated with soluble solids and fruit size, with explained phenotypic variance ranging from 9.02% to 26.35%. This study will provide a basis for SSR-based germplasm identification and further marker-assisted selection (MAS) of Chinese cherry.
1. Introduction
Chinese cherry (Cerasus pseudocerasus), belonging to the genus Cerasus of the family Rosaceae, is an important tetraploid fresh fruit crop (2n = 4x = 32) [,]. It originates in Southwest China and is widely distributed in the North and East China Plain, Qinling Mountains, Longmenshan Fault Zones, and Yun-Gui Plateau []. Its cultivation history dates back to 3000 years ago in China []. Chinese cherry is characterized by early bloom and maturity, and is full flavored, with rich nutritional ingredients and trace elements, wide adaptability, and intensive disease/pest resistance []. Therefore, the cherry industry is developing rapidly and has increasingly contributed to the rural vitalization of China.
During the past decade, we have carried out field investigation and assessment of cherry resources across its locality in China [,]. Some disadvantages—including small fruit size, high acid content, and short shelf-life—have hindered the development of Chinese cherry industry to a certain extent [,,]. Then we conducted intra- and inter-specific crossing by selecting excellent germplasm collections to breed new cultivars with early-matured, large, full-flavored fruit with a long shelf-life, and to accelerate genetic improvement of Chinese cherry. So far, we have obtained over 2000 individuals derived from five cross F1 populations of Chinese cherry [,]. Among them, two landraces (NZH and HF) and one semi-wild germplasm (PJHH) with obvious character differences (Table S1) as parents have bred outstanding offspring. It is urgent to identify its authenticity to speed up further application.
Marker-assisted identification/selection (MAS) is an active way to get DNA markers associated with valuable traits that accelerate the breeding process by replacing the selection with a phenotype to the selection at the DNA or gene level, which has been widely employed in fruit trees [,]. SSRs, or microsatellites, are short tandem repeats of one to six nucleotides, which are widely distributed in coding and non-coding regions of the whole genome [,]. To date, SSRs have been an effective tool in DNA-based diagnostics—especially marker-assisted selection and genetic determination—due to their desirable genetic attributes with multi-allelic nature, codominant inheritance, and high reproducibility []. Over the past few decades, with the public release of genomes of Rosaceae crops (GDR, https://www.rosaceae.org, accessed on 17 February 2022) [], large-scale SSR markers have been developed and applied for the marker assisted selection, genetic map construction, and trait-associated determination in the RosBREED project (https://www.rosbreed.org/, accessed on 17 February 2022) [,,,]. The availability of efficient and practical SSR markers is limited, and only 28 SSRs have been developed for the Chinese cherry [,].
Here, we developed novel genomic-SSR markers based on one high-quality whole genome and seven preliminarily assembled genome sequences, and evaluated their cross-species transferability and their utility in hybrid identification and trait association analysis by using 1236 samples of Chinese cherry and its relatives. Our objectives are: (i) to construct a genome-wide polymorphic SSR loci database for Chinese cherry; (ii) to develop novel SSR markers and evaluate their polymorphism and transferability within Chinese cherry and 17 relatives of Cerasus; (iii) to identify the authenticity of 1122 individuals from three cross F1 populations; and (iv) to analyze three fruit quality traits associated SSR markers using phenotypic data of 65 Chinese cherry landraces.
2. Materials and Methods
2.1. Plant Materials
In this study, eight genomes (unpublished data) were sequenced with pair-end sequencing method on Illumina Hiseq2000 for SSR marker development: LuYg (the whole genome); and whole-genome resequencing of CQhzz, CBJ4, CBJ7, CPZB2, CXC1, CYX4, and WQX13 with an average depth of 5.53×. Among them, sequenced samples whose names are marked in green/red and bold were listed in Table S1. The seven landraces exhibited rich diversity in important economic characters based on field investigation and assessment of cherry resources across China [,,]. A total of 1236 accessions were used for SSR marker evaluation and application, which contained 94 C. pseudocerasus accessions (92 landraces and 2 wild accessions), representing diverse genotypes, phenotypes, and geographical distributions [,,]; 17 accessions from 13 Subg. Cerasus and Subg. Microcerasus taxa (Table S2); and 1122 F1 individuals from HF × PJHH and reciprocal crosses of NZH and HF (Table S1). All of the F1 seedlings were planted in the orchard of Sichuan Agricultural University (Chengdu campus), China.
2.2. SSR Identification, Primer Design, and Selection
Taking the LuYg as the reference genome, seven draft genomes were constructed using Chromosome software []. SSR motifs were screened using the MISA Perl script (http://pgrc.ipk-gatersleben.de/misa, accessed on 17 February 2022) with the following parameters: repeat motifs of 2–6 bp and minimum repeat numbers of 6, 5, 5, 5, and 5 for di-, tri-, tetra-, penta-, and hexa-nucleotides, respectively. The sequences of SSR loci and their 600-bp flanking regions were extracted [] and used to carry out homology comparison analyses in BlastN.
Sixty-four SSR loci evenly distributed across eight linkage groups were selected from the polymorphic SSR database for primer design. Primer pairs were designed in Software Primer 6.0 with the following parameters: (1) the size of the primer was between 18 and 27 bp; (2) the size of the amplicon was from 100 to 400 bp; (3) the melting temperature ranged from 50 to 60 °C; and (4) the GC content ranged from 40% to 65%.
2.3. PCR Amplification and Gel Electrophoresis
Total genomic DNA was extracted from silica gel-dried leaf tissues following the modified cetyltrimethyl ammonium bromide (CTAB) method []. PCR amplifications were performed in 20 µL reactions following Zhang et al. []. PCR products were detected through 8% polyacrylamide gels after electrophoresis.
2.4. Data Analysis
Data scoring of SSR fragments was performed using Quantity One software (Bio-Rad, Hercules, CA, USA). DNA marker (20 bp, TAKARA) was used to estimate the molecular size of the SSR fragments. The number of alleles (Na), effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He), and allele frequency were calculated with POPGENE 32 []. Polymorphism information content (PIC) was calculated using PowerMarker 3.25 []. NTSYS-pc2.11 software [] was used to construct a neighbor-joining tree using an arithmetic average (UPGMA) algorithm based on Nei’s genetic distance (Nei et al., 1983).
2.5. Hybrids Identification
Five new SSR markers were selected for hybrid identification (Table S3) and the results of all primers were combined to confirm hybrid authenticity. The amplified bands of the hybrids were classified into three types: (I) paternal-specific bands; (II) maternal-specific bands; and (III) parental-shared bands. F1 seedlings with bands other than type I, II, or III were considered foreign origins. After excluding foreign origins, F1 seedlings with paternal-specific bands were considered true hybrids and individuals with the absence of paternal-specific bands were defined as maternal hybrids.
2.6. Association Analysis
Association analysis was conducted in TASSEL 3.0 software [] for total soluble solids (TSS), fruit longitudinal diameter (cm), and fruit transverse diameter (cm) of 65 Chinese cherry landraces (Table S2). Each specific band amplified by every SSR marker was analyzed as an independent locus. A general linear model (GLM) was employed, the Q matrix of population structure as a covariate was created using the model-based Bayesian clustering algorithm in STRUCTURE v2.3.4 []. Based on the ΔK statistic approach described by Evanno et al. [], the best K was calculated and visualized using the program STRUCTURE HARVESTER []. Adopting more stringent restrictions, the significance of association analysis results was classified into four grades (p-value < 0.01, <0.005, <0.002, and <0.001). The physical positions of associated markers were mapped on the genomes of Chinese cherry (Ppseudocerasus_Genome), sweet cherry (PAV_r1.0) [], and peach (Prunus_persica_v2.0) [], respectively.
3. Results
3.1. Development of SSR Markers
A total of 49,756 microsatellites were identified across the whole genome sequence of LuYg. Based on the comparative genomic analyses (Figure 1a), 25,779 polymorphic SSR loci with various motif repeats were detected, accounting for 51.81% of total SSR loci in the LuYg genome (Figure 1b). The repeats and positions of each differential SSRs in eight genomes were extracted consecutively, then the extracted information was reordered to form the differential SSR database. Among the eight linkage groups, the greatest number of differential SSRs (5762) was observed on LG1, and the least number (1409) was detected on LG7. Among polymorphic SSRs, 64 loci were randomly selected to design primers and were amplified in 11 Chinese cherry accessions. Sixty (93.75%) primer pairs produced clear and specific bands and 39 (60.94%) primer pairs were polymorphic (Table 1). The newly developed SSR markers were mapped to the genome of Chinese cherry sequenced on the Pacbio RS II platform (Figure 2).
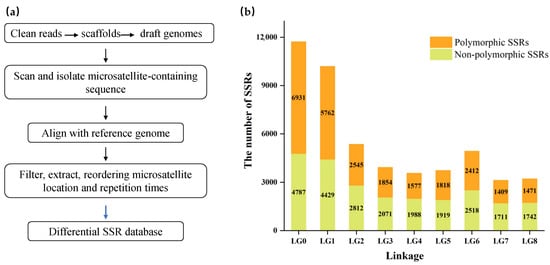
Figure 1.
Construction of differential SSR database. (a) Flow chart of SSR marker development; (b) The number of non-polymorphic/polymorphic SSRs across the Chinese cherry genome in differential SSR database. LG0: the number of differential SSRs from the reads unmapped on LG1–LG8 were counted into LG0.

Table 1.
Thirty-nine developed SSR markers and their transferability across Cerasus pseudocerasus and relatives.
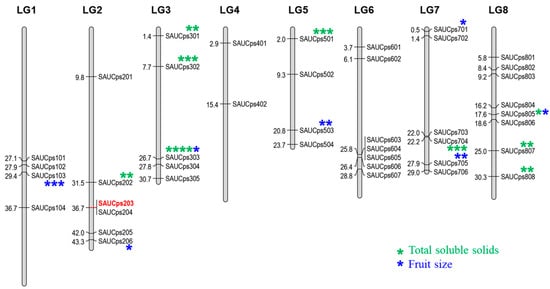
Figure 2.
Graphical distribution of new SSR markers among linkage groups of Chinese cherry. SAUCps203 represents the SSR markers with species-specificity. (*: p < 0.01; **: p < 0.005; ***: p < 0.002; ****: p < 0.001).
3.2. Characteristics of SSR Markers
3.2.1. Transferability in Chinese Cherry and Relatives
The transferability of 39 developed genomic SSR markers was evaluated in 94 Chinese cherry accessions (92 cultivated and 2 wild) and 13 Cerasus taxa (Table 1). Nineteen markers can be successfully amplified in all 94 Chinese cherry accessions, showing a 100% transferability rate. The remaining markers were applicable to 66–93 Chinese cherry accessions. Both SAUCps604 and 103 markers failed to amplify in two wild accessions.
All 39 SSR markers revealed cross-species transferability within Cerasus, and eight of them were applicable to all 13 taxa of Cerasus. These markers exhibited relatively high transferability ranging from 79.49% for C. mahaleb to 100.00% for C. avium × C. pseudocerasus among nine Subg. Cerasus taxa, while they showed lower transferability ranging from 53.85% for C. glandulosa to 74.36% for C. humilis among Subg. Microcerasus taxa. Therefore, the transferability of SSR markers was higher in the species more closely related to Chinese cherry.
3.2.2. Specificity in Chinese Cherry
Interestingly, we found a key SSR marker, SAUCps203, that was species-specific to Chinese cherry. All C. pseudocerasus accessions were successfully amplified, reproducing specific bands of approximately 260 bp (Figure 3). A specific band was also observed in the ‘Landing 2′, an interspecific hybrid of C. avium × C. pseudocerasus. No bands were found in any other accessions of Cerasus, Amygdalus, and Armeniaca (Figure 3).

Figure 3.
PCR profile of the SSR marker SAUCps203 with species-specificity in Cerasus pseudocerasus. Note: CER, Subgenus Cerasus; 1–94, C. pseudocerasus; 95–98, C. avium; 99, C. avium × C. pseudocerasus; 100, C. vulgaris × P./C. canescens and 101–102, C. vulgaris; 103–107, C. campanulate, C. yedoensis, C. serrulate, C. cerasoides, and C. mahaleb, respectively; MIC, Subgenus Microcerasus, including 108–111, C. humilis, C. glandulosa, C. japonica, and C. tomentosa, respectively; ARM, 112, Arm. mume; AMY: 113–114, Amy. persica and Amy. triloba, respectively (Table S2). The red arrow indicates the specific band.
3.2.3. Polymorphism in Chinese Cherry
A total of 143 allelic loci were detected among 94 Chinese cherry accessions using the 39 polymorphic markers, which meant that each marker corresponded to 3.6 allelic loci on average (ranging from 2 to 6), with an average major allele frequency (MAF) of 0.64 (ranging from 0.32 to 0.98), and mean polymorphic information content (PIC) of 0.415 (ranging from 0.039 to 0.728) (Table S4). Thirty-two newly developed SSR primer pairs exhibited moderate-to-high (PIC ≥ 0.25) levels of informativeness (Table S4). According to the UPGMA cluster analysis, the similarity coefficient ranged from 0.42 to 0.99 (Figure S1). Two different clusters were observed in the cluster tree. Cluster I mainly consisted of the Chinese cherry accessions from the Sichuan Basin and Yunnan-Guizhou Plateau, and cluster II mainly included accessions from the North China Plain and East China Plain.
3.3. Application of SSR Markers
3.3.1. Hybrid Identification
Five SSR markers succeeded in identifying a total of 1122 individuals, 1081 of which were true hybrid offsprings, 20 may be foreign origins, and the remaining 21 needed further identification due to the absence of paternal-specific bands (Table 2 and Figure 4). True hybrid offspring dominated extremely, ranging from 91.72% to 97.12% with an average of 96.34% among three cross F1 populations. A total of 204 and 103 true hybrids were identified by four SSR markers (SAUCps303, 102, 706, and 202) in reciprocal F1 populations between NZH and HF respectively. Totally, 744 out of 766 individuals were true hybrids of HF×PJHH identified by three markers (SAUCps706, 202, and 304). In addition, SSR markers greatly differed from each other in efficiency, ranging from 25.84% for SAUCps102 to 82.00% for SAUCps706. Especially, SAUCps706 showed the highest efficiency and universality, being able to identify all 903 true hybrids and 17 foreign origins in three cross F1 populations.

Table 2.
Identification of true hybrids and foreign origins of 1122 F1 individuals using five SSR markers.
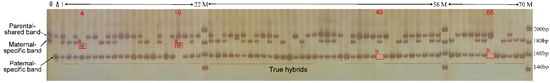
Figure 4.
Banding patters based upon SSR marker SAUCps706 among F1 individuals derived from the cross population of NZH×HF. ♀: female parent; ♂: male parent; M: DNA marker. 1–70 represented part of the F1 individuals, no. 4 and no. 19 showed foreign origins, and no. 49 and no. 65 were maternal-type individuals. a: Non-parental shared, maternal-specific band, or paternal-specific band; b: the absence of paternal-specific band.
3.3.2. Trait-Marker Association Analysis
Sixty-five Chinese cherry landraces were divided into two groups with a distinct maximum of delta K (K = 2) by 39 SSR markers with 126 polymorphic loci (Figures S2 and S3). Based on the GLM, 10 loci TSS-associated were mapped on linkage groups 2, 3, 5, 7, and 8; and 10 loci associated for fruit size were mapped on six linkage groups (1–3, 5, 7, and 8) (Table 3). Adopting stringent restrictions, eight loci, four loci, and one locus were significantly associated with TSS at the p = 0.005, 0.002, 0.001 levels, respectively, with phenotypic variance explained (R2) from 16.72% to 26.35%. Two and one loci were significantly associated with fruit size respectively at the p = 0.005 and p = 0.002 levels, with R2 ranging from 12.60% to 17.94%. TSS-associated markers SAUCps302–255 and 302–234 (5.97 Mb), SAUCps303–330 (18.92 Mb) were located in the first QTL qP-TSS3.1m (4.50–21.85 Mb) for TSS in sweet cherry genome [], and SAUCps303–330 (24.59 Mb) were close to the peak association signals Glu.2013.3.1 (24.09 Mb) for glucose content in peach []. Based on the genome annotation of Chinese cherry, six potential candidate genes were searched in the space defined by ±10 kb on either side of the significantly associated markers (Table 3). Only one gene, scaffold_7_1573, possibly regulate TSS accumulation by responding to salicylic acid. For fruit size, three genes (scaffold_8_1113, 1115, and 1116) on LG8 may response to auxin, scaffold_2_3446 regulate cell differentiation and scaffold_3_2691 may be involved in pectin biosynthetic process.

Table 3.
Three fruit traits associated with SSR loci and candidate genes.
4. Discussion
4.1. Efficient Development of SSR Markers
In recent years, tremendous efforts dedicated to NGS-based sequencing have expedited the generation of SSR-enriched DNA libraries and ESTs (expressed sequence tags) []. In spite of accessibility to numerous SSR loci of diverse Rosaceae crops [,,], significant efforts, time, and cost/resources are required []. The effective strategy used in this study is advantageous for screening informative SSR markers from large-scale markers by assessing their potential to detect fragment length polymorphism among different cultivated and wild Chinese cherry accessions. Similar strategies have been reported in rice [], wheat [], Nicotiana [], and pear []. The efficiency of novel SSR markers was ascribed to greater potential of detecting amplification (93.75%) and polymorphism (59.38%), and application value compared with previous development of SSR makers relied either on genome sequencing (polymorphism 9.50%, 19/200) [] or ESTs (polymorphism 5.00%, 8/160) [] of single Chinese cherry accessions. Therefore, the database containing 25,779 polymorphic SSR loci will serve as a vital genomic resource for driving genomics-assisted breeding applications and genetic enhancement in Chinese cherry. Although this strategy is of high efficiency and high throughput, the SSR marker prediction of polymorphism is still difficult to make completely consistent with the PCR results. This issue may be caused by the phenomenon of “sliding” [] or errors in sequencing data []. As sequencing quality improves, we believe that more crop sequencing data released for constructing polymorphic SSR databases will support more accurate predictions [].
4.2. Efficient Application of Novel Markers
Species identification is essential for large-scale biodiversity monitoring and conservation []. Along with the morphological similarities that prevail, Chinese cherry interspecies and variety level make species identification more difficult []. The monomorphic fragment (260 bp) obtained with SAUCps203 was common and unique in all C. pseudocerasus lineages, which can be used for fast and accurate identification of C. pseudocerasus. This finding contributes a valuable tool for C. pseudocerasus lineage protection and discrimination and can also be an aid to taxonomic study.
Chinese cherry is a perennial woody plant that needs more than three years to show vegetative and reproductive characteristics. It is necessary to identify the authenticity of F1 seedlings to shorten the period to breed true hybrids for future breeding programs. Here, over 1000 F1 individuals were identified as true hybrids by using only five SSR markers, which exhibited the great potential for accurate identification of large populations due to its codominant inheritance.
Fruit size and TSS content impact consumer acceptance and are considered as major selection criteria in cherry breeding. As glucose is the major sugar in cherry impacting TSS content [], we speculate that SAUCps303–330—close to glucose-associated peak signals in a previous study []—may serve as a potential functional marker for mining TSS-related genes. According to the genome annotation results of Chinese cherry, scaffold_2_3446 at 5.96 kb downstream of SAUCps206–171 is putatively homologous to the MSI4 gene which negatively regulates the cell cycle that may affect cell growth and division []. Additionally, scaffold_8_1115 is predicted as a homologous gene of SAUR20 [], which was speculated to increase fruit size by accelerating cell expansion in the mesocarp during the second rapid expansion period of Chinese cherry. The above results might contribute to the body of knowledge on the genetic control of important fruit traits in Chinese cherry.
5. Conclusions
This study constructed a database containing 25,779 polymorphic SSR loci of Chinese cherry based on comparative genomic analyses. Among 64 new markers, 32 were moderately to highly polymorphic and 38 exhibited high transferability across 13 Cerasus taxa. SAUCps203, species-specific to C. pseudocerasus, was of great value for C. pseudocerasus lineage discrimination and protection. The 1081 true hybrids identified by five SSR markers are valuable genetic material for further study. Twenty SSR loci were estimated to be significantly associated with TSS content and fruit size, exhibiting potential application in discovery of candidate genes. This work presents a major advance of Chinese cherry in the confirmation and identification of SSR markers in genomics-assisted breeding applications and provides a reference for the efficient identification of QTLs underlying complex traits in cherry.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8030222/s1, Table S1: F1 populations and their parents’ phenotype characters; Table S2: List of information for each accession in this study; Table S3: Information of five SSR markers for hybrids identification; Table S4: Characteristics of 39 microsatellite loci across 94 Chinese cherry accessions; Figure S1: Phenograms by UPGMA cluster analysis of 94 Chinese cherry accessions; Figure S2: Population structure distribution of 65 Chinese cherry accessions; Figure S3: Curve diagram of K value.
Author Contributions
Z.L. performed the experiments, analyzed the data, and drafted the manuscript. J.Z., designed the experiments and revised manuscript. Y.W., H.W., L.W., L.Z., M.X., W.H., S.Y., Q.C., T.C., Y.L., Y.Z. and H.T. provided the analysis and experiment support. Y.W. revised the manuscript. X.W. conceived the project and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (no. 31672114, no. 31801826), Sichuan Science and Technology Program (no. 2019JDTD0010, no. 2021065), Cherry Resources Sharing and Service Platform of Sichuan Province and Shuangzhi Project Innovation Team of Sichuan Agricultural University (no. P202107).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
The data used for the analysis in this study are available within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yü, D. Taxonomy of Fruit Trees in China; Agricultural Press: Beijing, China, 1979. [Google Scholar]
- Wang, Y.; Du, H.; Zhang, J.; Chen, T.; Chen, Q.; Tang, H.; Wang, X. Ploidy level of Chinese cherry (Cerasus pseudocerasus Lindl.) and comparative study on karyotypes with four Cerasus species. Sci. Hortic. 2018, 232, 46–51. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Y.; Chen, T.; Chen, Q.; Wang, L.; Liu, Z.; Wang, H.; Xie, R.; He, W.; Li, M.; et al. Evolution of Rosaceae plastomes highlights unique Cerasus diversification and independent origins of fruiting cherry. Front. Plant Sci. 2021, 12, 736053. [Google Scholar] [CrossRef]
- Yü, D.; Li, C. Flora of China; Science Press: Beijing, China, 1986. [Google Scholar]
- Chen, T.; Li, L.; Zhang, J.; Huang, Z.; Zhang, H.; Liu, Y.; Chen, Q.; Tang, H.; Wang, X. Investigation, collection and preliminary evaluation of genetic resources of Chinese cherry [Cerasus pseudocerasus (Lindl.) G. Don]. J. Fruit Sci. 2016, 33, 917–933. [Google Scholar]
- Liu, Y.; Chen, T.; Zhang, J.; Wang, J.; Wang, X. Genetic diversity analysis of Chinese cherry landraces (Prunus pseudocerasus) based on phenotypic traits. Acta Hortic. Sinica 2016, 43, 2119–2132. [Google Scholar]
- Huang, X.; Wang, X.; Chen, T.; Chen, J.; Tang, H. Research progress of germplasm diversity in Chinese cherry (Cerasus pseudocerasus). J. Fruit Sci. 2013, 30, 470–479. [Google Scholar]
- Wang, Y.; Wang, H.; Zhang, J.; Liu, Z.; Chen, Q.; He, W.; Yang, S.; Lin, Y.; Zhang, Y.; Li, M.; et al. Survey on intra-specific crossing and F1 seedling cultivation in seven combinations of Chinese cherry. Hort. J. 2022. [Google Scholar] [CrossRef]
- Du, H.; Wang, X.; Chen, T.; Zhang, H.; Du, T. Preliminary study on intraspecific hybridization of Cerasus pseudocerasus and interspecific hybridization with C. avium. Guihaia 2015, 35, 227–230. [Google Scholar]
- Calle, A.; Grimplet, J.; Le Dantec, L.; Wünsch, A. Identification and characterization of DAMs mutations associated with early blooming in sweet cherry, and validation of DNA-based markers for selection. Front. Plant Sci. 2021, 12, 621491. [Google Scholar] [CrossRef]
- Wu, B.; Shen, F.; Wang, X.; Zheng, W.; Xiao, C.; Deng, Y.; Wang, T.; Yu, H.; Zhou, Q.; Wang, Y.; et al. Role of MdERF3 and MdERF118 natural variations in apple flesh firmness/crispness retainability and development of QTL-based genomics-assisted prediction. Plant Biotechnol. J. 2021, 19, 1022–1037. [Google Scholar] [CrossRef]
- Jamali, S.H.; Cockram, J.; Hickey, L.T. Insights into deployment of DNA markers in plant variety protection and registration. Theor. Appl. Genet. 2019, 132, 1911–1929. [Google Scholar] [CrossRef]
- Hasanbegovic, J.; Hadziabulic, S.; Kurtovic, M.; Gasi, F.; Lazovic, B.; Dorbic, B.; Skender, A. Genetic characterization of almond (Prunus amygdalus L) using microsatellite markers in the area of Adriatic Sea. Turk. J. Agric. For. 2021, 45, 797–806. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the genome database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peace, C.P. DNA-informed breeding of rosaceous crops: Promises, progress and prospects. Hortic Res. 2017, 4, 17006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aranzana, M.J.; Decroocq, V.; Dirlewanger, E.; Eduardo, I.; Gao, Z.S.; Gasic, K.; Iezzoni, A.; Jung, S.; Peace, C.; Prieto, H.; et al. Prunus genetics and applications after de novo genome sequencing: Achievements and prospects. Hortic Res. 2019, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Iezzoni, A.F.; McFerson, J.; Luby, J.; Gasic, K.; Whitaker, V.; Bassil, N.; Yue, C.; Gallardo, K.; McCracken, V.; Coe, M.; et al. RosBREED: Bridging the chasm between discovery and application to enable DNA-informed breeding in Rosaceous crops. Hortic Res. 2020, 7, 177. [Google Scholar] [CrossRef]
- Tan, Q.; Li, S.; Zhang, Y.; Chen, M.; Wen, B.; Jiang, S.; Chen, X.; Fu, X.; Li, D.; Wu, H.; et al. Chromosome-level genome assemblies of five Prunus species and genome-wide association studies for key agronomic traits in peach. Hortic Res. 2021, 8, 231. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Zhu, Y.; Shao, X.; Guo, W. Development and validation of SSR markers based on transcriptomic data of Chinese cherry (Prunus pseudocerasus). Acta Hortic. Sinica 2016, 43, 1566–1576. [Google Scholar]
- Zhang, J.; Chen, T.; Wang, Y.; Chen, Q.; Sun, B.; Luo, Y.; Zhang, Y.; Tang, H.; Wang, X. Genetic diversity and domestication footprints of Chinese cherry [Cerasus pseudocerasus (Lindl.) G.Don] as revealed by nuclear microsatellites. Front. Plant Sci. 2018, 9, 238. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Hu, G.; Liu, Z.; Zhang, J.; Ma, L.; Tian, T.; Wang, H.; Chen, T.; Chen, Q.; He, W.; et al. Phenotyping in flower and main fruit traits of Chinese cherry [Cerasus pseudocerasus (Lindl.) G.Don]. Sci. Hortic. 2022, 296, 110920. [Google Scholar] [CrossRef]
- Tamazian, G.; Dobrynin, P.; Krasheninnikova, K.; Komissarov, A. Chromosomer: A reference-based genome arrangement tool for producing draft chromosome sequences. Gigascience 2016, 5, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Huang, X.; Zhang, J.; Chen, Q.; Liu, Y.; Tang, H.; Pan, D.; Wang, X. Genetic diversity and population structure patterns in Chinese cherry (Prunus pseudocerasus Lindl) landraces. Plant Mol. Biol. Rep. 2016, 34, 440–453. [Google Scholar] [CrossRef]
- Francis, C.Y.; Rong, C.Y.; Boyle, T. Popgene, Microsoft Window-Based Freeware for Population Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1999; pp. 1–31. [Google Scholar]
- Liu, J. PowerMarker: Integrated analysis environment for genetic marker data. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Rohlf, F.J. NTSYS-pc, Numerical Taxonomy and Multivariate Analysis System. Version 2.1.; State University of New York: New York, NY, USA, 2000. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.J.; Donnelly, P.J. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [Green Version]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Shirasawa, K.; Isuzugawa, K.; Ikenaga, M.; Saito, Y.; Yamamoto, T.; Hirakawa, H.; Isobe, S. The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. 2017, 24, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Verde, I.; Jenkins, J.; Dondini, L.; Micali, S.; Pagliarani, G.; Vendramin, E.; Paris, R.; Aramini, V.; Gazza, L.; Rossini, L.; et al. The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 2017, 18, 225. [Google Scholar] [CrossRef] [Green Version]
- Calle, A.; Wünsch, A. Multiple-population QTL mapping of maturity and fruit-quality traits reveals LG4 region as a breeding target in sweet cherry (Prunus avium L.). Hortic Res. 2020, 7, 127. [Google Scholar] [CrossRef] [PubMed]
- Cao, K.; Li, Y.; Deng, C.; Gardiner, S.E.; Zhu, G.; Fang, W.; Chen, C.; Wang, X.; Wang, L. Comparative population genomics identified genomic regions and candidate genes associated with fruit domestication traits in peach. Plant Biotechnol. J. 2019, 17, 1954–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Xiao, Y.; Mount, S.; Liu, Z. An Atlas of genomic resources for studying Rosaceae fruits and ornamentals. Front. Plant Sci. 2021, 12, 644881. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, C.; Lin, X.; Cui, B.; Wu, R.; Pang, X. De novo assembly and characterization of the fruit transcriptome of Chinese jujube (Ziziphus jujuba Mill.) using 454 pyrosequencing and the development of novel tri-nucleotide SSR markers. PLoS ONE 2014, 9, e106438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirakawa, H.; Shirasawa, K.; Kosugi, S.; Tashiro, K.; Nakayama, S.; Yamada, M.; Kohara, M.; Watanabe, A.; Kishida, Y.; Fujishiro, T.; et al. Dissection of the octoploid strawberry genome by deep sequencing of the genomes of Fragaria species. DNA Res. 2014, 21, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fang, Z.; Zhou, J.; Chen, H.; Hu, Z.; Gao, L.; Chen, L.; Ren, S.; Ma, H.; Lu, L.; et al. An accurate and efficient method for large-scale SSR genotyping and applications. Nucleic Acids Res. 2017, 45, e88. [Google Scholar] [CrossRef] [Green Version]
- Daware, A.; Das, S.; Srivastava, R.; Badoni, S.; Singh, A.K.; Agarwal, P.; Parida, S.K.; Tyagi, A.K. An efficient strategy combining SSR markers and advanced QTL-seq-driven QTL mapping unravels candidate genes regulating grain weight in rice. Front. Plant Sci. 2016, 7, 1535. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.; Wang, X.; Zhan, G.; Wei, G.; Zhou, X.; Zhao, J.; Huang, L.; Kang, Z. Genome-wide analysis of simple sequence repeats and efficient development of polymorphic SSR markers based on whole genome re-sequencing of multiple isolates of the wheat stripe rust fungus. PLoS ONE 2015, 10, e130362. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yang, S.; Chen, Y.; Zhang, S.; Zhao, Q.; Li, M.; Gao, Y.; Yang, L.; Bennetzen, J.L. Comparative genome-wide characterization leading to simple sequence repeat marker development for Nicotiana. BMC Genom. 2018, 19, 500. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Luo, J.; Wang, X.; Shi, C. A study on efficient screening of the primers for selecting polymorphic SSR markers based on the re-sequencing data in Pyrus. J. Fruit Sci. 2019, 36, 129–136. [Google Scholar]
- Primmer, C.R.; Ellegren, H. Patterns of molecular evolution in avian microsatellites. Mol. Biol. Evol. 1998, 15, 997–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copeland, N.; Jenkins, N.; Gilbert, D.; Eppig, J.; Maltais, L.; Miller, J.; Dietrich, W.; Weaver, A.; Lincoln, S.; Steen, R. A genetic linkage map of the mouse: Current applications and future prospects. Science 1993, 262, 57–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, X.; Song, J.; Zhu, Y.; Xu, H.; Huang, L.; Chen, S. Applying plant DNA barcodes for Rosaceae species identification. Cladistics 2011, 27, 165–170. [Google Scholar] [CrossRef]
- Gonçalves, A.C.; Campos, G.; Alves, G.; Garcia-Viguera, C.; Moreno, D.A.; Silva, L.R. Physical and phytochemical composition of 23 Portuguese sweet cherries as conditioned by variety (or genotype). Food Chem. 2021, 335, 127637. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Blanco, C.; Ruiz-García, L.; Martínez-Zapater, J.M.; Ausín, I.; Jarillo, J.A. Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat. Genet. 2004, 36, 162–166. [Google Scholar]
- Spartz, A.K.; Lee, S.H.; Wenger, J.P.; Gonzalez, N.; Itoh, H.; Inze, D.; Peer, W.A.; Murphy, A.S.; Overvoorde, P.J.; Gray, W.M. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012, 70, 978–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).