Systematic Identification of Long Non-Coding RNAs under Allelopathic Interference of Para-Hydroxybenzoic Acid in S. lycopersicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Sequencing and Identification of lncRNAs
2.3. Expression Analysis of Putative lncRNAs and Quantitative Reverse Transcription PCR (qRT-PCR) Validation
2.4. Target Gene Prediction and Coexpression Analysis of lncRNAs with PGs
2.5. Similarity Analysis of lncRNAs from S. lycopersicum and Arabidopsis thaliana
2.6. Prediction of Endogenous Target Mimics for miRNAs
2.7. GenBank Accession Numbers
3. Results
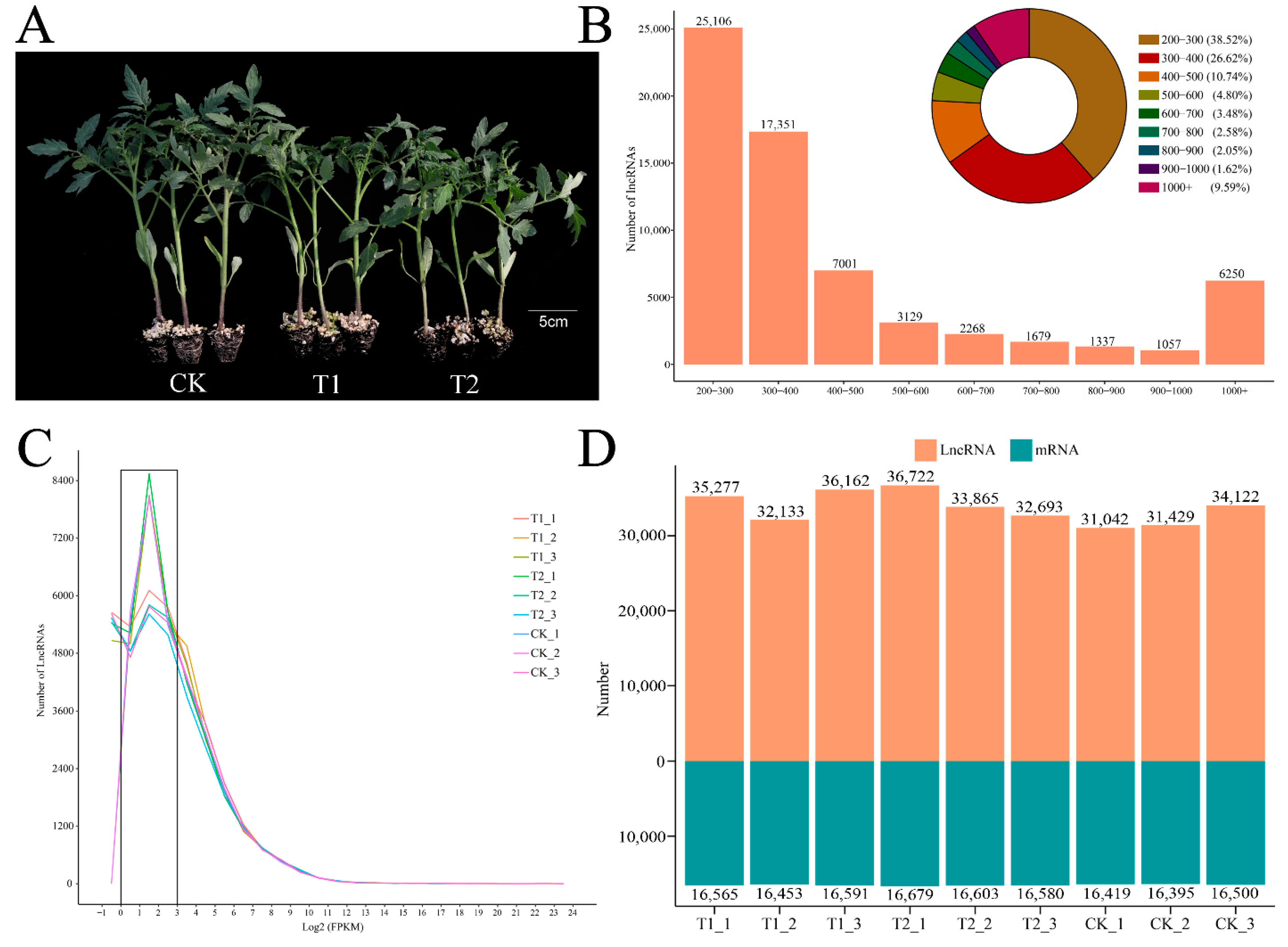
3.1. Identification of lncRNAs in S. lycopersicum
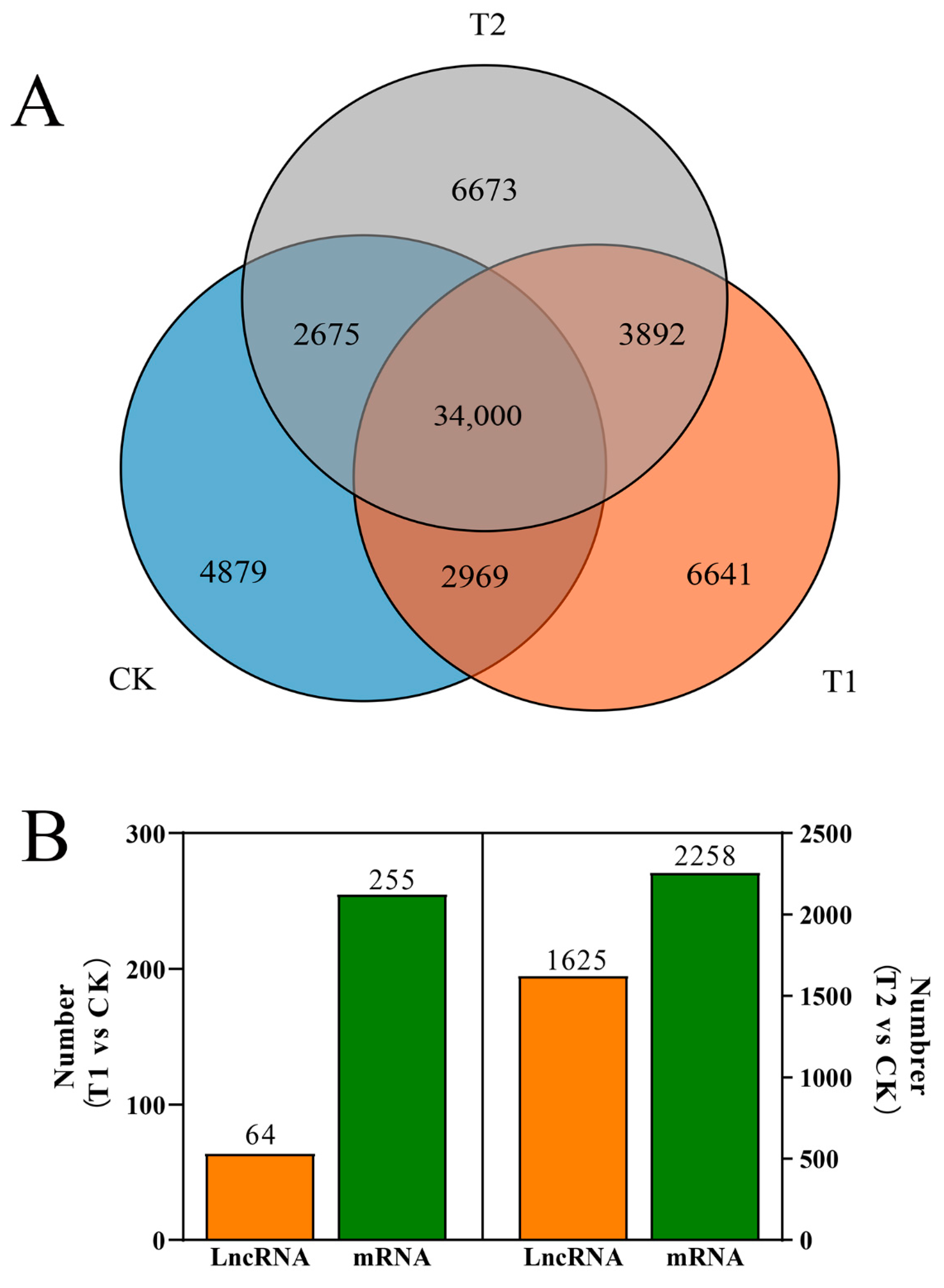
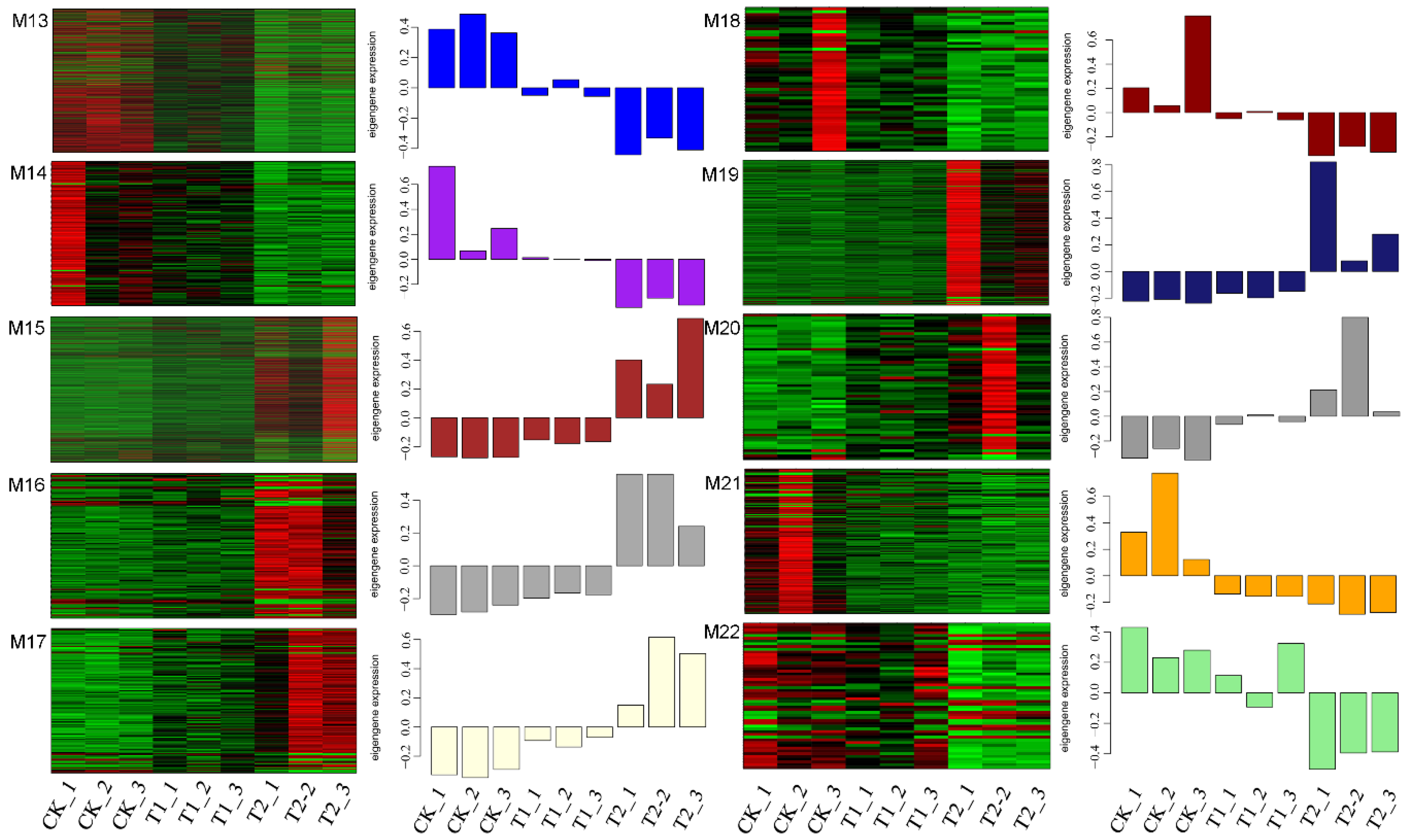
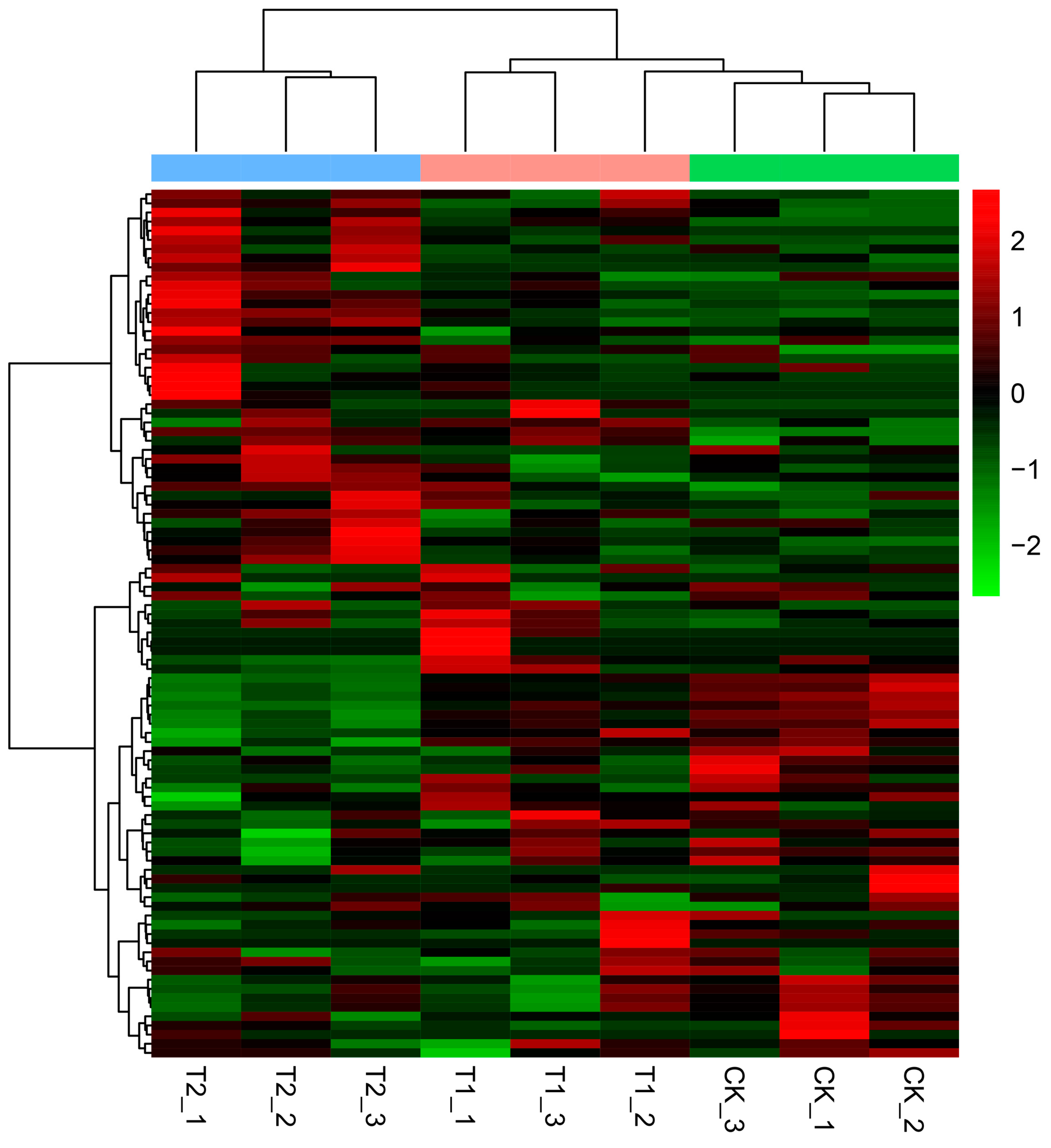
3.2. Expression of lncRNAs in S. lycopersicum Leaves under Different PHBA Treatments
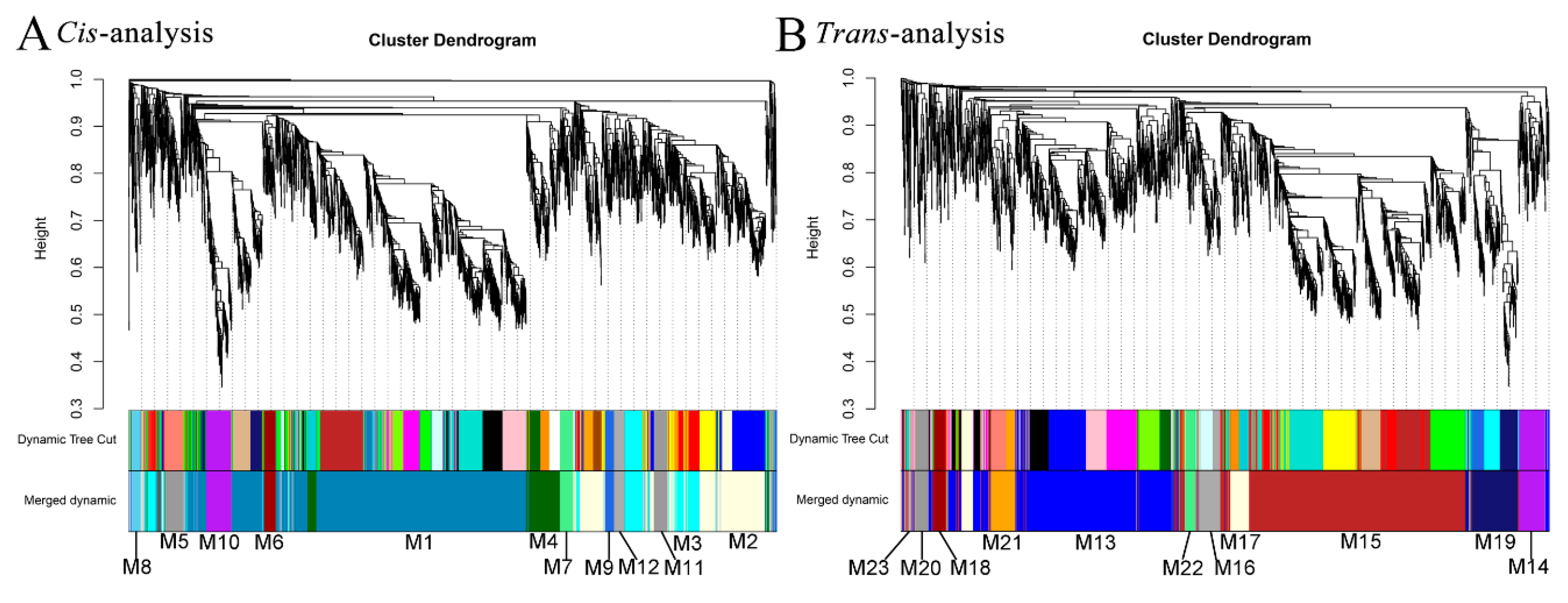
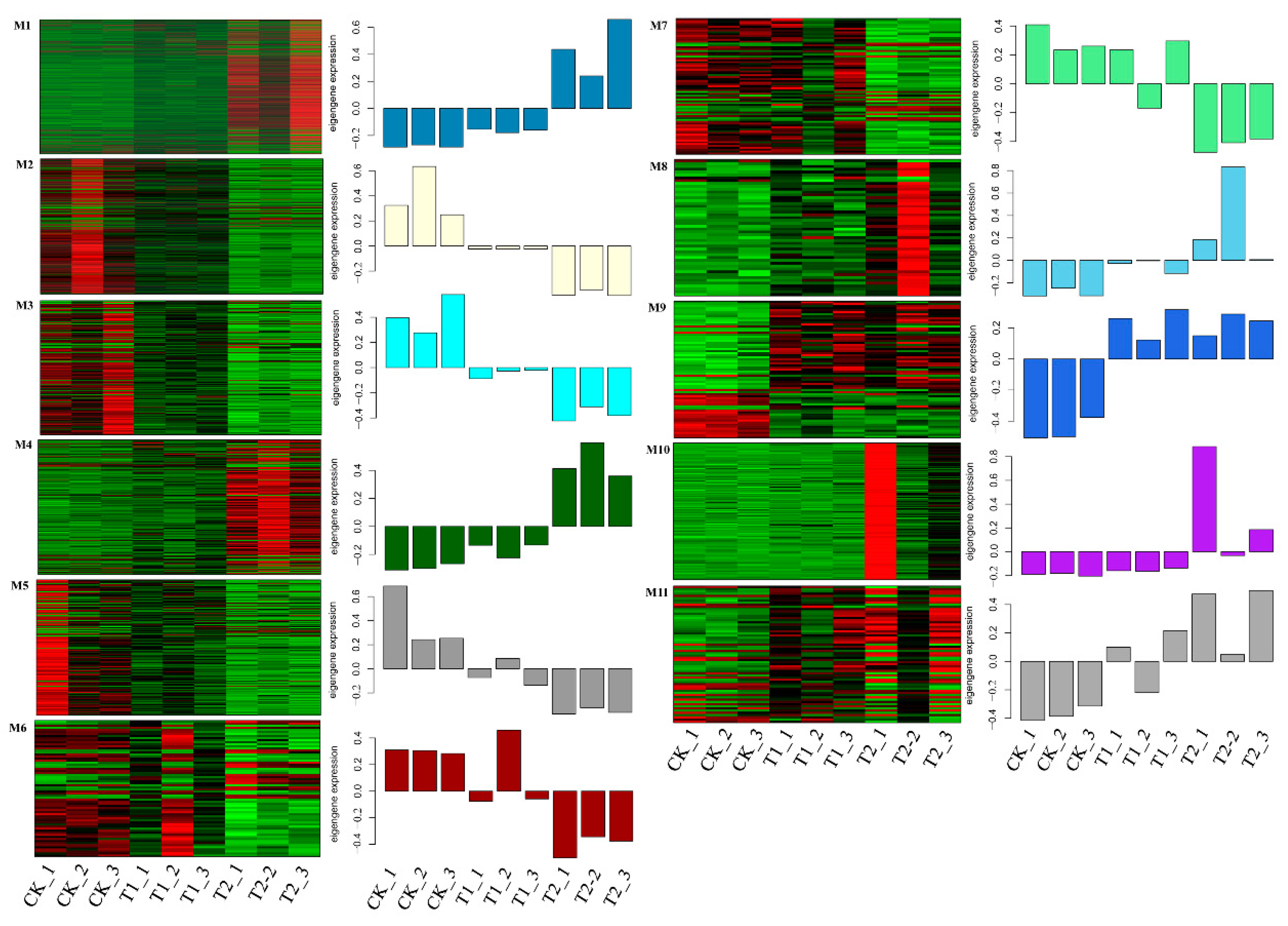
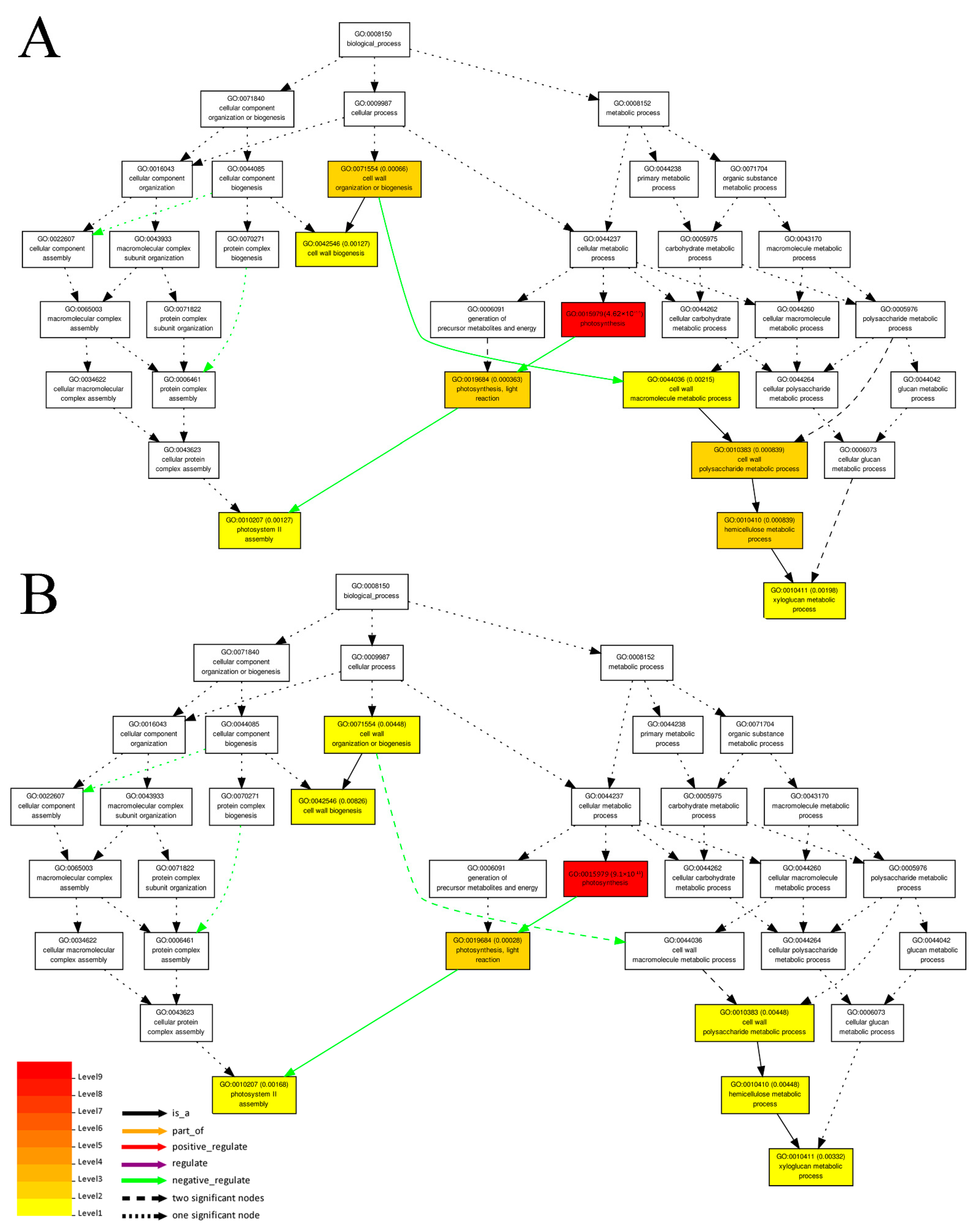
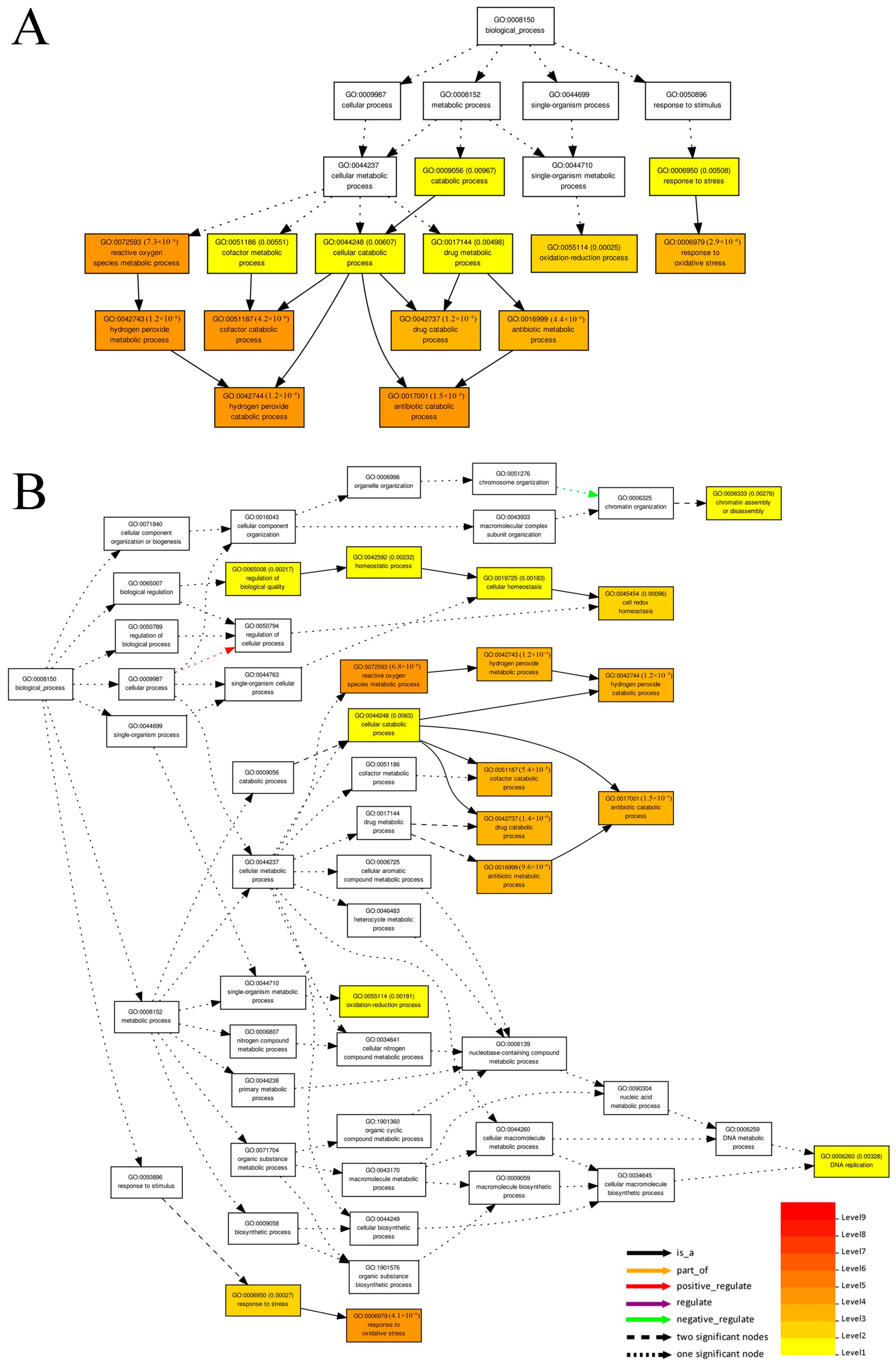
3.3. Prediction of Cis-Regulated Target PGs (CTPGs) of Lncrnas
3.4. Prediction of TTPGs of lncRNAs
3.5. Similarity Alignment and Conservation Analysis of lncRNAs in S. lycopersicum and Arabidopsis thaliana
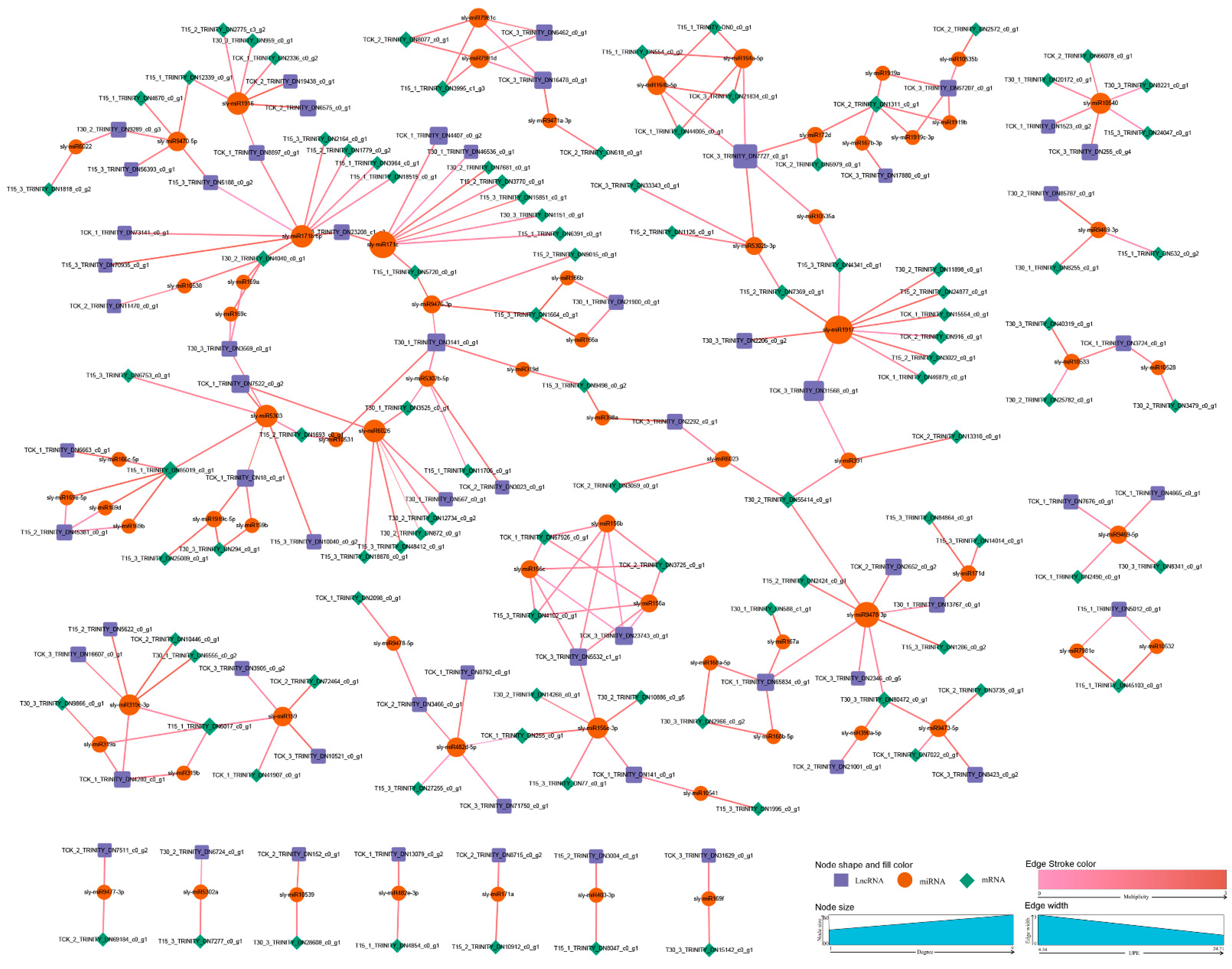
3.6. Prediction of lncRNAs as Endogenous Target Mimics of miRNAs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Crick, F.H.C.; Barnett, L.; Brenner, S.; Watts-Tobin, R.J. General Nature of the Genetic Code for Proteins. Nature 1961, 192, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Lu, X.; Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2014, 1839, 1097–1109. [Google Scholar] [CrossRef] [PubMed]
- Eddy, S.R. Non–coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Carninci, P.; Hayashizaki, Y. Noncoding RNA transcription beyond annotated genes. Curr. Opin. Genet. Dev. 2007, 17, 139–144. [Google Scholar] [CrossRef]
- Eddy, S.R. Computational genomics of noncoding RNA genes. Cell 2002, 109, 137–140. [Google Scholar] [CrossRef]
- Griffiths-Jones, S. Annotating noncoding RNA genes. Annu. Rev. Genomics Hum. Genet. 2007, 8, 279–298. [Google Scholar] [CrossRef]
- Zhu, Q.-H.; Wang, M.-B. Molecular functions of long non-coding RNAs in plants. Genes 2012, 3, 176–190. [Google Scholar] [CrossRef]
- Yamada, K.; Lim, J.; Dale, J.M.; Chen, H.; Shinn, P.; Palm, C.J.; Southwick, A.M.; Wu, H.C.; Kim, C.; Nguyen, M. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 2003, 302, 842–846. [Google Scholar] [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423. [Google Scholar] [CrossRef]
- Kong, R.; Zhang, E.-b.; Yin, D.-d.; You, L.-h.; Xu, T.-p.; Chen, W.-m.; Xia, R.; Wan, L.; Sun, M.; Wang, Z.-x. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol. Cancer 2015, 14, 82. [Google Scholar] [CrossRef]
- Wierzbicki, A.T. The role of long non-coding RNA in transcriptional gene silencing. Curr. Opin. Plant Biol. 2012, 15, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Sacco, L.D.; Baldassarre, A.; Masotti, A. Bioinformatics tools and novel challenges in long non-coding RNAs (lncRNAs) functional analysis. Int. J. Mol. Sci. 2012, 13, 97–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Ge, X.; Du, J.; Hu, J. Genome-wide Identification of Long Non-coding RNAs and Their Potential Functions in Poplar Growth and Phenylalanine Biosynthesis. Front. Genet. 2021, 12, 2113. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.-D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef]
- Shafiq, S.; Li, J.; Sun, Q. Functions of plants long non-coding RNAs. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2016, 1859, 155–162. [Google Scholar] [CrossRef]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.-H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef]
- Swiezewski, S.; Liu, F.; Magusin, A.; Dean, C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 2009, 462, 799. [Google Scholar] [CrossRef]
- Heo, J.B.; Sung, S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 2011, 331, 76–79. [Google Scholar] [CrossRef]
- Shin, H.; Shin, H.S.; Chen, R.; Harrison, M.J. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. Plant J. 2006, 45, 712–726. [Google Scholar] [CrossRef]
- Liu, C.; Muchhal, U.S.; Raghothama, K. Differential expression of TPS11, a phosphate starvation-induced gene in tomato. Plant Mol. Biol. 1997, 33, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.C.; Del Pozo, J.C.; Iglesias, J.; Rubio, V.; Solano, R.; De La Peña, A.; Leyva, A.; Paz-Ares, J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000, 24, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; García, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 2007, 39, 1033. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Chen, Y.Q. Long noncoding RNAs: New regulators in plant development. Biochem. Biophys. Res. Commun. 2013, 436, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-H. Roles of allelopathy in plant biodiversity and sustainable agriculture. Critical Reviews in Plant Sciences 1999, 18, 609–636. [Google Scholar] [CrossRef]
- EL-GAWAD, A.M.A. Ecology and allelopathic control of Brassica tournefortii in reclaimed areas of the Nile Delta, Egypt. Turk. J. Bot. 2014, 38, 347–357. [Google Scholar] [CrossRef]
- de Albuquerque, M.B.; dos Santos, R.C.; Lima, L.M.; de Albuquerque Melo Filho, P.; Nogueira, R.J.M.C.; Da Câmara, C.A.G.; de Rezende Ramos, A. Allelopathy, an alternative tool to improve cropping systems. A review. Agron. Sustain. Dev. 2011, 31, 379–395. [Google Scholar] [CrossRef]
- Cheng, Z.-h.; Xu, P. Lily (Lilium spp.) root exudates exhibit different allelopathies on four vegetable crops. Acta Agric. Scand. Sect. B–Soil Plant Sci. 2013, 63, 169–175. [Google Scholar]
- Hortal, S.; Bastida, F.; Moreno, J.L.; Armas, C.; García, C.; Pugnaire, F.I. Benefactor and allelopathic shrub species have different effects on the soil microbial community along an environmental severity gradient. Soil Biol. Biochem. 2015, 88, 48–57. [Google Scholar] [CrossRef]
- Gniazdowska, A.; Bogatek, R. Allelopathic interactions between plants. Multi site action of allelochemicals. Acta Physiol. Plant. 2005, 27, 395–407. [Google Scholar] [CrossRef]
- Nasrollahi, P.; Razavi, S.M.; Ghasemian, A.; Zahri, S. Physiological and Biochemical Responses of Lettuce to Thymol, as Allelochemical. Russ. J. Plant Physiol. 2018, 65, 598–603. [Google Scholar] [CrossRef]
- Cheng, F.; Cheng, Z. Research progress on the use of plant allelopathy in agriculture and the physiological and ecological mechanisms of allelopathy. Front. Plant Sci. 2015, 6, 1020. [Google Scholar] [CrossRef] [PubMed]
- Latif, S.; Chiapusio, G.; Weston, L.A. Allelopathy and the role of allelochemicals in plant defence. Adv. Bot. Res. 2017, 82, 19–54. [Google Scholar]
- Zhang, Q.; Zheng, X.-Y.; Lin, S.-X.; Gu, C.-Z.; Li, L.; Li, J.-Y.; Fang, C.-X.; He, H.-B. Transcriptome analysis reveals that barnyard grass exudates increase the allelopathic potential of allelopathic and non-allelopathic rice (Oryza sativa) accessions. Rice 2019, 12, 30. [Google Scholar] [CrossRef]
- Liang, X.; Hou, X.; Li, J.; Han, Y.; Zhang, Y.; Feng, N.; Du, J.; Zhang, W.; Zheng, D.; Fang, S. High-resolution DNA methylome reveals that demethylation enhances adaptability to continuous cropping comprehensive stress in soybean. BMC Plant Biol. 2019, 19, 79. [Google Scholar] [CrossRef]
- Fang, C.; Li, Y.; Li, C.; Li, B.; Ren, Y.; Zheng, H.; Zeng, X.; Shen, L.; Lin, W. Identification and comparative analysis of micro RNAs in barnyardgrass (E chinochloa crus-galli) in response to rice allelopathy. Plant Cell Environ. 2015, 38, 1368–1381. [Google Scholar] [CrossRef]
- Wang, J.; Meng, X.; Yuan, C.; Harrison, A.P.; Chen, M. The roles of cross-talk epigenetic patterns in Arabidopsis thaliana. Brief. Funct. Genom. 2015, 15, 278–287. [Google Scholar] [CrossRef]
- Xu, X.-W.; Zhou, X.-H.; Wang, R.-R.; Peng, W.-L.; An, Y.; Chen, L.-L. Functional analysis of long intergenic non-coding RNAs in phosphate-starved rice using competing endogenous RNA network. Sci. Rep. 2016, 6, 20715. [Google Scholar] [CrossRef]
- Lv, Y.; Liang, Z.; Ge, M.; Qi, W.; Zhang, T.; Lin, F.; Peng, Z.; Zhao, H. Genome-wide identification and functional prediction of nitrogen-responsive intergenic and intronic long non-coding RNAs in maize (Zea mays L.). BMC Genom. 2016, 17, 350. [Google Scholar] [CrossRef]
- Chen, M.; Wang, C.; Bao, H.; Chen, H.; Wang, Y. Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genom. 2016, 291, 1663–1680. [Google Scholar] [CrossRef]
- Wang, T.-Z.; Liu, M.; Zhao, M.-G.; Chen, R.; Zhang, W.-H. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Xin, J.W.; Chai, Z.X.; Zhang, C.F.; Yang, Y.M.; Ji, Q.M. Transcriptome analysis identified long non-coding RNAs involved in the adaption of yak to high-altitude environments. R. Soc. Open Sci. 2020, 7, 200625. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Limin, F.; Beifang, N.; Zhengwei, Z.; Sitao, W.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar]
- Costa, N.T.D.; Luz, A.W.A.; Henrique, B.P.; Maeda, S.P.T.; Silva, D.D.; Rossi, P.A. Pattern recognition analysis on long noncoding RNAs: A tool for prediction in plants. Brief. Bioinform. 2018, 2, 682–689. [Google Scholar]
- Urminder, S.; Niraj, K.; Mohan, S.; Rajkumar, R.; Garg, M. PLncPRO for prediction of long non-coding RNAs (lncRNAs) in plants and its application for discovery of abiotic stress-responsive lncRNAs in rice and chickpea. Nucleic Acids Res. 2017, 45, e183. [Google Scholar]
- Valentin, W.; Fabrice, L.; Benoît, H.; Guillaume, R.; Lætitia, L.; Tosso, L.; Vidhya, J.; Edouard, C.; Audrey, D.; Hannes, L. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nuclc Acids Res. 2017, 45, e57. [Google Scholar]
- Kent, W.J. Blat-the BLAST-like alignment tool. Genome Res. 2001, 12, 656–664. [Google Scholar]
- Huang, L.; Dong, H.; Zhou, D.; Li, M.; Liu, Y.; Zhang, F.; Feng, Y.; Yu, D.; Lin, S.; Cao, J. Systematic identification of long non-coding RNAs during pollen development and fertilization in Brassica rapa. Plant J. Cell Mol. Biol. 2018, 96, 203–222. [Google Scholar] [CrossRef]
- Wan, S.; Zhang, Y.; Duan, M.; Huang, L.; Yu, Y. Integrated Analysis of Long Non-coding RNAs (lncRNAs) and mRNAs Reveals the Regulatory Role of lncRNAs Associated With Salt Resistance in Camellia sinensis. Front. Plant Sci. 2020, 11, 218. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Method 2002, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Casero, D.; Sandoval, S.; Seet, C.S.; Scholes, J.; Zhu, Y.; Ha, V.L.; Luong, A.; Parekh, C.; Crooks, G.M. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat. Immunol. 2015, 16, 1282–1291. [Google Scholar] [CrossRef] [PubMed]
- Hakim, T.; Ivo, H. RNAplex: A fast tool for RNA-RNA interaction search. Bioinformatics 2008, 24, 2657–2663. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhu, J.; Liu, Y.; Chen, Y.; Li, Y.; Huang, L.; Chen, S.; Li, T.; Dang, Y.; Chen, T. Methamphetamine induces alterations in the long non-coding RNAs expression profile in the nucleus accumbens of the mouse. BMC Neuroence 2015, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 1–14. [Google Scholar] [CrossRef]
- Lv, J.; Liu, Z.; Yang, B.; Deng, M.; Zou, X. Systematic identification and characterization of long non-coding RNAs involved in cytoplasmic male sterility in pepper (Capsicum annuum L.). Plant Growth Regul. 2020, 91, 277–288. [Google Scholar] [CrossRef]
- Guo, L.L. Competing endogenous RNA networks and gastric cancer. World J. Gastroenterol. 2015, 21, 11680. [Google Scholar] [CrossRef]
- Xia, T.; Liao, Q.; Jiang, X.; Shao, Y.; Guo, J. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci. Rep. 2014, 4, 6088. [Google Scholar] [CrossRef]
- Edouard, S.; Luigi, F.; Suraj, J.; Marco, B.; Yang, K.Z.; Francesca, B.; Jacqueline, B.L.; Virginia, F.; Angenent, G.C.; Immink, R.G.H. Arabidopsis thaliana ambient temperature responsive lncRNAs. BMC Plant Biol. 2018, 18, 145. [Google Scholar]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef]
- Li, L.; Eichten, S.R.; Shimizu, R.; Petsch, K.; Yeh, C.T.; Wu, W.; Chettoor, A.M.; Givan, S.A.; Cole, R.A.; Fowler, J.E.; et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014, 8, e9585. [Google Scholar] [CrossRef] [PubMed]
- Keshi, M.; Wenshuo, S.; Mengyue, X.; Jiaxi, L.; Feixiong, Z. Genome-Wide Identification and Characterization of Long Non-Coding RNA in Wheat Roots in Response to Ca2+ Channel Blocker. Front. Plant Ence 2018, 9, 244. [Google Scholar]
- Aflitos, S.; Schijlen, E.; de Jong, H.; de Ridder, D.; Smit, S.; Finkers, R.; Wang, J.; Zhang, G.; Li, N.; 100 Tomato Genome Sequencing Consortium; et al. Exploring genetic variation in the tomato (Solanum section Lycopersicon) clade by whole-genome sequencing. Plant J. Cell Mol. Biol. 2015, 80, 136–148. [Google Scholar]
- Yan, P.; Luo, S.; Lu, J.Y.; Shen, X. Cis- and trans-acting lncRNAs in pluripotency and reprogramming. Curr. Opin. Genet. Dev. 2017, 46, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef] [PubMed]
- Vance, K.W.; Ponting, C.P. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014, 30, 348–355. [Google Scholar] [CrossRef]
- Wang, J.; Shen, Y.C.; Chen, Z.N.; Yuan, Z.C.; Wang, H.; Li, D.J.; Liu, K.; Wen, F.Q. Microarray profiling of lung long non-coding RNAs and mRNAs in lipopolysaccharide-induced acute lung injury mouse model. Biosci. Rep. 2019, 39, BSR20181634. [Google Scholar] [CrossRef]
- Guil, S.; Esteller, M. Cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol. 2012, 19, 1068. [Google Scholar] [CrossRef]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Rinn, J.L. A Large Intergenic Noncoding RNA Induced by p53 Mediates Global Gene Repression in the p53 Response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef]
- Li, F.; Wang, W.; Zhao, N.; Xiao, B.; Cao, P.; Wu, X.; Ye, C.; Shen, E.; Qiu, J.; Zhu, Q.H. Regulation of Nicotine Biosynthesis by an Endogenous Target Mimicry of MicroRNA in Tobacco. Plant Physiol. 2015, 169, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Somoza, I.; Weigel, D.; Franco-Zorilla, J.M.; García, J.A.; Paz-Ares, J. ceRNAs: miRNA target mimic mimics. Cell 2011, 147, 1431–1432. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, G.; Niu, Y.; Guo, J. Systematic Identification of Long Non-Coding RNAs under Allelopathic Interference of Para-Hydroxybenzoic Acid in S. lycopersicum. Horticulturae 2022, 8, 1134. https://doi.org/10.3390/horticulturae8121134
Liang G, Niu Y, Guo J. Systematic Identification of Long Non-Coding RNAs under Allelopathic Interference of Para-Hydroxybenzoic Acid in S. lycopersicum. Horticulturae. 2022; 8(12):1134. https://doi.org/10.3390/horticulturae8121134
Chicago/Turabian StyleLiang, Guoting, Yajie Niu, and Jing Guo. 2022. "Systematic Identification of Long Non-Coding RNAs under Allelopathic Interference of Para-Hydroxybenzoic Acid in S. lycopersicum" Horticulturae 8, no. 12: 1134. https://doi.org/10.3390/horticulturae8121134
APA StyleLiang, G., Niu, Y., & Guo, J. (2022). Systematic Identification of Long Non-Coding RNAs under Allelopathic Interference of Para-Hydroxybenzoic Acid in S. lycopersicum. Horticulturae, 8(12), 1134. https://doi.org/10.3390/horticulturae8121134



