Root and Rhizosphere Microbiome of Tomato Plants Grown in the Open Field in the South of West Siberia under Mineral Fertilization
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Experimental Setup
2.3. Soil Sampling and Chemical Analyses
2.4. Plant Sampling and Analyses
2.5. DNA Extraction and Sequencing
2.6. Bioinformatic Analysis
2.7. Statistical Analyses
3. Results
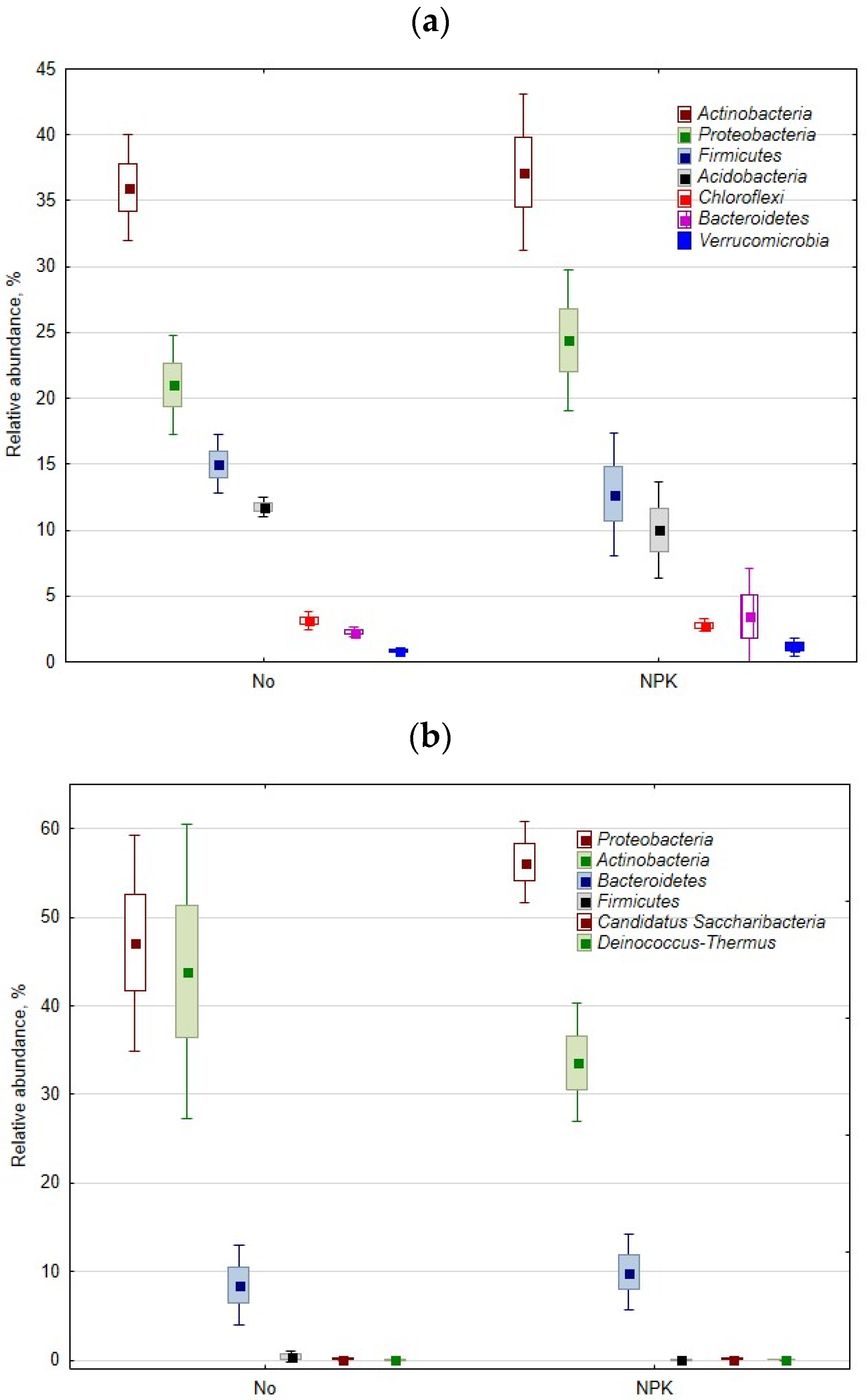
3.1. Rhizisphere and Root Bacteriobiome
3.1.1. General Pattern
3.1.2. The Effect of Mineral Fertilization on the Rhizosphere and Root Bacteriobiome
3.1.3. Alpha-Biodiversity in the Rhizosphere and Root Bacteriobiome
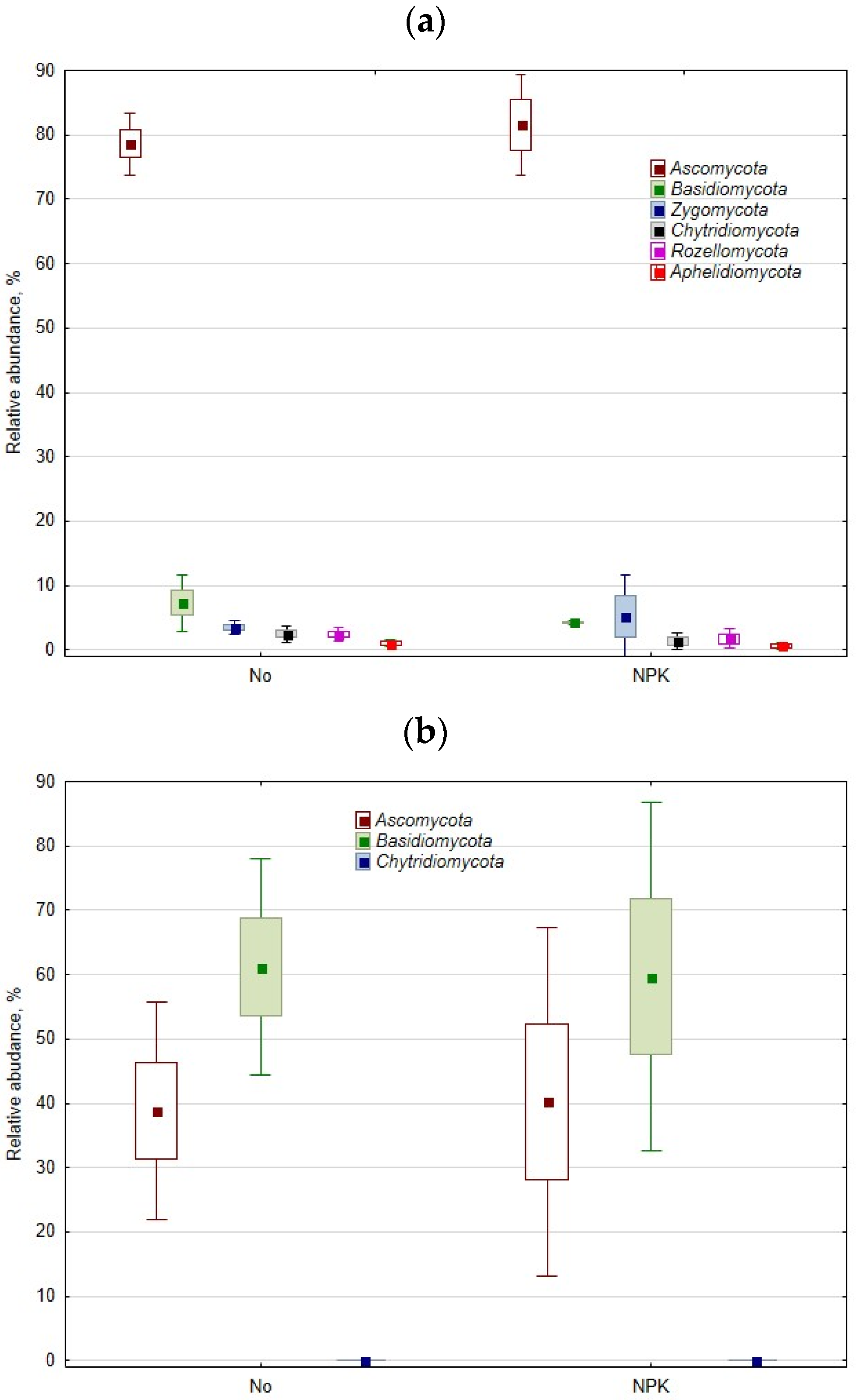
3.2. Rhizosphere and Root Mycobiome
3.2.1. General Pattern
3.2.2. The Effect of Mineral Fertilization on the Rhizosphere and Root Mycobiome
3.2.3. Alpha-Biodiversity in the Rhizosphere and Root Mycobiome
3.3. Tomato Production Properties
4. Discussion
4.1. Rhizosphere and Root Bacteriobiome
4.2. Rhizosphere and Root Mycobiome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Pozo, M.J.; Zabalgogeazcoa, I.; de Aldana, B.R.V.; Martinez-Medina, A. Untapping the potential of plant mycobiomes for applications in agriculture. Curr. Opin. Plant Biol. 2021, 60, 102034. [Google Scholar] [CrossRef]
- Trivedi, P.; Mattupalli, C.; Eversole, K.; Leach, J.E. Enabling sustainable agriculture through understanding and enhancement of microbiomes. New Phytol. 2021, 230, 2129–2147. [Google Scholar] [CrossRef]
- Baltrus, D.A. Adaptation, specialization, and coevolution within phytobiomes. Opin. Plant Biol. 2017, 38, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Papik, J.; Folkmanova, M.; Polivkova-Majorova, M.; Suman, J.; Uhlik, O. The invisible life inside plants: Deciphering the riddles of endophytic bacterial diversity. Biotechnol. Adv. 2020, 44, 107614. [Google Scholar] [CrossRef]
- Kracmarova, M.; Karpiskova, J.; Uhlik, O.; Strejcek, M.; Szakova, J.; Balik, J.; Demnerova, K.; Stiborova, H. Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization. Microorganisms 2020, 8, 1377. [Google Scholar] [CrossRef] [PubMed]
- Cangioli, L.; Mancini, M.; Napoli, M.; Fagorzi, C.; Orlandini, S.; Vaccaro, F.; Mengoni, A. Differential Response of Wheat Rhizosphere Bacterial Community to Plant Variety and Fertilization. Int. J. Mol. Sci. 2022, 23, 3616. [Google Scholar] [CrossRef]
- Mehlferber, E.C.; McCue, K.F.; Ferrel, J.E.; Koskella, B.; Khanna, R. Temporally Selective Modification of the Tomato Rhizosphere and Root Microbiome by Volcanic Ash Fertilizer Containing Micronutrients. Appl. Environ. Microbiol. 2022, 88, e0004922. [Google Scholar] [CrossRef]
- Food and Agriculture Organization of the United Nations. FAOSTAT. Data. Crops and Livestock Products. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 27 August 2022).
- Allard, S.M.; Walsh, C.S.; Wallis, A.E.; Ottesen, A.R.; Brown, E.W.; Micallef, S.A. Solanum lycopersicum (tomato) hosts robust phyllosphere and rhizosphere bacterial communities when grown in soil amended with various organic and synthetic fertilizers. Sci. Total Environ. 2016, 573, 555–563. [Google Scholar] [CrossRef]
- Dong, C.J.; Wang, L.L.; Li, Q.; Shang, Q.M. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef]
- IUSS Working Group. WRB, World Reference Base for Soil Resources 2014, Update 2015: International Soil Classification System for Naming Soils and Creating Legends for Soil Maps; FAO: Rome, Italy, 2015. [Google Scholar]
- Shishov, L.L.; Tonkonogov, V.D.; Lebedeva, I.I.; Gerasimoiva, M.I. (Eds.) Classification and Diagnostics of Soils in Russia, 1st ed.; Oykumena Pubs: Moscow, Russia, 2004. (In Russian) [Google Scholar]
- Carter, M.R.; Gregorich, E.G. (Eds.) Soil Sampling and Methods of Analysis, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2008. [Google Scholar]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Tyurin, M.; Kabilov, M.R.; Smirnova, N.; Tomilova, O.G.; Yaroslavtseva, O.; Alikina, T.; Glupov, V.V.; Kryukov, V.Y. Can Potato Plants Be Colonized with the Fungi Metarhizium and Beauveria under Their Natural Load in Agrosystems? Microorganisms 2021, 9, 1373. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon reads. bioRxiv 2016, 081257. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE USEARCH/UTAX Release for Fungi, Version 18.11.2018; UNITE Community: Tartu, Estonia, 2018. [CrossRef]
- Fauth, E.; Bernardo, J.; Camara, M.; Resetarits, W.J., Jr.; Van Buskirk, J.; McCollum, S.A. Simplifying the Jargon of Community Ecology: A Conceptual Approach. Am. Nat. 1996, 147, 282–286. [Google Scholar] [CrossRef]
- Hammer, O.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Hughes, J.B.; Hellmann, J.J. The Application of Rarefaction Techniques to Molecular Inventories of Microbial Diversity. Methods Enzym. 2005, 397, 292–308. [Google Scholar] [CrossRef]
- Lee, S.A.; Kim, Y.; Kim, J.M.; Chu, B.; Joa, J.H.; Sang, M.K.; Song, J.; Weon, H.Y. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci. Rep. 2019, 9, 9300. [Google Scholar] [CrossRef]
- Anzalone, A.; Mosca, A.; Dimaria, G.; Nicotra, D.; Tessitori, M.; Privitera, G.F.; Pulvirenti, A.; Leonardi, C.; Catara, V. Soil and Soilless Tomato Cultivation Promote Different Microbial Communities That Provide New Models for Future Crop Interventions. Int. J. Mol. Sci. 2022, 23, 8820. [Google Scholar] [CrossRef]
- López, S.; Pastorino, G.N.; Fernández-González, A.J.; Franco, M.; Fernández-López, M.; Balatti, P.A. The endosphere bacteriome of diseased and healthy tomato plants. Arch. Microbiol. 2020, 202, 2629–2642. [Google Scholar] [CrossRef] [PubMed]
- Zuluaga, M.; Lima Milani, K.M.; Azeredo Gonçalves, L.S.; Martinez de Oliveira, A.L. Diversity and plant growth-promoting functions of diazotrophic/N-scavenging bacteria isolated from the soils and rhizospheres of two species of Solanum. PLoS ONE 2020, 15, e0227422. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Colagiero, M.; Rosso, L.C.; Catalano, D.; Schena, L.; Ciancio, A. Response of Tomato Rhizosphere Bacteria to Root-Knot Nematodes, Fenamiphos and Sampling Time Shows Differential Effects on Low Level Taxa. Front. Microbiol. 2020, 11, 390. [Google Scholar] [CrossRef]
- Naumova, N.B.; Belanov, I.P.; Alikina, T.Y.; Kabilov, M.R. Undisturbed Soil Pedon under Birch Forest: Characterization of Microbiome in Genetic Horizons. Soil Syst. 2021, 5, 14. [Google Scholar] [CrossRef]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef]
- Khan, A.L.; Waqas, M.; Kang, S.M.; Al-Harrasi, A.; Hussain, J.; Al-Rawahi, A.; Al-Khiziri, S.; Ullah, I.; Ali, L.; Jung, H.Y.; et al. Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 2014, 52, 689–695. [Google Scholar] [CrossRef]
- Akter, S.; Lee, S.Y.; Moon, S.K.; Choi, C.; Balusamy, S.R.; Siddiqi, M.Z.; Ashrafudoulla, M.; Huq, M.A. Sphingomonas horti sp. nov., a novel bacterial species isolated from soil of a tomato garden. Arch. Microbiol. 2021, 203, 543–548. [Google Scholar] [CrossRef]
- Yoo, S.J.; Weon, H.Y.; Song, J.; Sang, M.K. Effects of Chryseobacterium soldanellicola T16E-39 and Bacillus siamensis T20E-257 on biocontrol against phytophthora blight and bacterial wilt and growth promotion in tomato plants. Int. J. Agric. Biol. 2020, 23, 534–540. [Google Scholar]
- Kwak, M.J.; Kong, H.G.; Choi, K.; Kwon, S.K.; Song, J.Y.; Lee, J.; Lee, P.A.; Choi, S.Y.; Seo, M.; Lee, H.J.; et al. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 2018, 36, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Tancos, M.A.; Chalupowicz, L.; Barash, I.; Manulis-Sasson, S.; Smart, C.D. Tomato fruit and seed colonization by Clavibacter michiganensis subsp. michiganensis through external and internal routes. Appl. Environ. Microbiol. 2013, 79, 6948–6957. [Google Scholar] [CrossRef] [PubMed]
- Dastogeer, K.; Yasuda, M.; Okazaki, S. Microbiome and pathobiome analyses reveal changes in community structure by foliar pathogen infection in rice. Front. Microbiol. 2022, 13, 949152. [Google Scholar] [CrossRef] [PubMed]
- Ftayeh, R.; von Tiedemann, A.; Koopmann, B.; Rudolph, K.; Abu-Ghorrah, M. First Record of Clavibacter michiganensis subsp. michiganensis Causing Canker of Tomato Plants in Syria. Plant Dis. 2008, 92, 649. [Google Scholar] [CrossRef] [PubMed]
- Ansari, M.; Taghavi, S.M.; Hamzehzarghani, H.; Valenzuela, M.; Siri, M.I.; Osdaghi, E. Multiple Introductions of Tomato Pathogen Clavibacter michiganensis subsp. michiganensis into Iran as Revealed by a Global-Scale Phylogeographic Analysis. Appl. Environ. Microbiol. 2019, 85, e02098-19. [Google Scholar] [CrossRef] [PubMed]
- Naumova, N.; Nechaeva, T.; Smirnova, N.; Fotev, Y.; Belousova, S. Effect of Sapropel Addition on Selected Soil Properties and Field Tomato Yield in South West Siberia. Asian J. Soil Sci. Plant Nutr. 2017, 1, 1–11. [Google Scholar] [CrossRef]
- Mechan Llontop, M.E.; Sharma, P.; Aguilera Flores, M.; Yang, S.; Pollok, J.; Tian, L.; Huang, C.; Rideout, S.; Heath, L.S.; Li, S.; et al. Strain-Level Identification of Bacterial Tomato Pathogens Directly from Metagenomic Sequences. Phytopathology 2020, 110, 768–779. [Google Scholar] [CrossRef]
- Osdaghi, E.; Rahimi, T.; Taghavi, S.M.; Ansari, M.; Zarei, S.; Portier, P.; Briand, M.; Jacques, M.A. Comparative Genomics and Phylogenetic Analyses Suggest Several Novel Species within the Genus Clavibacter, Including Nonpathogenic Tomato-Associated Strains. Appl. Environ. Microbiol. 2020, 86, e02873-19. [Google Scholar] [CrossRef]
- Jacques, M.A.; Durand, K.; Orgeur, G.; Balidas, S.; Fricot, C.; Bonneau, S.; Quillévéré, A.; Audusseau, C.; Olivier, V.; Grimault, V.; et al. Phylogenetic analysis and polyphasic characterization of Clavibacter michiganensis strains isolated from tomato seeds reveal that nonpathogenic strains are distinct from C. michiganensis subsp. michiganensis. Appl. Environ. Microbiol. 2012, 78, 8388–8402. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Jung, J.; Kim, J.S.; Taffner, J.; Berg, G.; Ryu, C.M. Archaea, tiny helpers of land plants. Comput. Struct. Biotechnol. J. 2020, 18, 2494–2500. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Tang, G.; Xu, X.; Nong, X.; Qi, S. Insights into Deep-Sea Sediment Fungal Communities from the East Indian Ocean Using Targeted Environmental Sequencing Combined with Traditional Cultivation. PLoS ONE 2014, 9, e109118. [Google Scholar] [CrossRef] [PubMed]
- Sen, K.; Sen, B.; Wang, G. Diversity, Abundance, and Ecological Roles of Planktonic Fungi in Marine Environments. J. Fungi 2022, 8, 491. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, X.; Almeida, L.C.S.d.S.; Pecoraro, L. Environmental Factors Affecting Diversity, Structure, and Temporal Variation of Airborne Fungal Communities in a Research and Teaching Building of Tianjin University, China. J. Fungi 2022, 8, 431. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.E.; Vannette, R.L.; Igwe, A.; Blundell, R.; Casteel, C.L.; Gaudin, A.C.M. Effects of agricultural management on rhizosphere microbial structure and function in processing tomato plants. Appl. Environ. Microbiol. 2019, 85, e01064-19. [Google Scholar] [CrossRef] [PubMed]
- Carlucci, A.; Raimondo, M.L.; Santos, J.; Phillips, A.J. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 2012, 28, 34–48. [Google Scholar] [CrossRef]
- Harrison Wright, A.; Ali, S.; Migicovsky, Z.; Douglas, G.M.; Yurgel, S.; Bunbury-Blanchette, A.; Franklin, J.; Adams, S.J.; Walker, A.K. A Characterization of a Cool-Climate Organic Vineyard’s Microbiome. Phytobiomes J. 2022, 6, 69–82. [Google Scholar] [CrossRef]
- Han, L.; Zhou, X.; Zhao, Y.; Wu, L.; Ping, X.; He, Y.; Peng, S.; He, X.; Du, Y. First report of Plectosphaerella plurivora causing root rot disease in Panax notoginseng in China. Phytopathol. Z. 2020, 168, 375–379. [Google Scholar] [CrossRef]
- Sosa, A.L.; Rosso, L.C.; Salusso, F.A.; Etcheverry, M.G.; Passone, M.A. Screening and identification of horticultural soil fungi for their evaluation against the plant parasitic nematode Nacobbus aberrans. World J. Microbiol. Biotechnol. 2018, 34, 63. [Google Scholar] [CrossRef]
- Bachmann, H.P.; Bobst, C.; Bütikofer, U.; Casey, M.G.; Dalla Torre, M.; Fröhlich-Wyder, M.T.; Fürst, M. Occurrence and significance of Fusarium domesticum alias Anticollanti on smear-ripened cheeses. LWT Food Sci. Technol. 2005, 38, 399–407. [Google Scholar] [CrossRef]
- Patel, R.; Mehta, K.; Prajapati, J.; Shukla, A.; Parmar, P.; Goswami, D.; Saraf, M. An anecdote of mechanics for Fusarium biocontrol by plant growth promoting microbes. Biol. Control 2022, 174, 105012. [Google Scholar] [CrossRef]
| Taxon | Rhizosphere | Roots | ||
|---|---|---|---|---|
| No | NPK | No | NPK | |
| Class level | ||||
| Actinobacteria | 13.8 ± 3.4 | 18.5 ± 8.3 | 43.8 ± 16.6 | 33.6 ± 6.7 |
| Alphaproteobacteria | 11.5 ± 2.6 | 13.8 ± 3.4 | 28.6 ± 6.8 a 1 | 41.9 ± 6.3 b |
| Thermoleophilia | 8.4 ± 0.9 | 7.0 ± 2.9 | 0.03 ± 0.04 | 0.01 ± 0.01 |
| Bacilli | 7.8 ±1.1 | 7.3 ± 2.2 | 0.3 ± 0.6 | 0.0 ± 0.1 |
| Acidimicrobiia | 7.8 ± 1.0 | 6.8 ± 1.9 | n.d.2 | n.d. |
| un. 3 Actinobacteria | 5.9 ± 1.0 | 4.9 ± 2.3 | 0.1 ± 0.1 | 0.0 ± 0.0 2 |
| Clostridia | 5.7 ± 1.1 | 4.3 ± 1.8 | 0.1 ± 0.1 | 0.0 ± 0.0 |
| Acidobacteria_Gp6 | 5.4 ± 0.9 | 4.1 ± 1.6 | n.d. | n.d. |
| Gammaproteobacteria | 1.9 ± 0.6 | 3.0 ± 1.5 | 9.0 ± 5.4 | 7.6 ± 4.2 |
| Betaproteobacteria | 4.7 ± 1.0 | 4.5 ± 1.3 | 9.4 ± 8.2 | 6.7 ± 1.7 |
| Deltaproteobacteria | 2.6 ± 0.4 | 2.7 ± 0.7 | n.d. | n.d. |
| Anaerolineae | 1.3 ± 0.5 | 0.8 ± 0.6 | n.d. | n.d. |
| Caldilineae | 1.2 ± 0.2 | 0.9 ± 0.3 | n.d. | n.d. |
| Cytophagia | 0.9 ± 0.2 | 1.1 ± 0.7 | 6.9 ± 3.8 | 5.7 ± 3.3 |
| Flavobacteriia | 0.4 ± 0.1 | 1.6 ± 2.8 | 1.3 ± 1.0 a | 3.6 ± 1.4 b |
| Order level | ||||
| Micrococcales | 4.9 ± 4.0 | 7.4 ± 6.0 | 40.8 ± 16.6 | 31.8 ± 6.1 |
| Rhizobiales | 8.8 ± 2.2 | 9.5 ± 2.6 | 16.8 ± 7.2 | 24.0 ± 4.0 |
| Bacillales | 7.8 ± 1.1 | 7.3 ± 2.2 | 0.6 ± 0.3 | 0.04 ± 0.07 |
| Acidimicrobiales | 7.8 ± 1.0 | 6.8 ± 1.9 | n.d. | n.d. |
| Gaiellales | 6.0 ± 0.9 | 4.6 ± 1.9 | n.d. | n.d. |
| Acidobacteria_Gp6 | 5.4 ± 0.9 | 4.1± 1.6 | n.d. | n.d. |
| Clostridiales | 5.2 ± 1.0 | 3.9 ± 1.5 | 0.00 ± 0.01 | 0.00 ± 0.00 |
| Sphingomonadales | 0.3 ± 0.2 | 0.6 ± 0.4 | 11.2 ± 1.0 a | 17.4 ± 3.9 b |
| Burkholderiales | 1.7 ± 0.4 | 1.6 ± 0.5 | 9.4 ± 8.2 | 6.7 ± 1.7 |
| Pseudomonadales | 0.1 ± 0.2 | 0.1 ± 0.2 | 8.1 ± 5.1 | 7.3 ± 4.3 |
| Cytophagales | 0.9 ± 0.2 | 1.1 ± 0.7 | 6.9 ± 3.8 | 5.7 ± 3.3 |
| Kineosporiales | n.d. | n.d. | 2.1 ± 1.5 | 1.1 ± 0.6 |
| Flavobacteriales | 0.4 ± 0.1 | 1.6 ± 2.8 | 1.3 ± 1.0 a | 3.6 ± 1.4 b |
| Family level | ||||
| Gaiellaceae | 6.0 ± 0.9 | 4.6 ± 1.9 | n.d. | n.d. |
| Acidobacteria_Gp6 | 5.4 ± 0.9 | 4.1 ± 1.6 | n.d. | n.d. |
| un. Rhizobiales | 5.0 ± 0.9 | 4.7 ± 1.0 | n.d. | n.d. |
| Ilumatobacteraceae | 4.3 ± 0.7 | 3.5 ± 1.1 | n.d. | n.d. |
| Micrococcaceae | 3.6 ± 3.4 | 3.7 ± 3.6 | 0.3 ± 0.3 | 0.0 ± 0.0 |
| Hyphomicrobiaceae | 2.9 ± 0.7 | 2.4 ± 0.5 | n.d. | n.d. |
| Acidobacteria_Gp16 | 2.2 ± 0.7 | 2.5 ± 1.1 | n.d. | n.d. |
| Iamiaceae | 2.2 ± 0.2 | 2.0 ± 0.5 | n.d. | n.d. |
| Planococcaceae | 1.7 ± 0.4 | 1.6 ± 0.5 | n.d. | n.d. |
| Bacillaceae1 | 1.6 ± 0.1 | 1.3 ± 0.5 | n.d. | n.d. |
| Nocardioidaceae | 1.6 ± 1.4 | 4.9 ± 4.8 | 0.1 ± 0.2 | 0.03 ± 0.03 |
| Caldilineaceae | 1.2 ± 0.2 | 0.9 ± 0.3 | n.d. | n.d. |
| Paenibacillaceae1 | 1.0 ± 0.1 | 1.0 ± 0.4 | 0.03 ± 0.04 | 0.01 ± 0.01 |
| Clostridiaceae1 | 1.0 ± 0.2 | 0.8 ± 0.4 | n.d. | n.d. |
| Rhodobacteraceae | 1.0 ± 0.2 | 1.9 ± 1.1 | 0.2 ± 0.2 | 0.1 ± 0.1 |
| Microbacteriaceae | 0.5 ± 0.3 | 1.6 ± 1.9 | 40.0 ± 17.1 | 31.6 ± 6.2 |
| Sphingomonadaceae | 0.2 ± 0.1 | 0.4 ± 0.2 | 11.2 ± 1.0 a | 17.1 ± 3.5 b |
| Methylobacteriaceae | 0.1 ± 0.2 | 0.1 ± 0.3 | 8.9 ± 4.0 | 13.4 ± 5.0 |
| Pseudomonadaceae | 0.1 ± 0.2 | 0.1 ± 0.2 | 8.1 ± 5.1 | 7.3 ± 4.3 |
| Oxalobacteraceae | 0.04 ± 0.03 | 0.10 ± 0.11 | 6.7 ± 7.4 | 3.5 ± 0.7 |
| Hymenobacteraceae | 0.2 ± 0.1 | 0.1 ± 0.1 | 6.4 ± 3.4 | 5.1 ± 3.1 |
| Rhizobiaceae | n.d. | n.d. | 4.5 ± 1.8 | 4.4 ± 1.9 |
| Aurantimonadaceae | n.d. | n.d. | 3.2 ± 2.1 | 5.9 ± 3.2 |
| Comamonadaceae | n.d. | n.d. | 2.7 ± 1.4 | 3.2 ± 1.2 |
| Kineosporiaceae | n.d. | n.d. | 2.1 ± 1.5 | 1.1 ± 0.6 |
| Weeksellaceae | 0.01 ± 0.01 | 0.01 ± 0.00 | 1.3 ± 1.0 a | 3.6 ± 1.4 b |
| Genus level | ||||
| Gaiella | 6.0 ± 0.9 | 4.6 ± 1.9 | n.d. | n.d. |
| Acidobacteria_Gp6 | 5.8 ± 1.0 | 4.3 ± 1.7 | n.d. | n.d. |
| un. Rhizobiales | 5.0 ± 0.9 | 4.7 ± 1.0 | 0.1 ± 0.2 | 0.0 ± 0.0 |
| Clavibacter | n.d. | n.d. | 36.3 ±18.8 | 23.9 ± 6.2 |
| Sphingomonas | 0.1 ± 0.1 | 0.2 ± 0.1 | 11.1 ± 1.0 a | 17.1 ± 3.4 b |
| Methylobacterium | n.d. | n.d. | 8.9 ± 4.0 | 13.4 ± 5.0 |
| Pseudomonas | 0.1 ± 0.2 | 0.1 ± 0.2 | 8.1 ± 5.1 | 7.3 ± 4.3 |
| Massilia | n.d. | n.d. | 6.6 ± 7.4 | 3.5 ± 0.6 |
| Hymenobacter | n.d. | n.d. | 6.4 ± 3.4 | 5.1 ± 3.1 |
| Agrobacterium | n.d. | n.d. | 4.5 ± 1.8 | 4.4 ± 1.9 |
| Aureimonas | n.d. | n.d. | 3.2 ± 2.1 | 5.9 ± 3.2 |
| Rathayibacter | n.d. | n.d. | 2.9 ± 1.8 a | 6.3 ± 3.5 b |
| un. 2 Comamonadaceae | 0.2 ± 0.1 | 0.2 ± 0.2 | 2.7 ± 1.3 | 3.2 ± 1.2 |
| Kineococcus | n.d. | n.d. | 1.9 ± 1.5 | 1.0 ± 1.2 |
| Chryseobacterium | n.d. | n.d. | 1.3 ± 1.0 a | 3.6 ± 1.4 b |
| No. | OTU | Rhizosphere | Roots | ||
|---|---|---|---|---|---|
| No | NPK | No | NPK | ||
| 4 | Clavibacter sp. | n.d. 1 | n.d. | 36.3 ± 18.8 | 23.9 ± 6.2 |
| 7 | Pseudarthrobacter | 3.6 ± 3.4 | 3.6 ± 3.5 | n.d. 1 | n.d. |
| 9 | Sphingomonas sp. | n.d. | n.d. | 7.2 ± 3.0 | 7.4 ± 3.5 |
| 15 | Methylobacterium sp. | n.d. | n.d. | 8.4 ± 4.4 | 13.1 ± 4.9 |
| 16 | Aureimonas sp. | n.d. | n.d. | 2.9 ± 2.2 | 4.9 ± 3.8 |
| 26 | Pseudomonas sp. | n.d. | n.d. | 6.5 ± 3.1 | 6.5 ± 3.2 |
| 27 | un. 2 Rhizobiales | 3.6 ± 0.6 | 3.0 ± 0.6 | n.d. | n.d. |
| 28 | Agrobacterium sp. | n.d. | n.d. | 4.5 ± 1.8 | 4.4 ± 1.9 |
| 29 | Rathayibacter sp. | n.d. | n.d. | 2.9 ± 1.8 a 3 | 6.3 ± 3.5 b |
| 31 | Chryseobacterium sp. | n.d. | n.d. | 1.2 ±1.0 a | 3.6 ± 1.4 b |
| 43 | Sphingomonas sp. | n.d. | n.d. | 0.9 ± 1.0 a | 3.4 ± 1.5 b |
| 56 | un. Hyphomicrobiaceae | 1.5 ± 0.3 | 1.3 ± 0.3 | n.d. | n.d. |
| 57 | un. Actinobacteria | 2.2 ± 0.5 | 1.5 ± 0.9 | n.d. | n.d. |
| 58 | Sphingomonas sp. | n.d. | n.d. | 1.6 ± 1.0 a | 4.3 ± 1.2 b |
| 60 | un. Comamonadaceae | n.d. | n.d. | 2.1 ±1.0 | 1.8 ± 0.5 |
| 65 | Kineococcus sp. | n.d. | n.d. | 1.9 ± 1.5 | 1.0 ± 0.5 |
| 68 | Massilia sp. | n.d. | n.d. | 6.6 ± 7.4 | 3.5 ± 0.6 |
| 84 | un. Acidobacteria_Gp6 | 1.6 ± 0.3 b | 1.1 ± 0.2 a | n.d. | n.d. |
| 88 | un. Gaiella | 1.7 ± 0.2 | 1.3 ± 0.8 | n.d. | n.d. |
| 95 | un. Nocardioides | 0.5 ± 0.4 | 1.5 ± 1.4 | n.d. | n.d. |
| 101 | Hymenobacter sp. | n.d. | n.d. | 5.0 ± 4.0 | 3.7 ± 2.9 |
| 107 | un. Actinobacteria | 1.3 ± 0.3 | 0.9 ± 0.4 | n.d. | n.d. |
| 122 | un. Nocardioides | 0.4 ± 0.4 | 1.2 ± 1.1 | n.d. | n.d. |
| 188 | un. Desertimonas | 1.0 ± 0.2 | 0.7 ± 0.3 | n.d. | n.d. |
| 1027 | Gaiella occulta | 1.0 ± 0.2 | 0.8 ± 0.2 | n.d. | n.d. |
| 1099 | Hymenobacter sp. | n.d. | n.d. | 1.0 ± 1.0 | 0.6 ± 0.4 |
| 3987 | Sphingomonas sp. | n.d. | n.d. | 1.0 ± 0.7 | 1.4 ± 0.9 |
| 6043 | Aureimonas sp. | n.d. | n.d. | 0.3 ± 0.3 | 1.0 ± 1.3 |
| Taxon | Rhizosphere | Roots | ||
|---|---|---|---|---|
| No | NPK | No | NPK | |
| Richness | 1338 ± 167 | 1381 ± 203 | 90 ± 47 | 76 ± 13 |
| Chao-1 | 1909 ± 218 | 1923 ± 234 | 100 ± 42 | 84 ± 15 |
| Simpson (1-D) | 0.99 ± 0.00 | 0.99 ± 0.00 | 0.80 ± 0.13 | 0.89 ± 0.03 |
| Shannon | 6.0 ± 0.2 | 6.0 ± 0.2 | 2.4 ± 0.6 | 2.7 ± 0.1 |
| Evenness | 0.31 ± 0.00 | 0.29 ± 0.04 | 0.14 ± 0.03 | 0.21 ± 0.03 |
| Equitability | 0.84 ± 0.01 | 0.83 ± 0.02 | 0.55 ± 0.09 | 0.63± 0.02 |
| Dominance (D) | 0.01 ± 0.00 | 0.01 ± 0.00 | 0.20 ± 0.13 | 0.11 ± 0.03 |
| Berger-Parker | 0.05 ± 0.01 | 0.05 ± 0.02 | 0.36 ± 0.19 | 0.24 ± 0.06 |
| Taxon | Rhizosphere | Roots | ||
|---|---|---|---|---|
| No | NPK | No | NPK | |
| Class level | ||||
| Tremellomycetes | 3.0 ± 3.9 | 1.7 ± 0.7 | 55.6 ± 16.3 | 57.3 ± 26.7 |
| Dothideomycetes | 13.4 ± 7.9 | 20.1 ± 1.6 | 34.3 ± 17.8 | 30.3 ± 24.8 |
| Microbotryomycetes | 0.3 ± 0.2 | 0.3 ± 0.1 | 4.0 ± 3.0 | 1.6 ± 1.1 |
| Cystobasidiomycetes | 0.05 ± 0.03 | 0.09 ± 0.12 | 1.1 ± 1.6 | 0.3 ± 0.3 |
| Leotiomycetes | 8.2 ± 1.4 | 5.5 ± 3.1 | 0.1 ± 0.1 | 0.4 ±0.6 |
| Sordariomycetes | 42.6 ± 6.2 | 44.8 ± 7.1 | 0.1 ± 0.3 | 1.1 ± 2.4 |
| Pezizomycetes | 4.5 ± 1.3 | 3.6 ± 0.9 | n.d. 1 | n.d. |
| Eurotiomycetes | 3.0 ± 0.6 | 2.5 ± 0.9 | 0.003 ± 0.002 | 0.04 ± 0.04 |
| Agaricomycetes | 3.6 ± 3.5 | 2.0 ± 0.9 | 0.1 ± 0.1 | 0.02 ± 0.02 |
| Aphelidiomycetes | 1.0 ± 0.6 | 0.6 ± 0.5 | n.d. | n.d. |
| Order level | ||||
| Tremellales | 0.7 ± 0.5 | 0.8 ± 0.4 | 51.9 ± 14.8 | 53.4 ± 24.6 |
| Pleosporales | 4.0 ± 1.7 | 13.0 ± 11.2 | 21.3 ± 12.2 | 23.1 ± 24.0 |
| Capnodiales | 0.8 ± 1.8 | 1.8 ± 3.7 | 12.6 ± 8.3 | 6.9 ± 3.7 |
| Cystofilobasidiales | 0.3 ± 0.2 | 0.2 ± 0.1 | 3.7 ± 2.7 | 3.9 ± 4.2 |
| Leucosporidiales | 0.05 ± 0.08 | 0.01 ± 0.00 | 3.2 ± 3.0 | 1.1 ± 0.9 |
| Cystobasidiomycetes_is | 0.004 ± 0.003 | 0.01 ± 0.01 | 1.0 ± 1.6 | 0.2 ± 0.1 |
| Helotiales | 7.2 ± 1.2 | 4.3 ± 2.8 | 0.1 ± 0.1 | 0.4 ± 0.6 |
| Glomerellales | 0.5 ± 0.3 | 1.0 ± 0.3 | 0.0 ± 0.0 1 | 1.1 ± 2.4 |
| Hypocreales | 13.0 ± 2.2 | 19.1 ± 9.1 | 0.1 ± 0.3 | 0.001 ± 0.002 |
| Microascales | 15.7 ± 5.9 | 10.0 ± 5.7 | n.d. | n.d. |
| Mortierellales | 4.0 ± 1.2 | 5.1 ± 6.6 | n.d. | n.d. |
| Pezizales | 4.3 ± 1.4 | 3.5 ±0.9 | n.d. | n.d. |
| Sordariales | 4.4 ± 1.5 | 3.1 ±1.6 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Sordariomycetidae_is 2 | 5.2 ± 0.7 | 8.9 ±4.0 | n.d. | n.d. |
| Dothideomycetes_is | 4.4 ± 1.6 | 3.5 ± 2.2 | n.d. | n.d. |
| Eurotiales | 1.7 ± 0.7 | 1.7 ± 1.1 | n.d. | n.d. |
| Agaricales | 2.3 ± 3.4 | 0.5 ± 0.2 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| Coniochaetales | 2.0 ± 0.4 | 1.4 ± 1.0 | n.d. | n.d. |
| Onygenales | 1.1 ± 0.5 | 0.6 ± 0.4 | n.d. | n.d. |
| Family level | ||||
| Pleosporaceae | 0.2 ± 0.1 | 1.8 ± 3.1 | 17.4 ± 12.0 | 20.7 ± 22.4 |
| un. Tremellales | 0.0 ± 0.0 | 0.0 ± 0.0 | 15.9 ± 7.6 | 23.1 ± 13.2 |
| Tremellaceae | 0.5 ± 0.4 | 0.5 ± 0.2 | 14.9 ± 17.7 | 8.0 ± 6.2 |
| Bulleribasidiaceae | 0.2 ± 0.3 | 0.3 ± 0.5 | 14.4 ± 10.1 | 17.7 ± 16.1 |
| Mycosphaerellaceae | 0.8 ± 1.8 | 1.8 ± 3.7 | 12.6 ± 8.3 | 6.9 ± 3.7 |
| Tremellales_is | 0.0 ± 0.0 | 0.0 ± 0.0 | 6.4 ± 4.3 | 4.3 ± 5.8 |
| Cystofilobasidiaceae | 0.2 ± 0.2 | 0.1 ± 0.1 | 4.1 ± 2.7 | 3.6 ± 4.1 |
| Leucosporidiaceae | 0.0 ± 0.1 | 0.0 ± 0.0 | 3.6 ± 3.0 | 1.1 ± 0.9 |
| Symmetrosporaceae | 0.0 ± 0.0 | 0.0 ± 0.0 | 2.1 ± 1.6 | 0.1 ± 0.1 |
| Phaeosphaeriaceae | 0.1 ± 0.1 | 0.0 ± 0.0 | 0.9 ± 0.8 | 0.7 ± 0.5 |
| Sclerotiniaceae | 1.0 ± 0.5 | 0.8 ± 0.5 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Microascaceae | 10.2 ± 3.6 | 7.0 ± 3.5 | n.d. | n.d. |
| Nectriaceae | 6.2 ± 1.2 | 9.3 ± 5.2 | 0.03 ± 0.08 | 0.00 ± 0.00 |
| Plectosphaerellaceae | 5.5 ± 0.9 a 3 | 9.9 ± 3.9 b | 0.0 ± 0.0 | 1.1 ± 2.4 |
| Mortierellaceae | 3.9 ± 1.2 | 5.1 ± 6.6 | n.d. | n.d. |
| Pseudeurotiaceae | 2.9 ± 1.1 | 2.3 ±1.5 | n.d. | n.d. |
| Psathyrellaceae | 1.9 ± 3.4 | 0.3 ± 0.1 | 0.01 ± 0.01 | 0.01 ± 0.00 |
| Ascodesmidaceae | 1.9 ± 0.6 | 1.4 ± 0.9 | n.d. | n.d. |
| Clavicipitaceae | 1.7 ± 1.0 | 0.9 ± 0.7 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| Trichosporonaceae | 1.6 ± 3.5 | 0.4 ± 0.5 | n.d. | n.d. |
| Lasiosphaeriaceae | 1.6 ± 0.5 b 3 | 0.8 ± 0.5 a | n.d. | n.d. |
| Chaetomiaceae | 1.6 ± 0.5 | 1.7 ± 0.6 | 0.01 ± 0.00 | 0.01 ± 0.00 |
| Pyronemataceae | 1.5 ± 0.9 | 0.9 ± 0.4 | n.d. | n.d. |
| Aspergillaceae | 0.7 ± 0.3 | 1.2 ± 1.2 | n.d. | n.d. |
| Didymellaceae | 1.0 ± 0.6 | 8.8 ± 11.9 | 2.1 ±1.7 | 1.0 ± 0.8 |
| Trichocomaceae | 1.0 ± 0.5 | 0.5 ± 0.4 | n.d. | n.d. |
| Sclerotiniaceae | 1.0 ± 0.5 | 0.8 ± 0.6 | 0.00 ± 0.00 | 0.00 ± 0.00 |
| un. 4 GS16 | 1.0 ± 0.5 | 0.6 ± 0.5 | n.d. | n.d. |
| Ascobolaceae | 0.8 ± 0.4 | 1.0 ± 0.4 | n.d. | n.d. |
| Genus level | ||||
| Alternaria | 0.01 ± 0.01 | 1.6 ± 3.2 | 17.3 ±12.0 | 20.7 ± 22.4 |
| un. Tremellales | 0.00 ± 0.00 | 0.00 ± 0.00 | 15.9 ± 7.6 | 23.1 ± 13.2 |
| Cryptococcus | 0.5 ± 0.4 | 0.5 ± 0.2 | 14.2 ± 17.9 | 7.4 ± 5.9 |
| Vishniacozyma | 0.2 ± 0.4 | 0.3 ± 0.5 | 13.9 ± 10.1 | 17.2 ± 16.4 |
| Davidiella | 0.8 ± 1.8 | 1.8 ± 3.7 | 12.4 ± 8.3 | 6.8 ± 3.7 |
| Dioszegia | 0.00 ± 0.00 | 0.00 ± 0.01 | 6.9 ± 4.7 | 4.8 ± 6.3 |
| Cystofilobasidium | 0.00 ± 0.00 | 0.00 ± 0.00 | 3.6 ± 2.7 | 3.6 ± 4.1 |
| Leucosporidium | 0.01 ± 0.01 | 0.01 ± 0.00 | 3.2 ± 3.0 | 1.1 ± 0.9 |
| un. Didymellaceae | 0.2 ± 0.2 | 0.4 ± 0.4 | 1.3 ± 1.6 | 0.7 ± 0.5 |
| Botryotinia | 1.0 ± 0.5 | 0.8 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| Lectera | 0.3 ± 0.2 | 0.3 ± 0.2 | 0.0 ± 0.0 | 1.1 ± 2.4 |
| Wardomyces | 5.2 ± 2.4 | 2.6 ± 2.5 | n.d. | n.d. |
| Tetracladium | 4.9 ± 1.4 | 2.6 ± 1.7 | n.d. | n.d. |
| Plectosphaerella | 4.0 ± 0.8 a | 8.7 ± 4.0 b | n.d. | n.d. |
| Mortierella | 4.0 ± 1.2 | 5.1 ± 6.6 | n.d. | n.d. |
| Gibberella | 3.7 ± 1.1 | 6.1 ± 4.6 | 0.0 ± 0.1 | 0.0 ± 0.0 |
| Pseudogymnoascus | 1.9 ± 1.1 | 1.7 ± 1.1 | n.d. | n.d. |
| Metarhizium | 1.6 ± 1.0 | 0.8 ± 0.6 | n.d. | n.d. |
| Apiotrichum | 1.6 ± 3.5 | 0.4 ± 0.5 | n.d. | n.d. |
| Parasola | 1.5 ± 3.3 | 0.0 ± 0.0 | n.d. | n.d. |
| Cephaliophora | 1.4 ± 0.5 | 1.1 ± 0.8 | n.d. | n.d. |
| Gibellulopsis | 1.1 ± 0.3 | 0.9 ± 0.2 | n.d. | n.d. |
| Dokmaia | 1.1 ± 0.6 | 1.8 ± 0.5 | 0.0 ± 0.1 | 0.0 ± 0.0 |
| un. GS16 | 1.0 ± 0.6 | 0.6 ± 0.5 | n.d. | n.d. |
| Didymella | 0.5 ± 0.5 | 8.3 ± 12.0 | 0.2 ± 0.4 | 0.5 ± 0.5 |
| Fusarium | 0.7 ± 0.3 a | 2.0 ± 1.1 b | 0.000 ± 0.001 | 0.000 ± 0.001 |
| Penicillium | 0.6 ± 0.4 | 1.2 ± 1.2 | n.d. | n.d. |
| Emericellopsis | 1.0 ± 0.9 | 0.8 ± 0.7 | n.d. | n.d. |
| Ascobolus | 1.0 ± 0.4 | 0.7 ± 0.3 | n.d. | n.d. |
| No. | OTU | Rhizosphere | Roots | ||
|---|---|---|---|---|---|
| No | NPK | No | NPK | ||
| 1 | un. 1 Alternaria | 0.0 ± 0.0 2 | 1.5 ± 3.0 | 5.8 ± 12.1 | 19.7 ± 21.7 |
| 2 | un. Tremellales | n.d. 2 | n.d. | 11.9 ± 7.8 | 17.3 ± 12.5 |
| 3 | un. Ascomycota | 3.9 ± 7.0 | 1.1 ± 1.4 | 4.1 ± 2.0 | 8.4 ± 7.4 |
| 4 | Davidiella sp. | 0.8 ± 1.8 | 1.8 ± 3.7 | 12.4 ± 8.3 | 6.8 ± 3.7 |
| 7 | Vishniacozyma victoriae | n.d. 2 | n.d. | 6.4 ± 2.6 | 13.9 ± 13.8 |
| 8 | Cryptococcus sp. | n.d. | n.d. | 9.0 ± 19.1 | 0.3 ±0.6 |
| 10 | Dioszegia crocea | n.d. | n.d. | 6.2 ± 4.4 | 4.3 ± 5.8 |
| 13 | un. Tremellales | n.d. | n.d. | 3.9 ± 2.5 | 5.7 ± 4.1 |
| 14 | Phoma exigua | 0.5 ± 0.5 | 8.3 ± 11.9 | n.d. | n.d. |
| 15 | Gibberella sp. | 2.0 ± 0.7 | 2.2 ± 1.8 | n.d. | n.d. |
| 18 | Cystofilobasidium macerans | n.d. | n.d. | 2.6 ± 1.2 | 3.8 ± 1.7 |
| 19 | Plectosphaerella cucumerina | 2.5 ± 0.5 | 3.0 ± 0.4 | n.d. | n.d. |
| 21 | Mortierella minutissima | 3.1 ± 1.0 | 4.8 ± 6.7 | n.d. | n.d. |
| 24 | Leucosporidium sp. | n.d. | n.d. | 3.2 ± 2.9 | 1.0 ± 0.9 |
| 25 | un. Hypocreales | 2.1 ± 2.3 | 1.1 ±0.6 | n.d. | n.d. |
| 27 | un. Alternaria | 0.0 ± 0.0 | 0.1 ±0.2 | 1.3 ±1.4 | 0.9 ± 0.7 |
| 31 | Wardomyces inflatus | 5.1 ± 2.4 | 2.6 ± 2.5 | n.d. | n.d. |
| 33 | Tetracladium sp. | 1.8 ± 0.5 | 1.3 ± 1.0 | n.d. | n.d. |
| 34 | un. Microascaceae | 7.1 ± 2.6 | 4.4 ± 2.6 | n.d. | n.d. |
| 35 | un. Ascomycota | 3.9 ± 3.3 | 1.8 ± 0.7 | n.d. | n.d. |
| 39 | Cryptococcus chernovii | n.d. | n.d. | 2.1 ±1.7 | 1.7 ±2.0 |
| 40 | Cryptococcus festucosus | n.d. | n.d. | 0.5 ± 0.2 | 1.6 ± 1.8 |
| 46 | Dokmaia monthadangii | 1.0 ± 0.6 | 1.7 ± 0.5 | n.d. | n.d. |
| 47 | Tetracladium maxilliforme | 2.8 ± 1.2 | 1.1 ± 0.9 | n.d. | n.d. |
| 57 | Botryotinia sp. | 1.0 ± 0.5 | 0.8 ± 0.6 | 0.0 ± 0.0 | 0.0 ± 0.0 |
| 65 | Fusarium cerealis | 1.3 ± 0.6 | 1.5 ± 1.3 | n.d. | n.d. |
| 66 | Gibellulopsis nigrescens | 1.1 ± 0.3 | 0.9 ± 0.2 | n.d. | n.d. |
| 67 | Cephaliophora sp. | 1.4 ± 0.5 | 1.0 ± 0.7 | n.d. | n.d. |
| 70 | Metarhizium sp. | 1.4 ± 1.0 | 0.7 ± 0.5 | n.d. | n.d. |
| 76 | un. Dothideomycetes | 1.1 ± 0.5 | 1.1 ± 0.8 | n.d. | n.d. |
| 78 | Pseudogymnoascus sp. | 1.0 ± 0.3 | 1.1 ± 0.6 | n.d. | n.d. |
| 82 | Cryptococcus tephrensis | 0.0 ± 0.0 | 0.1 ±0.1 | 1.9 ± 2.2 | 2.3 ± 2.4 |
| 85 | un. Hypocreales | 0.0 ± 0.1 | 3.0 ± 5.6 | n.d. | n.d. |
| 89 | Ascobolus sp. | 0.7 ± 0.3 | 1.0 ± 0.4 | n.d. | n.d. |
| 90 | un. Coniochaetales | 1.0 ± 0.3 | 0.9 ± 0.7 | n.d. | n.d. |
| 101 | Apiotrichum dulcitum | 1.6 ± 3.5 | 0.4 ± 0.5 | n.d. | n.d. |
| 103 | Emericellopsis microspora | 0.8 ± 0.7 | 1.0 ± 0.9 | n.d. | n.d. |
| 118 | Symmetrospora coprosmae | n.d. | n.d. | 1.0 ± 1.6 | 0.2 ± 0.1 |
| 142 | Fusarium domesticum | 0.2 ± 0.2 a3 | 1.1 ± 1.1 b | n.d. | n.d. |
| 152 | Lectera capsica | 0.3 ± 0.2 | 0.3 ±0.2 | 0.0 ± 0.0 | 1.1 ± 2.4 |
| 175 | Plectosphaerella plurivora | 1.4 ± 0.5 a | 5.1 ± 3.7 b | n.d. | n.d. |
| 190 | Parasola kuehneri | 1.5 ± 3.3 | 0.0 ± 0.0 | n.d. | n.d. |
| 643 | Vishniacozyma heimaeyensis | 0.2 ± 0.3 | 0.3 ± 0.5 | 7.4 ± 10.1 | 3.2 ± 2.5 |
| 1095 | un. Didymellaceae | 0.2 ± 0.2 | 0.4 ± 0.4 | 1.3 ± 1.5 | 0.7 ± 0.5 |
| 3321 | Cryptococcus magnus | n.d. | n.d. | 0.3 ± 0.1 | 1.1± 1.3 |
| Taxon | Rhizosphere | Roots | ||
|---|---|---|---|---|
| No | NPK | No | NPK | |
| Richness | 928 ± 55 | 790 ± 174 | 171 ± 40 | 144 ± 34 |
| Chao-1 | 1044 ± 73 | 909 ± 146 | 203 ±52 | 179 ± 40 |
| Simpson (1-D) | 0.97 ± 0.01 | 0.95 ± 0.04 | 0.85 ± 0.04 | 0.81 ±0.10 |
| Shannon | 4.8 ± 0.1 | 4.4 ±0.7 | 2.5 ± 0.2 | 2.3 ± 0.4 |
| Evenness | 0.13 ± 0.02 | 0.11 ± 0.04 | 0.08 ± 0.01 | 0.08 ±0.03 |
| Equitability | 0.70 ± 0.02 | 0.66 ± 0.09 | 0.49 ± 0.03 | 0.47 ±0.07 |
| Dominance (D) | 0.03 ± 0.01 | 0.04 ± 0.03 | 0.15 ± 0.04 | 0.19 ±0.10 |
| Berger-Parker | 0.10 ± 0.03 | 0.14 ±0.08 | 0.31 ± 0.09 | 0.34 ± 0.16 |
| No | NPK | p-Value | |
|---|---|---|---|
| Fruits, pcs/plant | 30 ± 12 | 31 ± 16 | 0.773 |
| Yield (Y), kg/plant | 1.28 ± 0.48 | 1.65 ± 0.92 | 0.312 |
| Average fruit mass, g | 43 ± 10 | 49 ± 13 | 0,261 |
| Aboveground phytomass 1 (A), g/plant | 224 ± 173 | 340 ± 275 | 0.334 |
| Belowground phytomass (B), g/plant | 15.6 ± 7.2 | 27.8 ± 17.1 | 0.226 |
| A/B | 13.1 ± 6.1 | 11.1 ± 6.0 | 0.625 |
| Total phytomass, g/plant | 1.66 ± 0.67 | 2.24 ± 1.28 | 0.279 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naumova, N.; Baturina, O.; Nechaeva, T.; Kabilov, M. Root and Rhizosphere Microbiome of Tomato Plants Grown in the Open Field in the South of West Siberia under Mineral Fertilization. Horticulturae 2022, 8, 1051. https://doi.org/10.3390/horticulturae8111051
Naumova N, Baturina O, Nechaeva T, Kabilov M. Root and Rhizosphere Microbiome of Tomato Plants Grown in the Open Field in the South of West Siberia under Mineral Fertilization. Horticulturae. 2022; 8(11):1051. https://doi.org/10.3390/horticulturae8111051
Chicago/Turabian StyleNaumova, Natalia, Olga Baturina, Taisia Nechaeva, and Marsel Kabilov. 2022. "Root and Rhizosphere Microbiome of Tomato Plants Grown in the Open Field in the South of West Siberia under Mineral Fertilization" Horticulturae 8, no. 11: 1051. https://doi.org/10.3390/horticulturae8111051
APA StyleNaumova, N., Baturina, O., Nechaeva, T., & Kabilov, M. (2022). Root and Rhizosphere Microbiome of Tomato Plants Grown in the Open Field in the South of West Siberia under Mineral Fertilization. Horticulturae, 8(11), 1051. https://doi.org/10.3390/horticulturae8111051







