Identification of QTL for Plant Architecture and Flowering Performance Traits in a Multi-Environment Evaluation of a Petunia axillaris × P. exserta Recombinant Inbred Line Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Data Analysis
2.3. Marker Development, Linkage Map Construction, and QTL Mapping
3. Results
3.1. Trait Analysis
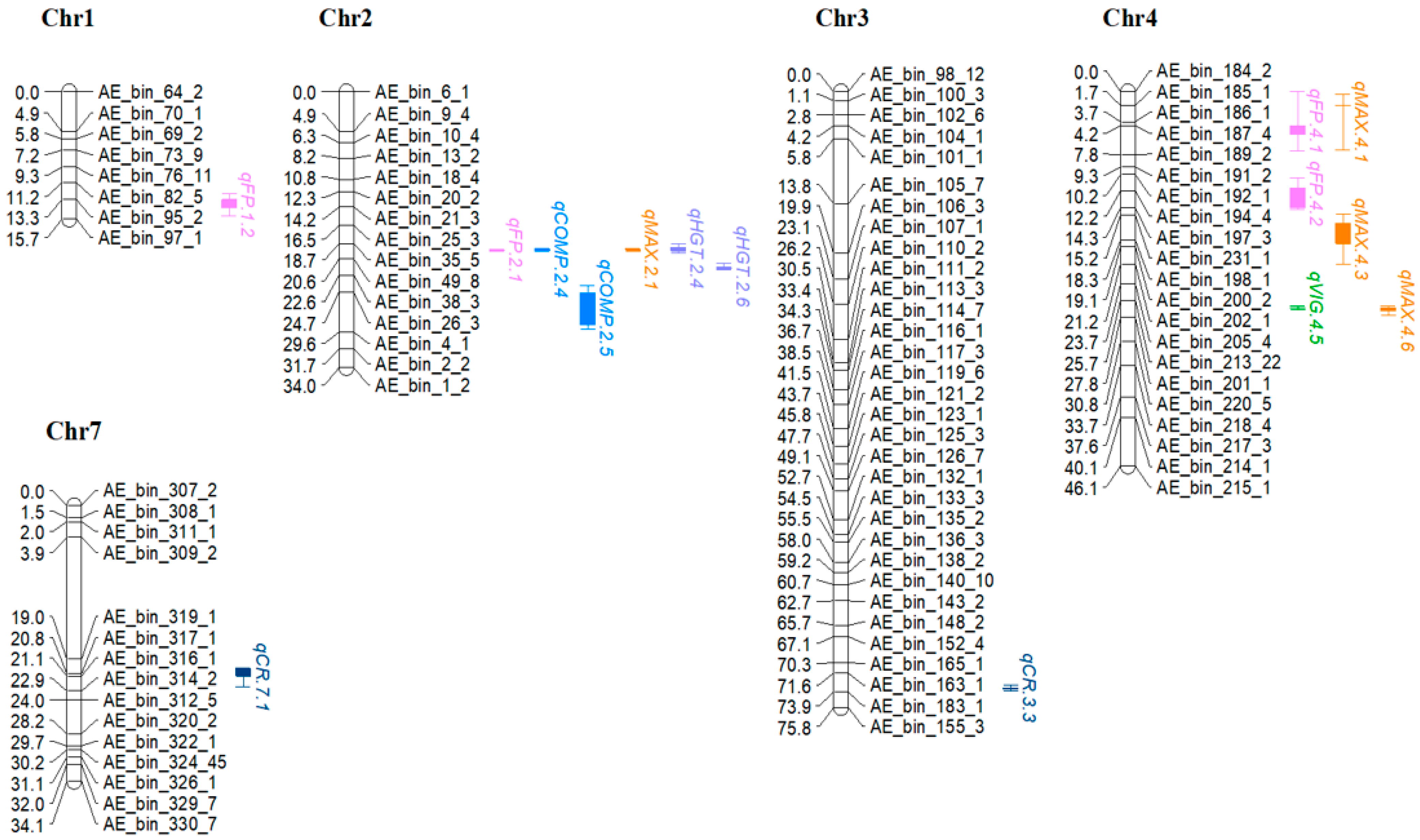
3.2. QTL Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- United States Department of Agriculture—National Agricultural Statistics Service. Floriculture Crops 2020 Summary; United States Department of Agriculture—National Agricultural Statistics Service: Washington, DC, USA, 2021; p. 63. [Google Scholar]
- Guo, Y.; Warner, R.M. Dissecting genetic diversity and genomic background of Petunia cultivars with contrasting growth habits. Hortic. Res. 2020, 7, 155. [Google Scholar] [CrossRef] [PubMed]
- Bombarely, A.; Moser, M.; Amrad, A.; Bao, M.; Bapaume, L.; Barry, C.S.; Bliek, M.; Boersma, M.R.; Borghi, L.; Bruggmann, R.; et al. Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2016, 2, 16074. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Ando, T.; Iida, S.-i.; Suzuki, A.; Buto, K.-i.; Tsukamoto, T.; Hashimoto, G.; Marchesi, E. Cross compatibility of Petunia cultivars and P. axillaris with native taxa of Petunia in relation to their chromosome number. J. Jpn. Soc. Hortic. Sci. 1996, 65, 625–634. [Google Scholar] [CrossRef][Green Version]
- Watanabe, H.; Ando, T.; Tsukamoto, T.; Hashimoto, G.; Marchesi, E. Cross-compatibility of Petunia exserta with other Petunia taxa. J. Jpn. Soc. Hortic. Sci. 2001, 70, 33–40. [Google Scholar] [CrossRef]
- Warner, R.M.; Walworth, A.E. Quantitative inheritance of crop timing traits in interspecific hybrid Petunia populations and interactions with crop quality parameters. J. Hered. 2010, 101, 308–316. [Google Scholar] [CrossRef]
- Griesbach, R.J. Petunia. In Flower Breeding and Genetics: Issues, Challenges and Opportunities for the 21st Century; Anderson, N.O., Ed.; Springer: Dordrecht, The Netherlands, 2007; pp. 301–336. [Google Scholar]
- Devicente, M.C.; Tanksley, S.D. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 1993, 134, 585–596. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Nelson, J.C. Advanced backcross QTL analysis: A method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor. Appl. Genet. 1996, 92, 191–203. [Google Scholar] [CrossRef]
- Guo, Y.; Lin, W.K.; Chen, Q.; Vallejo, V.A.; Warner, R.M. Genetic determinants of crop timing and quality traits in two interspecific Petunia recombinant inbred line populations. Sci. Rep. 2017, 7, 3200. [Google Scholar] [CrossRef]
- Chen, Q.C.; Guo, Y.; Warner, R.M. Identification of quantitative trait loci for component traits of flowering capacity across temperature in. G3 (Bethesda) 2019, 9, 3601–3610. [Google Scholar] [CrossRef]
- Cao, Z.; Guo, Y.; Yang, Q.; He, Y.; Fetouh, M.I.; Warner, R.M.; Deng, Z. Genome-wide search for quantitative trait loci controlling important plant and flower traits in petunia using an interspecific recombinant inbred population of Petunia axillaris and Petunia exserta. G3 (Bethesda) 2018, 8, 2309–2317. [Google Scholar] [CrossRef]
- Cao, Z.; Guo, Y.F.; Yang, Q.; He, Y.H.; Fetouh, M.I.; Warner, R.M.; Deng, Z.N. Genome-wide identification of quantitative trait loci for important plant and flower traits in petunia using a high-density linkage map and an interspecific recombinant inbred population derived from Petunia integrifolia and P. axillaris. Hortic. Res. 2019, 6, 27. [Google Scholar] [CrossRef] [PubMed]
- Warner, R.M. Temperature and photoperiod influence flowering and morphology of four Petunia spp. Hortscience 2010, 45, 365–368. [Google Scholar] [CrossRef]
- Devlaming, P.; Vaneekeres, J.E.M.; Wiering, H. A gene for flower color fading in Petunia hybrida. Theor. Appl. Genet. 1982, 61, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Quattrocchio, F.; Verweij, W.; Kroon, A.; Spelt, C.; Mol, J.; Koes, R. PH4 of petunia is an R2R3 MYB protein that activates vacuolar acidification through interactions with basic-helix-loop-helix transcription factors of the anthocyanin pathway. Plant Cell 2006, 18, 1274–1291. [Google Scholar] [CrossRef] [PubMed]
- Griesbach, R.J.; Stehmann, J.R.; Meyer, F. Anthocyanins in the “red” flowers of Petunia exserta. Phytochemistry 1999, 51, 525–528. [Google Scholar] [CrossRef]
- Berardi, A.E.; Esfeld, K.; Jäggi, L.; Mandel, T.; Cannarozzi, G.M.; Kuhlemeier, C. Complex evolution of novel red floral color in Petunia. Plant Cell 2021, 33, 2273–2295. [Google Scholar] [CrossRef]
- Peel, M.C.; Finlayson, B.L.; McMahon, T.A. Updated world map of the Koppen-Geiger climate classification. Hydrol. Earth Syst. Sci. 2007, 11, 1633–1644. [Google Scholar] [CrossRef]
- Fehr, W.R. Heritability. In Principles of Cultivar Development, Vol 1: Theory and Technique; Macmillan: New York, NY, USA, 1987. [Google Scholar]
- Xu, S.Z. Genetic mapping and genomic selection using recombination breakpoint data. Genetics 2013, 195, 1103–1115. [Google Scholar] [CrossRef]
- Bossolini, E.; Klahre, U.; Brandenburg, A.; Reinhardt, D.; Kuhlemeier, C. High resolution linkage maps of the model organism Petunia reveal substantial synteny decay with the related genome of tomato. Genome 2011, 54, 327–340. [Google Scholar] [CrossRef]
- Wang, S.; Basten, C.J.; Zeng, Z.B. Windows QTL Cartographer 2.5. Available online: https://brcwebportal.cos.ncsu.edu/qtlcart/WQTLCart.htm (accessed on 15 September 2022).
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R.E. Estimates of genetic and environmental variability in soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Vallejo, V.A.; Tychonievich, J.; Lin, W.-K.; Wangchu, L.; Barry, C.S.; Warner, R.M. Identification of QTL for crop timing and quality traits in an interspecific Petunia population. Mol. Breed. 2015, 35, 2. [Google Scholar] [CrossRef]
- Erwin, J.E.; Heins, R.D. Thermomorphogenic responses in stem and leaf development. Hortscience 1995, 30, 940–949. [Google Scholar] [CrossRef]
- Kaczperski, M.P.; Carlson, W.H.; Karlsson, M.G. Growth and development of Petunia x hybrida as a function of temperature and irradiance. J. Am. Soc. Hortic. Sci. 1991, 116, 232–237. [Google Scholar] [CrossRef]
- Hodges, L.; Suratman, M.N.; Brandle, J.R.; Hubbard, K.G. Growth and yield of snap beans as affected by wind protection and microclimate changes due to shelterbelts and planting dates. Hortscience 2004, 39, 996–1004. [Google Scholar] [CrossRef]
- Spelt, C.; Quattrocchio, F.; Mol, J.; Koes, R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell 2002, 14, 2121–2135. [Google Scholar] [CrossRef]
- Strommer, J.; Peters, J.L.; Gerats, T. Genetic recombination and mapping in petunia. In Petunia; Springer: New York, NY, USA, 2009; pp. 325–341. [Google Scholar]
| Trait Description (Units) | Trait Abbreviation | QTL Abbreviation |
|---|---|---|
| Floral coverage of plant canopy (%) | Flow | FP |
| Plant vigor (1–9 scale) | Vigor | VIG |
| Plant compactness (1–9 scale) | Comp | COM |
| Plant height (cm) | Height | HGT |
| Plant maximum width (cm) | MaxWid | MAX |
| Plant minimum width (cm) | MinWid | MIN |
| Flower color retention (1–9 scale) | ColorRet | CR |
| RILs | Parents | |||||||
|---|---|---|---|---|---|---|---|---|
| Trait z | n y | Mean | Sd | Min | Max | PA | PE | t-Test |
| Gilroy, CA (CA1) | ||||||||
| Flow | 538 | 52.9 | 25.81 | 0 | 90 | 53.3 | 66.7 | xns |
| Vigor | 539 | 4.9 | 1.44 | 1 | 9 | 6.3 | 4.3 | * |
| Comp | 539 | 4.1 | 1.08 | 1 | 7 | 3.3 | 2.3 | ns |
| Height | 539 | 42.9 | 9.86 | 13 | 69 | 41.3 | 42.0 | ns |
| MaxWid | 539 | 105.9 | 27.39 | 11 | 213 | 163.7 | 80.1 | * |
| MinWid | 446 | 87.6 | 25.68 | 10 | 163 | 121.3 | 71.3 | * |
| ColorRet | 331 | 4.0 | 1.87 | 1 | 8 | wna | 7.7 | |
| Buellton, CA (CA2) | ||||||||
| Flow | 591 | 54.4 | 22.55 | 0 | 90 | 50.0 | 73.3 | ns |
| Vigor | 595 | 4.3 | 1.22 | 1 | 8 | 5.7 | 4.0 | * |
| Comp | 595 | 4.1 | 1.11 | 1 | 8 | 3.3 | 5.3 | * |
| Height | 595 | 40.2 | 8.80 | 13 | 78 | 40.0 | 39.0 | ns |
| MaxWid | 595 | 98.7 | 24.65 | 13 | 173 | 160.5 | 76.0 | * |
| MinWid | 569 | 82.8 | 23.81 | 8 | 170 | 116.1 | 71.0 | * |
| ColorRet | 440 | 4.88 | 1.92 | 1 | 9 | na | 8.0 | |
| Huntersville, NC | ||||||||
| Flow | 397 | 41.11 | 21.22 | 0 | 80 | 45.0 | 55.0 | ns |
| Vigor | 397 | 5.92 | 2.21 | 1 | 9 | 8.0 | 7.0 | ns |
| Comp | 397 | 1.46 | 0.83 | 1 | 5 | 1.0 | 2.0 | ns |
| Height | 397 | 45.26 | 14.86 | 12 | 79 | 52.0 | 58.5 | ns |
| MaxWid | 397 | 111.71 | 35.16 | 7 | 196 | 157.0 | 115.0 | * |
| Bellefonte, PA | ||||||||
| Flow | 584 | 45.99 | 20.74 | 0 | 80 | 30.0 | 40.0 | ns |
| Vigor | 584 | 5.52 | 2.18 | 1 | 9 | 6.0 | 4.7 | ns |
| Comp | 584 | 3.49 | 2.09 | 1 | 9 | 2.0 | 2.7 | ns |
| Height | 584 | 46.42 | 15.42 | 3 | 79 | 54.7 | 44.0 | ns |
| MaxWid | 584 | 116.11 | 34.02 | 8 | 176 | 130.3 | 98.3 | * |
| ColorRet | 429 | 5.72 | 1.86 | 1 | 9 | na | 7.0 | |
| Trait z | Flow | Vigor | Comp | Height | MaxWid | MinWid |
|---|---|---|---|---|---|---|
| All locations | ||||||
| Vigor | 0.49 ** y | |||||
| Comp | 0.12 ** | −0.32 ** | ||||
| Height | 0.34 ** | 0.65 ** | −0.21 ** | |||
| MaxWid | 0.42 ** | 0.73 ** | −0.32 ** | 0.62 ** | ||
| MinWid | 0.42 ** | 0.76 ** | 0.18 ** | 0.45 ** | 0.82 ** | |
| ColorRet | −0.19 ** | 0.07 * | −0.16 ** | 0.14 ** | −0.03 | −0.17 ** |
| Gilroy, CA (CA1) | ||||||
| Vigor | 0.61 ** | |||||
| Comp | 0.50 ** | 0.42 ** | ||||
| Height | 0.40 ** | 0.55 ** | 0.52 ** | |||
| MaxWid | 0.44 ** | 0.76 ** | 0.26 ** | 0.52 ** | ||
| MinWid | 0.47 ** | 0.77 ** | 0.29 ** | 0.49 ** | 0.83 ** | |
| ColorRet | −0.14 ** | −0.11 | −0.07 | 0.03 | −0.15 * | −0.14 * |
| Buellton, CA (CA2) | ||||||
| Vigor | 0.43 ** | |||||
| Comp | 0.34 ** | 0.23 ** | ||||
| Height | 0.11 * | 0.45 ** | 0.26 ** | |||
| MaxWid | 0.36 ** | 0.76 ** | 0.08 * | 0.43 ** | ||
| MinWid | 0.38 ** | 0.74 ** | 0.10 * | 0.40 ** | 0.81 ** | |
| ColorRet | −0.15 * | −0.17 ** | −0.01 | 0.12 * | −0.20 ** | −0.15 * |
| Bellefonte, PA | ||||||
| Vigor | 0.69 ** | |||||
| Comp | −0.37 ** | −0.51 ** | ||||
| Height | 0.55 ** | 0.76 ** | −0.47 ** | |||
| MaxWid | 0.66 ** | 0.76 ** | −0.68 ** | 0.70 ** | ||
| ColorRet | −0.09 | 0.11 * | 0.04 | 0.07 | −0.11 * | |
| Huntersville, NC | ||||||
| Vigor | 0.74 ** | |||||
| Comp | −0.34 ** | −0.47 ** | ||||
| Height | 0.50 ** | 0.59 ** | −0.44 ** | |||
| MaxWid | 0.48 ** | 0.64 ** | −0.56 ** | 0.64 ** | ||
| Trait z | H2 | ||||
|---|---|---|---|---|---|
| All locations | Buellton, CA | Gilroy, CA | Bellefonte, PA | Huntersville, NC | |
| Flow | 0.75 | 0.65 | 0.49 | 0.97 | 0.86 |
| Vigor | 0.77 | 0.77 | 0.55 | 0.96 | 0.88 |
| Comp | 0.74 | 0.60 | 0.45 | 0.93 | 0.90 |
| Height | 0.79 | 0.79 | 0.58 | 0.92 | 0.76 |
| MaxWid | 0.75 | 0.76 | 0.53 | 0.95 | 0.85 |
| MinWid | 0.68 | 0.72 | 0.40 | ||
| ColorRet | 0.79 | 0.71 | 0.67 | 0.99 | |
| Trait z | QTL | Chr | Nearest Marker | Loc. | Posit. (cm) | Interval (cm) y | LOD x | LOD Threshold w | A v | %VE u |
|---|---|---|---|---|---|---|---|---|---|---|
| Flow | qFP.1.1 | 1 | AE_bin_78_1 | CA1 | 8.81 | 6.8–9.1 | 2.76 | 2.66 | −4.47 | 6.07 |
| Flow | qFP.1.2 | 1 | AE_bin_95_2 | CA2 | 13.31 | 13.3–15.3 | 7.36 | 2.44 | −6.65 | 14.20 |
| Flow | AE_bin_95_2 | NC | 14.31 | 12.6–15.3 | 2.99 | 2.59 | −4.85 | 5.88 | ||
| Flow | qFP.2.1 | 2 | AE_bin_58_12 | NC | 19.51 | 19.4–19.7 | 11.45 | 2.59 | −10.27 | 23.81 |
| Flow | AE_bin_61_3 | PA | 19.61 | 19.4–19.7 | 8.56 | 2.63 | −10.97 | 18.95 | ||
| Flow | qFP.2.2 | 2 | AE_bin_52_48 | CA2 | 20.21 | 19.9–21.2 | 2.94 | 2.44 | −4.22 | 5.21 |
| Flow | qFP.2.3 | 2 | AE_bin_4_1 | NC | 27.71 | 25.7–28.8 | 5.17 | 2.59 | −10.77 | 17.56 |
| Flow | qFP.3.1 | 3 | AE_bin_112_12 | NC | 31.41 | 27.9–33.8 | 2.84 | 2.59 | 5.07 | 4.92 |
| Flow | qFP.4.1 | 4 | AE_bin_187_4 | PA | 4.21 | 0–6.2 | 4.31 | 2.63 | 6.28 | 8.91 |
| Flow | AE_bin_187_4 | CA2 | 5.21 | 1.5–7.3 | 5.80 | 2.44 | 6.00 | 11.37 | ||
| Flow | qFP.4.2 | 4 | AE_bin_195_4 | NC | 11.91 | 10.6–14.3 | 3.07 | 2.59 | 4.76 | 5.65 |
| Flow | AE_bin_197_3 | CA2 | 14.31 | 12.8–14.5 | 4.81 | 2.44 | 5.35 | 9.02 | ||
| Flow | qFP.4.3 | 4 | AE_bin_207_2 | CA1 | 25.01 | 24.1–25.4 | 3.04 | 2.66 | 5.10 | 6.77 |
| Flow | qFP.6.1 | 6 | AE_bin_236_1 | CA2 | 10.11 | 8.1–13.2 | 3.04 | 2.44 | −4.39 | 5.53 |
| Vigor | qVIG.2.1 | 2 | AE_bin_61_3 | PA | 19.61 | 19.4–19.7 | 12.56 | 2.74 | −1.09 | 22.34 |
| Vigor | qVIG.2.2 | 2 | AE_bin_52_48 | NC | 20.01 | 19.9–20.5 | 17.41 | 2.65 | −1.22 | 30.72 |
| Vigor | qVIG.2.3 | 2 | AE_bin_44_4 | PA | 21.61 | 21.1–21.8 | 12.82 | 2.74 | −1.10 | 22.61 |
| Vigor | qVIG.2.4 | 2 | AE_bin_4_1 | PA | 28.71 | 26.4–30.5 | 5.17 | 2.74 | −1.15 | 15.47 |
| Vigor | qVIG.3.1 | 3 | AE_bin_128_16_310_2 | CA1 | 48.51 | 48.1–48.9 | 2.59 | 2.54 | 0.22 | 4.19 |
| Vigor | qVIG.4.1 | 4 | AE_bin_184_2 | NC | 0.01 | 0–1.0 | 8.68 | 2.65 | 0.79 | 13.85 |
| Vigor | qVIG.4.2 | 4 | AE_bin_187_4 | CA2 | 4.21 | 2.6–6.6 | 11.28 | 2.65 | 0.55 | 26.01 |
| Vigor | qVIG.4.3 | 4 | AE_bin_197_3 | CA2 | 14.31 | 12.2–14.4 | 11.79 | 2.65 | 0.56 | 27.08 |
| Vigor | qVIG.4.4 | 4 | AE_bin_198_1 | CA1 | 17.21 | 15.2–18.3 | 6.27 | 2.54 | 0.44 | 14.05 |
| Vigor | qVIG.4.5 | 4 | AE_bin_229_48 | CA1 | 26.61 | 26.4–26.9 | 10.86 | 2.54 | 0.51 | 19.71 |
| Vigor | PA | 26.61 | 26.4–27.1 | 3.05 | 2.74 | 0.51 | 4.62 | |||
| Vigor | qVIG.4.6 | 4 | AE_bin_201_1 | CA2 | 27.81 | 27.4–29.9 | 5.19 | 2.65 | 0.37 | 9.37 |
| Vigor | qVIG.6.1 | 6 | AE_bin_239_2 | NC | 21.01 | 15.3–23.4 | 3.81 | 2.65 | −0.65 | 5.41 |
| Vigor | qVIG.6.2 | 6 | AE_bin_252_5 | NC | 35.31 | 35.1–35.5 | 3.81 | 2.65 | 0.68 | 5.48 |
| Vigor | qVIG.7.1 | 7 | AE_bin_320_2 | CA1 | 28.21 | 24.6–29.2 | 2.69 | 2.54 | 0.24 | 5.09 |
| Comp | qCOMP.1.1 | 1 | AE_bin_74_1 | CA1 | 8.01 | 6.3–8.8 | 2.63 | 2.52 | −0.18 | 5.28 |
| Comp | qCOMP.1.2 | 1 | AE_bin_91_6 | CA2 | 11.51 | 10.9–11.6 | 8.25 | 2.48 | −0.32 | 14.04 |
| Comp | qCOMP.1.3 | 1 | AE_bin_95_2 | CA1 | 14.31 | 12.6–15.3 | 2.69 | 2.52 | −0.19 | 5.91 |
| Comp | qCOMP.2.1 | 2 | AE_bin_6_1 | CA2 | 0.01 | 0–3.4 | 2.59 | 2.48 | −0.20 | 4.23 |
| Comp | qCOMP.2.2 | 2 | AE_bin_16_7 | CA2 | 10.21 | 9.5–10.8 | 6.50 | 2.48 | −0.32 | 12.92 |
| Comp | qCOMP.2.3 | 2 | AE_bin_31_10 | NC | 18.21 | 18.0–19.0 | 6.65 | 2.49 | 0.34 | 15.28 |
| Comp | qCOMP.2.4 | 2 | AE_bin_55_14 | CA2 | 19.41 | 19.3–19.4 | 8.19 | 2.48 | 0.36 | 13.71 |
| Comp | AE_bin_58_12 | PA | 19.51 | 19.3–19.6 | 7.42 | 2.68 | 0.81 | 14.84 | ||
| Comp | qCOMP.2.5 | 2 | AE_bin_28_3 | NC | 24.31 | 23.9–26.7 | 7.31 | 2.49 | 0.36 | 16.67 |
| Comp | AE_bin_4_1 | PA | 28.71 | 26.3–29.3 | 3.20 | 2.68 | 1.05 | 14.70 | ||
| Comp | qCOMP.2.6 | 2 | AE_bin_3_202_229_1 | CA1 | 32.61 | 31.7–33.6 | 3.61 | 2.52 | −0.30 | 7.17 |
| Comp | qCOMP.4.1 | 4 | AE_bin_206_4 | PA | 24.11 | 23.3–24.9 | 5.59 | 2.68 | −0.67 | 10.81 |
| Comp | qCOMP.5.1 | 5 | AE_bin_290_8 | PA | 10.31 | 8.4–11.6 | 2.72 | 2.68 | −0.45 | 5.06 |
| Comp | qCOMP.5.2 | 5 | AE_bin_295_2 | CA1 | 15.01 | 13.9–17.6 | 4.50 | 2.52 | 0.24 | 9.62 |
| Comp | qCOMP.7.1 | 7 | AE_bin_315_1 | CA2 | 22.41 | 20.5–24.1 | 3.18 | 2.48 | −0.19 | 4.88 |
| ColorRet | qCR.2.1 | 2 | AE_bin_15_11 | PA | 9.81 | 8.9–10.5 | 4.00 | 2.68 | −0.65 | 9.55 |
| ColorRet | qCR.2.2 | 2 | AE_bin_50_8 | PA | 19.21 | 19.1–19.3 | 7.10 | 2.68 | −0.95 | 15.95 |
| ColorRet | qCR.2.3 | 2 | AE_bin_4_1 | PA | 27.71 | 25.2–28.7 | 3.74 | 2.68 | −1.10 | 14.13 |
| ColorRet | qCR.3.1 | 3 | AE_bin_165_1 | PA | 69.71 | 68.0–70.3 | 3.26 | 2.68 | −0.53 | 7.87 |
| ColorRet | qCR.3.2 | 3 | AE_bin_163_1 | CA2 | 71.61 | 71.2–72.1 | 4.11 | 2.59 | −0.58 | 9.50 |
| ColorRet | qCR.3.3 | 3 | AE_bin_180_16 | CA2 | 73.41 | 73.1–73.6 | 4.71 | 2.59 | −0.61 | 10.72 |
| ColorRet | AE_bin_177_199_237_1 | CA1 | 73.51 | 73.3–73.7 | 6.20 | 2.66 | −0.72 | 14.87 | ||
| ColorRet | qCR.6.1 | 6 | AE_bin_244_6 | PA | 25.61 | 23.4–26.6 | 4.59 | 2.68 | −0.66 | 9.90 |
| ColorRet | qCR.6.2 | 6 | AE_bin_263_50_86_2 | PA | 32.01 | 30.6–32.3 | 3.94 | 2.68 | −0.59 | 8.30 |
| ColorRet | qCR.7.1 | 7 | AE_bin_318_1 | PA | 20.21 | 20.1–22.1 | 4.34 | 2.68 | −0.57 | 9.58 |
| ColorRet | AE_bin_316_1 | CA2 | 21.11 | 20.8–22.4 | 5.92 | 2.59 | −0.68 | 15.97 | ||
| ColorRet | qCR.7.2 | 7 | AE_bin_324_45 | PA | 30.21 | 29.4–30.8 | 4.87 | 2.68 | −0.60 | 10.33 |
| ColorRet | qCR.7.3 | 7 | AE_bin_329_7 | CA2 | 32.01 | 31.1–33.0 | 5.97 | 2.59 | −0.65 | 14.46 |
| MaxWid | qMAX.1.1 | 1 | AE_bin_64_2 | CA1 | 0.01 | 0–1.2 | 3.83 | 2.58 | 5.46 | 6.82 |
| MaxWid | qMAX.1.2 | 1 | AE_bin_82_5 | CA1 | 11.31 | 11.0–11.5 | 4.90 | 2.58 | 6.13 | 8.49 |
| MaxWid | qMAX.2.1 | 2 | AE_bin_55_14 | NC | 19.41 | 19.3–19.4 | 17.23 | 2.54 | −20.75 | 36.88 |
| MaxWid | AE_bin_58_12 | PA | 19.51 | 19.3–19.7 | 11.62 | 2.71 | −15.60 | 19.27 | ||
| MaxWid | qMAX.2.2 | 2 | AE_bin_27_2 | NC | 24.11 | 23.4–24.3 | 13.88 | 2.54 | −20.03 | 31.34 |
| MaxWid | qMAX.4.1 | 4 | AE_bin_185_1 | CA2 | 1.71 | 0.3–7.2 | 5.03 | 2.68 | 6.97 | 8.51 |
| MaxWid | PA | 1.71 | 0.8–3.7 | 6.46 | 2.71 | 11.47 | 9.73 | |||
| MaxWid | qMAX.4.2 | 4 | AE_bin_193_1 | CA2 | 11.11 | 10.9–11.2 | 10.16 | 2.68 | 10.17 | 22.27 |
| MaxWid | qMAX.4.3 | 4 | AE_bin_231_1 | CA1 | 16.21 | 15.1–17.4 | 8.96 | 2.58 | 9.34 | 19.43 |
| MaxWid | AE_bin_198_1 | CA2 | 17.21 | 15.9–21 | 5.97 | 2.68 | 8.22 | 12.08 | ||
| MaxWid | AE_bin_199_3 | PA | 18.71 | 17.9–21.2 | 4.46 | 2.71 | 9.29 | 6.52 | ||
| MaxWid | qMAX.4.4 | 4 | AE_bin_207_2 | CA1 | 25.01 | 24.6–25.6 | 10.47 | 2.58 | 9.45 | 19.44 |
| MaxWid | qMAX.4.5 | 4 | AE_bin_210_117_218_4_2_1 | CA2 | 26.01 | 25.9–26.1 | 5.51 | 2.68 | 7.28 | 9.68 |
| MaxWid | qMAX.4.6 | 4 | AE_bin_229_48 | CA1 | 26.61 | 26.4–26.9 | 10.94 | 2.58 | 9.56 | 20.08 |
| MaxWid | AE_bin_226_1 | PA | 27.01 | 26.4–27.5 | 4.54 | 2.71 | 9.15 | 6.65 | ||
| MinWid | qMIN.4.1 | 4 | AE_bin_187_4 | CA2 | 5.21 | 4.9–7.3 | 5.84 | 2.65 | 7.40 | 11.26 |
| MinWid | qMIN.4.2 | 4 | AE_bin_231_1 | CA1 | 16.21 | 16.0–17.5 | 8.02 | 2.63 | 9.43 | 20.64 |
| MinWid | qMIN.4.3 | 4 | AE_bin_200_2 | CA2 | 19.11 | 18.7–20.8 | 7.44 | 2.65 | 8.09 | 13.61 |
| MinWid | qMIN.4.4 | 4 | AE_bin_207_2 | CA1 | 25.01 | 24.9–25.3 | 10.67 | 2.63 | 9.99 | 23.37 |
| MinWid | qMIN.4.5 | 4 | AE_bin_229_48 | CA1 | 26.61 | 26.4–27.4 | 10.66 | 2.63 | 10.00 | 23.31 |
| MinWid | qMIN.7.1 | 7 | AE_bin_218_4 | CA1 | 33.01 | 31.0–34.0 | 3.26 | 2.63 | 5.48 | 6.84 |
| Height | qHGT.2.1 | 2 | AE_bin_7_5 | PA | 2.91 | 0–4.9 | 2.73 | 2.61 | −4.20 | 6.85 |
| Height | qHGT.2.2 | 2 | AE_bin_17_3 | PA | 11.01 | 10.8–11.4 | 5.73 | 2.61 | −5.45 | 11.27 |
| Height | qHGT.2.3 | 2 | AE_bin_30_1 | NC | 17.91 | 16.8–18.2 | 6.96 | 2.84 | −5.98 | 14.89 |
| Height | qHGT.2.4 | 2 | AE_bin_50_8 | PA | 19.21 | 18.7–19.4 | 14.01 | 2.61 | −7.92 | 24.56 |
| Height | AE_bin_61_3 | NC | 19.61 | 19.3–19.9 | 7.89 | 2.84 | −6.02 | 16.23 | ||
| Height | qHGT.2.5 | 2 | AE_bin_49_8 | CA1 | 20.61 | 20.2–20.7 | 4.04 | 2.60 | −2.21 | 7.48 |
| Height | qHGT.2.6 | 2 | AE_bin_44_4 | PA | 21.61 | 21.1–21.8 | 14.88 | 2.61 | −8.13 | 25.71 |
| Height | AE_bin_43_2 | CA2 | 21.81 | 21.6–21.9 | 9.96 | 2.67 | −3.20 | 15.92 | ||
| Height | qHGT.2.7 | 2 | AE_bin_27_2 | NC | 24.11 | 22.8–24.3 | 6.74 | 2.84 | −5.95 | 14.33 |
| Height | qHGT.2.8 | 2 | AE_bin_4_1 | PA | 28.71 | 26.9–31.3 | 8.80 | 2.61 | −9.78 | 23.59 |
| Height | qHGT.2.9 | 2 | AE_bin_3_202_229_2 | CA2 | 32.41 | 32.2–32.6 | 6.88 | 2.67 | −3.54 | 11.38 |
| Height | qHGT.4.1 | 4 | AE_bin_187_4 | PA | 4.21 | 4.0–6.7 | 3.89 | 2.61 | 4.83 | 9.19 |
| Height | qHGT.4.2 | 4 | AE_bin_206_4 | CA2 | 24.11 | 23.3–24.8 | 6.27 | 2.67 | 2.52 | 8.72 |
| Height | qHGT.4.3 | 4 | AE_bin_229_48 | CA1 | 26.61 | 26.4–27.1 | 5.59 | 2.60 | 2.49 | 9.39 |
| Height | qHGT.5.1 | 5 | AE_bin_288_1 | CA1 | 7.01 | 5.0–8.4 | 4.29 | 2.60 | 2.24 | 8.26 |
| Height | qHGT.5.2 | 5 | AE_bin_299_1 | CA2 | 13.31 | 12.9–13.4 | 7.21 | 2.67 | 2.51 | 10.41 |
| Height | qHGT.7.1 | 7 | AE_bin_314_2 | CA2 | 22.91 | 22.1–24.5 | 2.84 | 2.67 | −1.47 | 3.53 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Q.C.; Warner, R.M. Identification of QTL for Plant Architecture and Flowering Performance Traits in a Multi-Environment Evaluation of a Petunia axillaris × P. exserta Recombinant Inbred Line Population. Horticulturae 2022, 8, 1006. https://doi.org/10.3390/horticulturae8111006
Chen QC, Warner RM. Identification of QTL for Plant Architecture and Flowering Performance Traits in a Multi-Environment Evaluation of a Petunia axillaris × P. exserta Recombinant Inbred Line Population. Horticulturae. 2022; 8(11):1006. https://doi.org/10.3390/horticulturae8111006
Chicago/Turabian StyleChen, QiuXia C., and Ryan M. Warner. 2022. "Identification of QTL for Plant Architecture and Flowering Performance Traits in a Multi-Environment Evaluation of a Petunia axillaris × P. exserta Recombinant Inbred Line Population" Horticulturae 8, no. 11: 1006. https://doi.org/10.3390/horticulturae8111006
APA StyleChen, Q. C., & Warner, R. M. (2022). Identification of QTL for Plant Architecture and Flowering Performance Traits in a Multi-Environment Evaluation of a Petunia axillaris × P. exserta Recombinant Inbred Line Population. Horticulturae, 8(11), 1006. https://doi.org/10.3390/horticulturae8111006






