Detection of a Point Mutation (G143A) in Cyt b of Corynespora cassiicola That Confers Pyraclostrobin Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Pathogens and Plants
2.2. Determination of Pyraclostrobin Sensitivity
2.3. Genomic DNA Extraction and Analysis of the Cyt b Gene
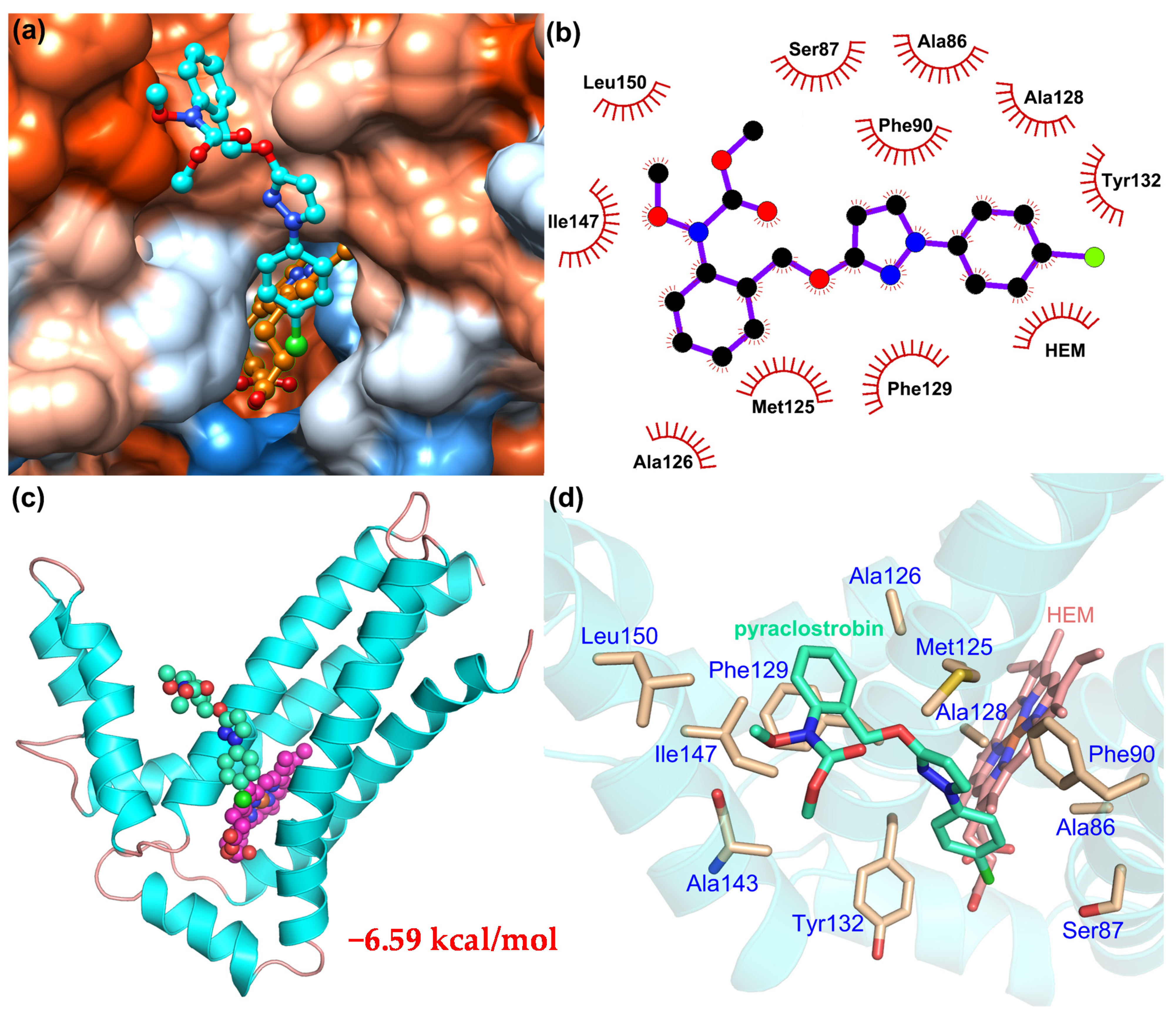
2.4. Molecular Docking of Pyraclostrobin in Cyt b of C. cassiicola
2.5. LAMP Primer Design
2.6. LAMP Reaction Mixtures
2.7. Optimization of LAMP Reaction Time and Temperature
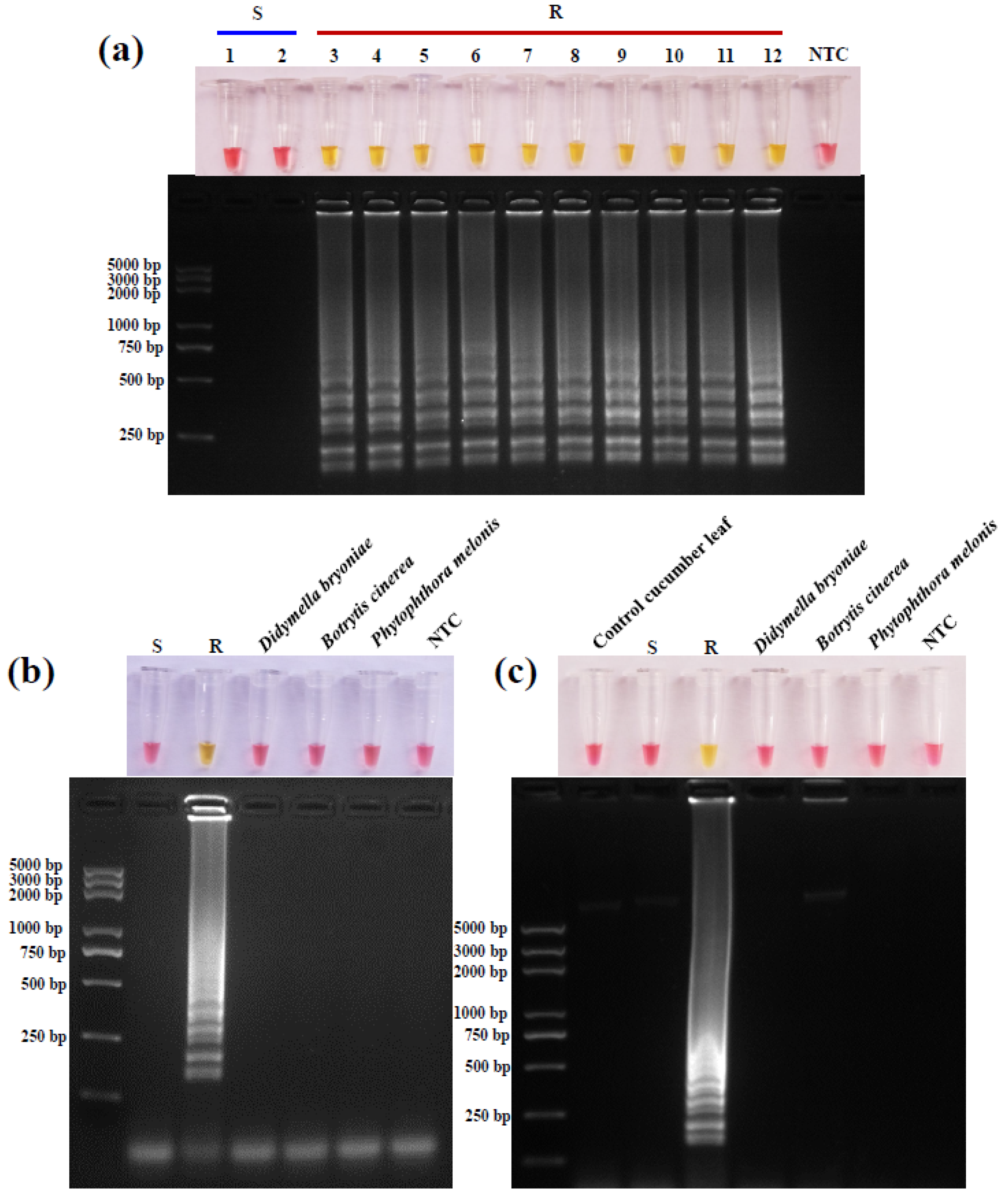
2.8. LAMP Assay Specificity
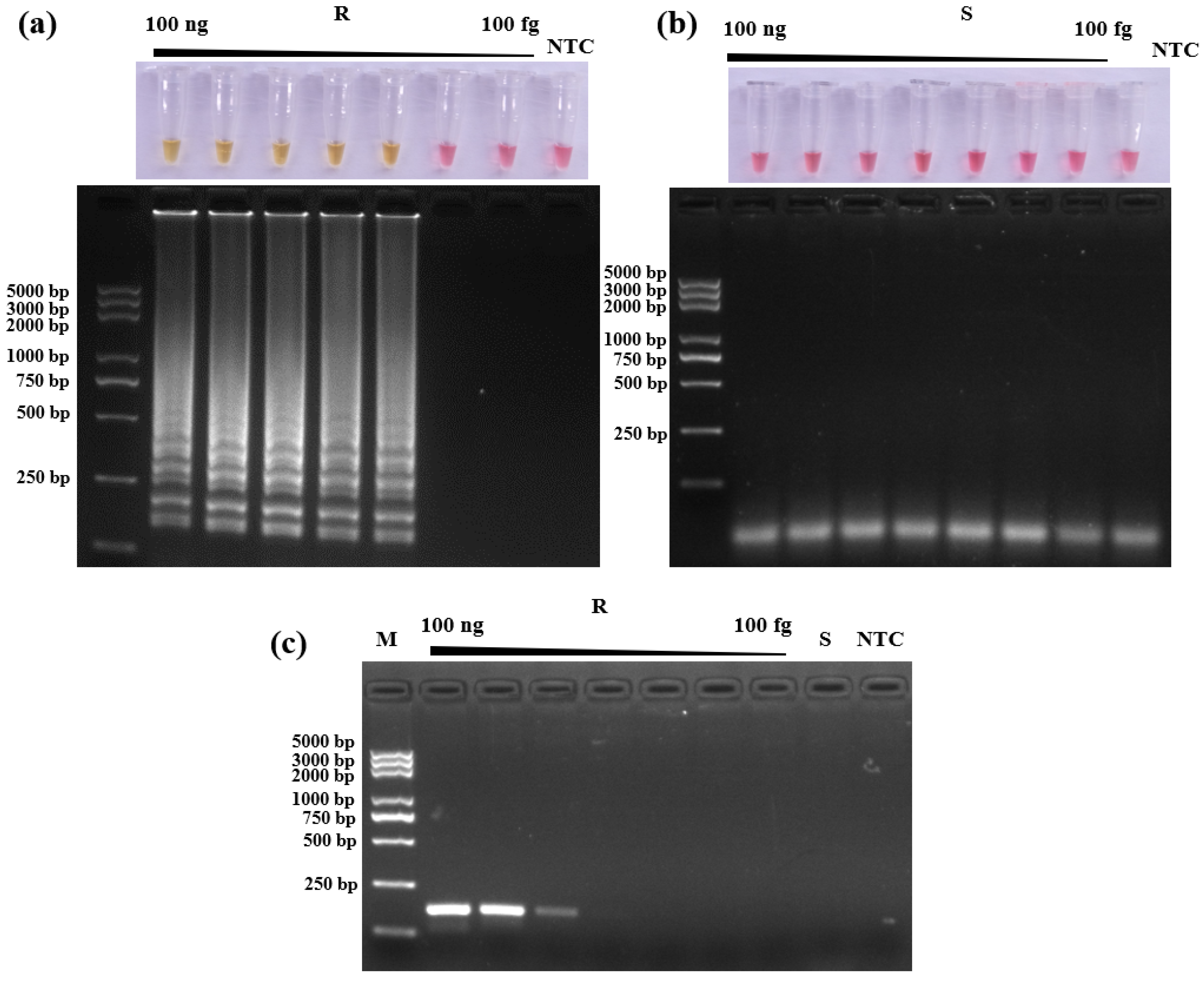
2.9. LAMP Assay Detection Limit
3. Results
3.1. Pyraclostrobin Sensitivity of C. cassiicola Isolates and Sequence of Cyt b Gene
3.2. Pyraclostrobin Docking in the Cyt b of C. cassiicola
3.3. LAMP Reaction Optimization
3.4. Detection Limit of LAMP Assay and Conventional PCR
3.5. LAMP Method Specificity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miyamoto, T.; Ishii, H.; Seko, T.; Kobori, S.; Tomita, Y. Occurrence of Corynespora cassiicola isolates resistant to boscalid on cucumber in Ibaraki Prefecture, Japan. Plant Pathol. 2010, 58, 1144–1151. [Google Scholar] [CrossRef]
- Li, T.; Xiu, Q.; Zhang, J.; Wang, J.X.; Duan, Y.B.; Zhou, M.G. Pharmacological characteristics and efficacy of fluazinam against Corynespora cassiicola, causing cucumber target spot in greenhouses. Plant Dis. 2020, 104, 2449–2454. [Google Scholar] [CrossRef]
- Carris, L.M.; Glawe, D.A.; Gray, L.E. Isolation of the soybean pathogens Corynespora cassiicola and Phialophora gregata from cysts of Heterodera glycines in Illinois. Mycologia 1986, 78, 503–506. [Google Scholar] [CrossRef]
- Dixon, L.J.; Schlub, R.L.; Pernezny, K.; Datnoff, L.E. Host specialization and phylogenetic diversity of Corynespora cassiicola. Phytopathology 2009, 99, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.X.; Ge, Y.P.; Shen, Y.N.; Li, M.; Zhang, X.; Chen, H.; Deng, S.; de Hoog, G.S.; Liu, W.D. Phaeohyphomycosis caused by a plant pathogen, Corynespora cassiicola. Med. Mycol. 2011, 49, 657–661. [Google Scholar] [CrossRef]
- Yamada, H.; Takahashi, N.; Hori, N.; Asano, Y.; Mochizuki, K.; Ohkusu, K.; Nishimura, K. Rare case of fungal keratitis caused by Corynespora cassiicola. J. Infect Chemother. 2013, 19, 1167–1169. [Google Scholar] [CrossRef]
- Smith, L.J. Host Range, Phylogenetic, and Pathogenic Diversity of Corynespora cassiicola (Berk. & Curt.) Wei. Ph.D. Thesis, University of Florida, Gainesville, FL, USA, 2008. [Google Scholar]
- Déon, M.; Scomparin, A.; Tixier, A.; Mattos, C.R.R.; Leroy, T.; Seguin, M.; Roeckel-Drevet, P.; Pujade-Renaud, V. First characterization of endophytic Corynespora cassiicola isolates with variant cassiicolin genes recovered from rubber trees in Brazil. Fungal Divers 2012, 54, 87–99. [Google Scholar] [CrossRef]
- Date, H.; Kataoka, E.; Tanina, K.; Sasaki, S.; Inoue, K.; Nasu, H.; Kasuyama, S. Sensitivity of Corynespora cassicola, causal agent of Corynespora leaf spot of cucumber, to thiophanate-methyl, diethofencarb and azoxystrobin. Jpn. J. Phytopathol. 2004, 70, 10–13. [Google Scholar] [CrossRef]
- Miyamoto, T.; Ishii, H.; Stammler, G.; Koch, A.; Ogawara, T.; Tomita, Y.; Fountaine, J.M.; Ushio, S.; Seko, T.; Kobori, S. Distribution and molecular characterization of Corynespora cassiicola isolates resistant to boscalid. Plant Pathol. 2010, 59, 873–881. [Google Scholar] [CrossRef]
- Ishii, H.; Miyamoto, T.; Ushio, S.; Kakishima, M. Lack of cross-resistance to a novel succinate dehydrogenase inhibitor, fluopyram, in highly boscalid-resistant isolates of Corynespora cassiicola and Podosphaera xanthii. Pest Manag. Sci. 2011, 67, 474–482. [Google Scholar] [CrossRef]
- Duan, Y.; Xin, W.; Lu, F.; Li, T.; Li, M.; Wu, J.; Wang, J.; Zhou, M. Benzimidazole- and QoI-resistance in Corynespora cassiicola populations from greenhouse-cultivated cucumber: An emerging problem in China. Pestic. Biochem. Phys. 2019, 153, 95–105. [Google Scholar] [CrossRef]
- Mackenzie, K.J.; Xavier, K.V.; Wen, A.; Timilsina, S.; Adkison, H.M.; Dufault, N.S.; Vallad, G.E. Widespread QoI fungicide resistance revealed among Corynespora cassiicola tomato isolates in Florida. Plant Dis. 2019, 104, 893–903. [Google Scholar] [CrossRef] [PubMed]
- Xavier, S.A.; de Mello, F.E.; da Silva, H.P.; Canteri, M.G.; Koga, L.J.; Lopes, I.O.N.; Godoy, C.V. Microtiter method to monitor Corynespora cassiicola and sensitivity of the pathogen to carbendazim, prothioconazole and pyraclostrobin. Crop Prot. 2021, 144, 105554. [Google Scholar] [CrossRef]
- Dufour, M.C.; Fontaine, S.; Montarry, J.; Corio-Costet, M.F. Assessment of fungicide resistance and pathogen diversity in Erysiphe necator using quantitative real-time PCR assays. Pest Manag. Sci. 2011, 67, 60–69. [Google Scholar] [CrossRef]
- Furuya, S.; Suzuki, S.; Kobayashi, H.; Saito, S.; Takayanagi, T. Rapid method for detecting resistance to a QoI fungicide in Plasmopara viticola populations. Pest Manag. Sci. 2010, 65, 840–843. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Aoki, Y.; Saito, S.; Suzuki, S. Characterisation of heteroplasmic status at codon 143 of the Botrytis cinerea cytochrome b gene in a semi-quantitative AS-PCR assay. Pest Manag. Sci. 2015, 71, 467–477. [Google Scholar] [CrossRef]
- Gobeil-Richard, M.; Tremblay, D.M.; Beaulieu, C.; der Heyden, H.V.; Carisse, O. A pyrosequencing-based method to quantify genetic substitutions associated with resistance to succinate dehydrogenase inhibitor fungicides in Botrytis spp. populations. Pest Manag. Sci. 2016, 72, 566–573. [Google Scholar] [CrossRef]
- Massi, F.; Torriani, S.F.F.; Borghi, L.; Toffolatti, S.L. Fungicide resistance evolution and detection in plant pathogens: Plasmopara viticola as a case study. Microorganisms 2021, 9, 119. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Fukuta, S.; Nimi, Y.; Ohishi, K.; Yoshimura, Y.; Anai, N.; Hotta, M.; Fukaya, M.; Kato, T.; Oya, T.; Kanbe, M. Development of reverse transcription loop-mediated isothermal amplification (RT-LAMP) method for detection of two viruses and chrysanthemum stunt viroid. Annu. Rep. Kansai Plant Prot. Soc. 2005, 47, 31–36. [Google Scholar] [CrossRef]
- Safavieh, M.; Ahmed, M.U.; Ng, A.; Zourob, M. High-throughput real-time electrochemical monitoring of LAMP for pathogenic bacteria detection. Biosens. Bioelectron. 2014, 58, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Niessen, L. Current state and future perspectives of loop-mediated isothermal amplification (LAMP)-based diagnosis of filamentous fungi and yeasts. Appl. Microbiol. Biotechnol. 2015, 99, 553–574. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Hieno, A.; Kusunoki, M.; Suga, H.; Kageyama, K. LAMP detection of four plant-pathogenic oomycetes and its application in lettuce fields. Plant Dis. 2018, 103, 298–307. [Google Scholar] [CrossRef]
- Hu, X.R.; Dai, D.J.; Wang, H.D.; Zhang, C.Q. Rapid on-site evaluation of the development of resistance to quinone outside inhibitors in Botrytis cinerea. Sci. Rep. 2017, 7, 13861. [Google Scholar] [CrossRef]
- Chen, S.; Schnabel, G.; Yuan, H.; Luo, C. LAMP detection of the genetic element ‘Mona’ associated with DMI resistance in Monilinia fructicola. Pest Manag. Sci. 2019, 75, 779–786. [Google Scholar] [CrossRef]
- Fan, F.; Yin, W.X.; Li, G.Q.; Lin, Y.; Luo, C.X. Development of a LAMP method for detecting SDHI fungicide resistance in Botrytis cinerea. Plant Dis. 2018, 102, 1612–1618. [Google Scholar] [CrossRef]
- Vielba-Fernández, A.; de Vicente, A.; Pérez-García, A.; Fernández-Ortuño, D. Monitoring methyl benzimidazole carbamate-resistant isolates of the cucurbit powdery mildew pathogen, Podosphaera xanthii, using loop-mediated isothermal amplification. Plant Dis. 2019, 103, 1515–1524. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, L.; Li, H.; Gao, Y.; Mu, W.; Liu, F. Development of a LAMP method for detecting the N75S mutant in SDHI-resistant Corynespora cassiicola. Anal. Biochem. 2020, 597, 113687. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, S.; Lu, X.; Pang, Z.; Cai, M.; Liu, X. Assessing the risk that Phytophthora melonis can develop a point mutation (V1109L) in CesA3 conferring resistance to carboxylic acid amide fungicides. PLoS ONE 2012, 7, e42069. [Google Scholar] [CrossRef]
- Lange, C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. Eur. Mol. Biol. Organ. 2001, 20, 6591–6600. [Google Scholar] [CrossRef] [PubMed]
- Sierotzki, H.; Kraus, N.; Assemat, P.; Stanger, C.; Cleere, S.; Windass, J.; Gisi, U. Evaluation of resistance to QoI fungicides in Plasmopara viticola populations in Europe. In Modern Fungicides and Antifungal Compounds IV; Dehne, H.W., Gisi, U., Kuck, K.H., Russell, P.E., Lyr, H., Eds.; BCPC Publishing: Alton, UK, 2005; pp. 73–80. [Google Scholar]
- Genet, J.L.; Jaworska, G.; Deparis, F. Effect of dose rate and mixtures of fungicides on selection for QoI resistance in populations of Plasmopara viticola. Pest Manag. Sci. 2006, 62, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Lesemann, S.S.; Schimpke, S.; Dunemann, F.; Deising, H.B. Mitochondrial heteroplasmy for the cytochrome b gene controls the level of strobilurin resistance in the apple powdery mildew fungus Podosphaera leucotricha (Ell.&Ev.) E.S. Salmon. J. Plant Dis. Protect. 2006, 113, 259–266. [Google Scholar]
- Ishii, H.; Yano, K.; Date, H.; Furuta, A.; Sagehashi, Y.; Yamaguchi, T.; Sugiyama, T.; Nishimura, K.; Hasama, W. Molecular characterization and diagnosis of QoI resistance in cucumber and eggplant fungal pathogens. Phytopathology 2007, 97, 1458–1466. [Google Scholar] [CrossRef] [PubMed]
- Esser, L.; Quinn, B.; Li, Y.F.; Zhang, M.; Elberry, M.; Yu, L.; Yu, C.A.; Xia, D. Crystallographic studies of quinol oxidation site inhibitors: A modified classification of inhibitors for the cytochrome bc1 complex. J. Mol. Biol. 2004, 341, 281–302. [Google Scholar] [CrossRef]
- Sierotzki, H.; Frey, R.; Wullschleger, J.; Palermo, S.; Karlin, S.; Godwin, J.; Gisi, U. Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag. Sci. 2007, 63, 225–233. [Google Scholar] [CrossRef]
- Sierotzki, H. Respiration Inhibitors: Complex III. In Fungicide Resistance in Plant Pathogens; Ishii, H., Hollomon, D.W., Eds.; Springer Publishing: Tokyo, Japan, 2015; pp. 119–144. [Google Scholar]
- Klosowski, A.C.; de Mio, L.L.M.; Miessner, S.; Rodrigues, R.; Stammler, G. Detection of the F129L mutation in the cytochrome b gene in Phakopsora pachyrhizi. Pest Manag. Sci. 2016, 72, 1211–1215. [Google Scholar] [CrossRef] [PubMed]



| Primer Name | Sequence (5′-3′) a | Use |
|---|---|---|
| F3 | CCTGGGTTATGTTTTACCATAC | Forward outer primer for LAMP |
| B3 | AAGTGCATTAGTGCTAAAGC | Backward outer primer for LAMP |
| BIP | TTGAGGAGGTTTCTCTGTTAACAATGCTAATACGAAAGGTAAAACGAAAT | Backward inner primer for LAMP |
| FIP1 | ACTCAAGGGATGGCACTCATTAGGTCAAATGTCCTTATGACC | Forward inner primers to detect the G143A mutants of Cyt b gene in C. cassiicola for LAMP |
| FIP2 | ACTCAAGGGATGGCACTCATTAGGTCAAATGTCCTTATGAAC | |
| FIP3 | ACTCAAGGGATGGCACTCATTAGGTCAAATGTCCTTATGATC | |
| F | GTCAAATGTCCTTATGATC | To amplify partial fragments (163 bp) of Cyt b gene of C. cassiicola |
| R | GCTAATACGAAAGGTAAAACGAAAT | |
| CcCytb-F | GCGAATTCCTATTTAGTTGATTC | PCR primers to amplify the partial Cytb from C. cassiicola [12] |
| CcCytb-R | GGTTACCTGATCCAGCTGTATC |
| Source | Pyraclostrobin | ||
|---|---|---|---|
| N † | Range of EC50 (μg/mL) | Mean ± SD ⁑ (μg/mL) | |
| Lanling County, Linyi City | 25 | 1.98–8.68 | 5.09 ± 2.25 ns |
| Xin County, Liaochen City | 16 | 1.67–6.94 | 4.74 ± 1.58 |
| Daiyue District, Taian City | 22 | 1.74–8.74 | 5.09 ± 1.79 |
| Laicheng Distrist, Laiwu City | 4 | 1.95–4.69 | 3.71 ± 2.20 |
| Pingyuan County, Dezhou City | 12 | 1.68–8.82 | 3.92 ± 2.04 |
| Shouguang City, Weifang City | 26 | 1.99–8.00 | 5.32 ± 1.92 |
| Jiyang County, Jinan City | 12 | 1.70–6.21 | 3.68 ± 1.71 |
| Zhoucun County, Zibo City | 14 | 1.88–8.69 | 4.43 ± 1.95 |
| Cumulative | 131 | 1.67–8.82 | 4.74 * |
| Duan’s lab § | 2 | 0.006–0.01 | 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Li, C.; Li, G.; Zhu, J.; Liu, F.; Jiang, L.; Mu, W.; Liu, X. Detection of a Point Mutation (G143A) in Cyt b of Corynespora cassiicola That Confers Pyraclostrobin Resistance. Horticulturae 2021, 7, 155. https://doi.org/10.3390/horticulturae7060155
Li X, Li C, Li G, Zhu J, Liu F, Jiang L, Mu W, Liu X. Detection of a Point Mutation (G143A) in Cyt b of Corynespora cassiicola That Confers Pyraclostrobin Resistance. Horticulturae. 2021; 7(6):155. https://doi.org/10.3390/horticulturae7060155
Chicago/Turabian StyleLi, Xiuhuan, Chengcheng Li, Guixiang Li, Jiamei Zhu, Feng Liu, Lin Jiang, Wei Mu, and Xili Liu. 2021. "Detection of a Point Mutation (G143A) in Cyt b of Corynespora cassiicola That Confers Pyraclostrobin Resistance" Horticulturae 7, no. 6: 155. https://doi.org/10.3390/horticulturae7060155
APA StyleLi, X., Li, C., Li, G., Zhu, J., Liu, F., Jiang, L., Mu, W., & Liu, X. (2021). Detection of a Point Mutation (G143A) in Cyt b of Corynespora cassiicola That Confers Pyraclostrobin Resistance. Horticulturae, 7(6), 155. https://doi.org/10.3390/horticulturae7060155






