A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance
Abstract
1. Introduction
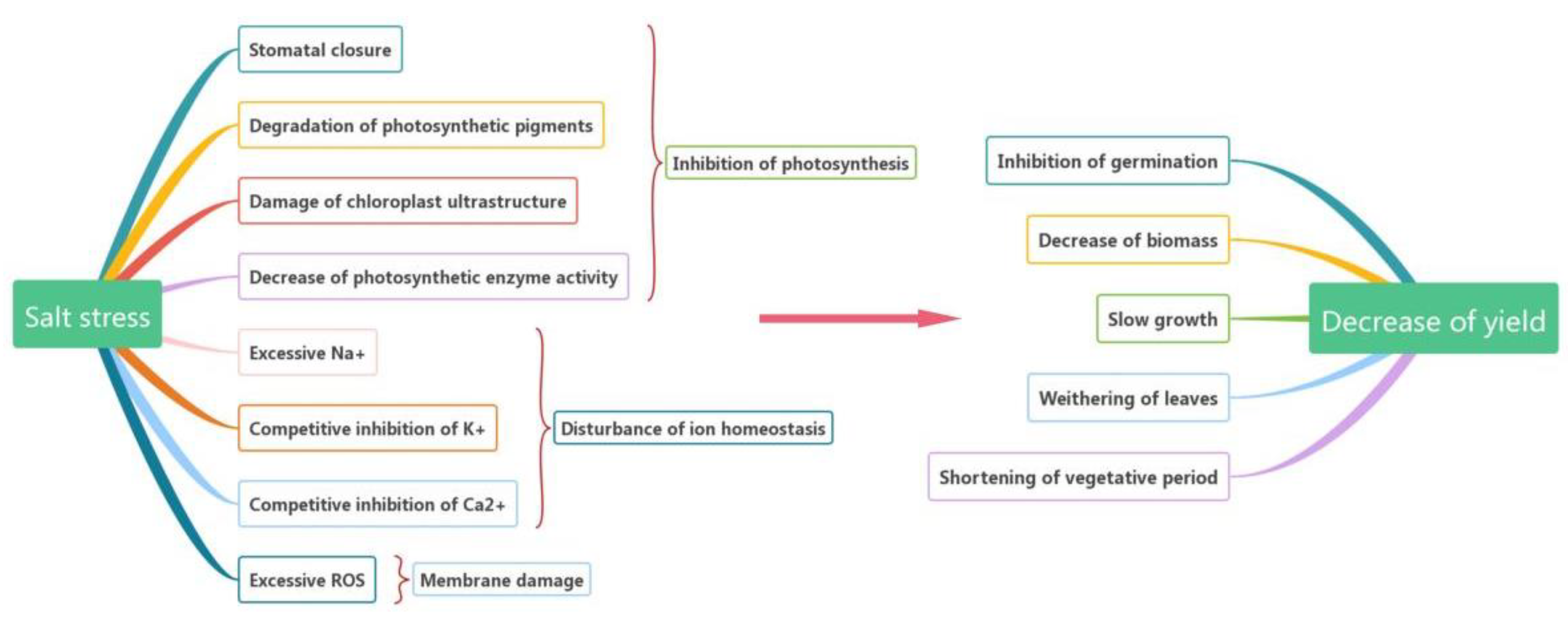
2. Effects of Salt Stress on Plants
2.1. Inhibition of Photosynthesis
2.2. Disturbance of Ion Homeostasis
2.3. Membrane Damage
2.4. Indicators for Salt Stress in Plants
3. Physiological Mechanisms of Plant Tolerance under Salt Stress
3.1. Osmotic Adjustment
3.1.1. Organic Substances
3.1.2. Inorganic Ions
3.2. Scavenging of ROS
3.2.1. Enzymatic Antioxidants
3.2.2. Non-Enzymatic Antioxidants
3.3. Other Physiological Regulation under Salt Stress
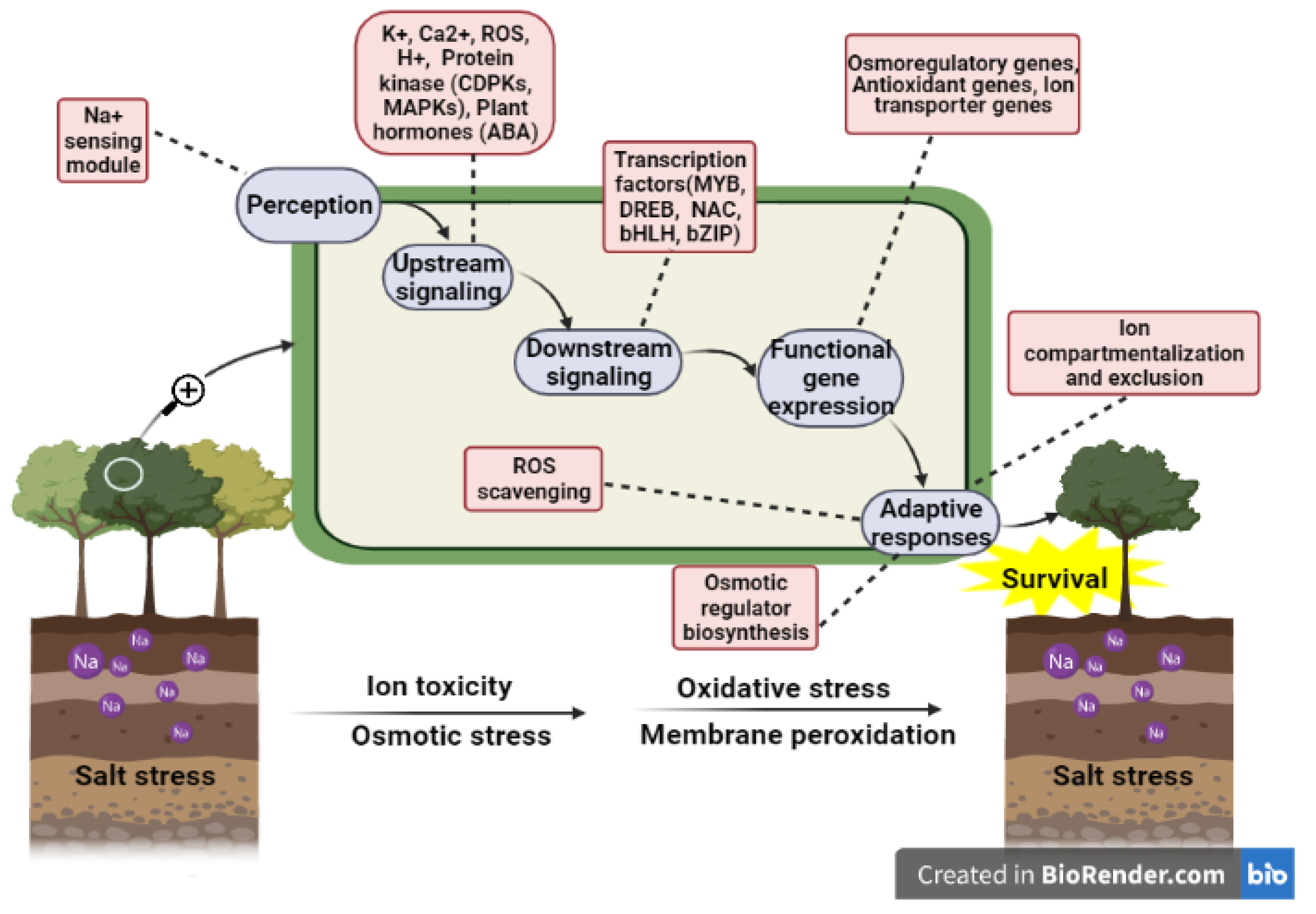
4. Salt Stress Signal Transduction System
4.1. Ca2+-Dependent Signal Transduction Pathway
4.1.1. SOS Pathway
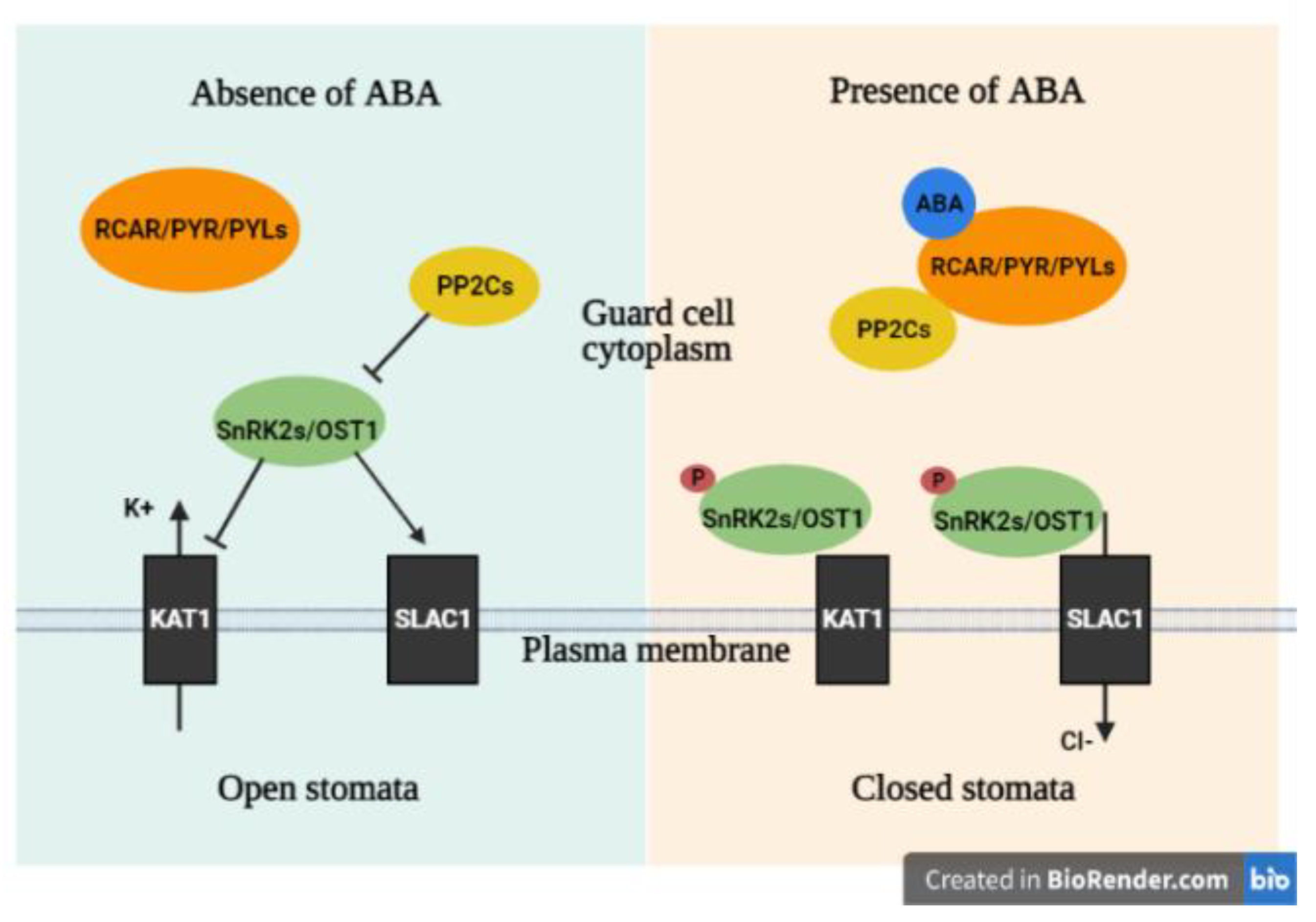
4.1.2. ABA Pathway
4.1.3. CDPK Pathway
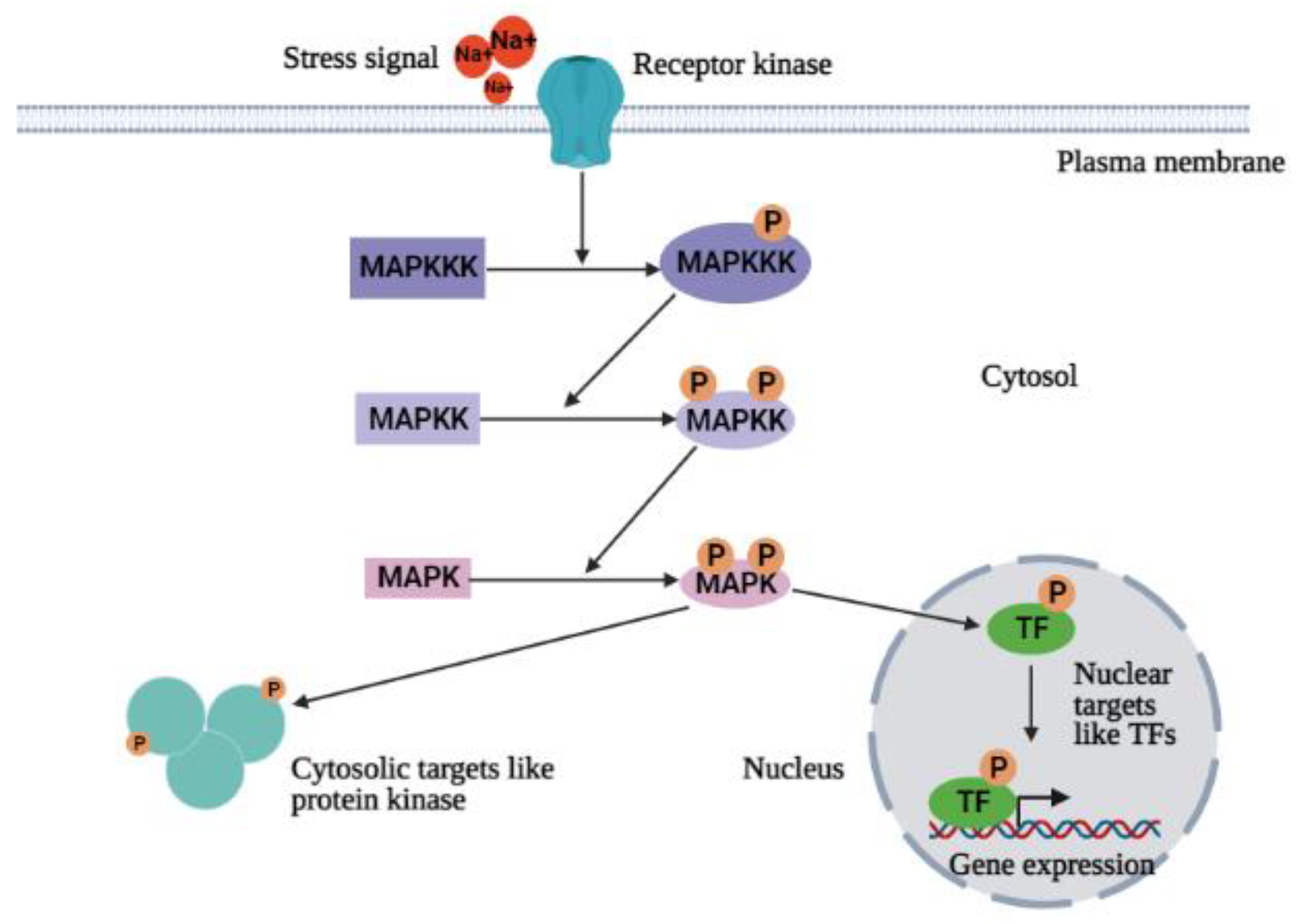
4.2. Ca2+-Independent Signal Transduction Pathway (MAPKs Cascade)
5. Salt Tolerance Related Genes
5.1. Osmotic Adjustment Related Genes
5.2. Ion Transporter Related Genes
5.3. Antioxidant-Related Genes
5.4. Signal Transduction-Related Genes
5.5. Regulatory Genes
| Name of Gene | Origin of Gene | Transgenic Plant | Role of Gene | Reference |
|---|---|---|---|---|
| P5CS1 | Phaseolus vulgaris | Arabidopsis thaliana | rate-limiting enzyme in proline biosynthesis | [134] |
| P5CS2 | Phaseolus vulgaris | Arabidopsis thaliana | rate-limiting enzyme in proline biosynthesis | [134] |
| LrAMADH1 | Lycium ruthenicum Murr | null | catalyze betaine aldehyde to betaine | [136] |
| SlBADH | Suaeda liaodonggensis | Solanum lycopersicum | catalyze betaine aldehyde to betaine | [137] |
| AtTPPD | Arabidopsis thaliana | Arabidopsis thaliana | catalyze dephosphorylation of trehalose 6-phosphate to form trehalose | [139] |
| HvHKT1;1 | Hordeum vulgare | Arabidopsis thaliana | transport Na+ and mediates the absorption of Na+ when the external K + is deficient | [145] |
| HvHKT1;5 | Hordeum vulgare | null | negatively transport Na+ in Barley | [146] |
| OsHKT1 | Oryza sativa | null | transport Na+ | [147] |
| OsHKT2 | Oryza sativa | null | transport Na+ | [147] |
| OsVHA | Oryza sativa | null | transport Na+ | [147] |
| AtNHX1 | Arabidopsis thaliana | Arachis hypogaea, Torenia fournieri | vacuolar Na+/H+ antiporter | [149] |
| LfNHX1 | Leptochloa fusca | null | vacuolar Na+/H+ antiporter | [150] |
| IhNHX1 | Iris halophila | null | vacuolar Na+/H+ antiporter | [151] |
| HcNHX1 | Halostachys caspica | null | vacuolar Na+/H+ antiporter | [152] |
| PgNHX1 | Pennisetum alopecuroides | Brassica juncea | vacuolar Na+/H+ antiporter | [155] |
| VrNHX1 | Vigna radiata | Arabidopsis thaliana | vacuolar Na+/H+ antiporter | [156] |
| PtVP1.1 | Populus trichocarpa | Populus trichocarpa | vacuolar H+ phosphorylase | [157] |
| TaTVP1 | Triticum aestivuml | Arabidopsis thaliana | vacuolar H+ phosphorylase | [158] |
| AVP1 | Arabidopsis thaliana | Solanum lycopersicum | vacuolar H+ phosphorylase | [159] |
| ChVDE | Cerasus humilis | Arabidopsis thaliana | violax-anthin de-epoxidase that catalyze the transformation of different CAR | [167] |
| OsNAC45 | Oryza sativa | null | transcription factor participate in different stress responses and ABA signal response | [176] |
| OsMADS25 | Oryza sativa | null | transcription factor involved in ABA-mediated regulatory pathways and ROS scavenging | [177] |
| AtCDPK27 | Arabidopsis thaliana | null | membrane-localized protein kinase in CDPKs signaling | [179] |
| AtCPK12 | Arabidopsis thaliana | null | protein kinase in CDPKs signaling | [180] |
| PtMAPKK4 | Populus trichocarpa | Nicotiana tabacum | protein kinase in MAPKs signaling | [184] |
| VvMKK2 | Vitis vinifera | Arabidopsis thaliana | protein kinase in MAPKs signaling | [185] |
| VvMKK4 | Vitis vinifera | Arabidopsis thaliana | protein kinase in MAPKs signaling | [185] |
| ZmMKK4 | Zea mays | Arabidopsis thaliana | protein kinase in MAPKs signaling | [186] |
| ZmMPK5 | Zea mays | Nicotiana tabacum | protein kinase in MAPKs signaling | [187] |
| GhMAP3K40 | Gossypium herbaceum | Nicotiana benthamiana | protein kinase in MAPKs signaling | [188] |
| GhMPK2 | Gossypium herbaceum | Nicotiana tabacum | protein kinase in MAPKs signaling | [189] |
| GmbZIP2 | Glycine max | null | transcription factor involved in salt stress response | [190] |
| AtbHLH122 | Arabidopsis thaliana | null | increased salt tolerance by regulating salt responsive gene AtKUP2 | [191] |
| AtWRKY33 | Arabidopsis thaliana | null | increased salt tolerance by regulating salt responsive gene AtKUP2 | [191] |
| AtMYB20 | Arabidopsis thaliana | null | negatively regulates type 2C serine/threonine protein phosphatases | [192] |
| ThDREB | Tamarix hispida | Nicotiana tabacum | transcription factor involved in stress responses | [193] |
| SlMYB102 | Solanum lycopersicum | null | transcription factor involved in stress responses | [194] |
| AtGSTF8 | Arabidopsis thaliana | null | function in the root fine-tuning the redox homeostasis | [196] |
| AtGSTU19 | Arabidopsis thaliana | null | function in the root fine-tuning the redox homeostasis | [196] |
| PpSARK | Physcomitrella patens | null | senescence-associated receptor-like kinase related to ABA | [175] |
| GmTIP2;3 | Glycine max | null | a tonoplast intrinsic protein related to osmotic regulation | [140] |
| VvNAC17 | Vitis vinifera | Arabidopsis thaliana | up-regulates the expression of ABA and stress-related genes | [195] |
6. Improvement Techniques for Increasing Plant Salt Tolerance
6.1. Non-Genetic Improvement Techniques
6.2. Genetic Improvement Techniques
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ludwig, M.; Wilmes, P.; Schrader, S. Measuring soil sustainability via soil resilience. Sci. Total Environ. 2018, 626, 1484–1493. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Diaz-Vivancos, P.; Alvarez, S.; Fernandez-Garcia, N.; Sanchez-Blanco, M.J.; Hernandez, J.A. Nacl-Induced Physiological and Biochemical Adaptative Mechanisms in the Ornamental Myrtus Communis L. Plants. J. Plant Physiol. 2015, 183, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant Responses to Salt Stress: Adaptive Mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef]
- Acosta-Motos, J.R.; Diaz-Vivancos, P.; Alvarez, S.; Fernandez-Garcia, N.; Sanchez-Blanco, M.J.; Hernandez, J.A. Physiological and Biochemical Mechanisms of the Ornamental Eugenia Myrtifolia L. Plants for Coping with NaCl Stress and Recovery. Planta 2015, 242, 829–846. [Google Scholar] [CrossRef]
- Barba-Espín, G.; Clemente-Moreno, M.J.; Álvarez, S.; García-Legaz, M.F.; Hernandez, J.A.; Diaz-Vivancos, P. Salicylic acid negatively affects the response to salt stress in pea plants. Plant Biol. 2011, 13, 909–917. [Google Scholar] [CrossRef]
- Hernandez, J.A.; Ferrer, M.A.; Jimenez, A.; Barcelo, A.R.; Sevilla, F. Antioxidant Systems and O2.−/H2O2 Production in the Apoplast of Pea Leaves. Its Relation with Salt-Induced Necrotic Lesions in Minor Veins. Plant Physiol. 2001, 127, 817–831. [Google Scholar] [CrossRef]
- Sudhir, P.; Murthy, S. Effects of salt stress on basic processes of photosynthesis. Photosynthetica 2004, 42, 481–486. [Google Scholar] [CrossRef]
- Gong, X.; Chao, L.; Zhou, M.; Hong, M.; Luo, L.; Wang, L.; Ying, W.; Jingwei, C.; Songjie, G.; Fashui, H. Oxidative damages of maize seedlings caused by exposure to a combination of potassium deficiency and salt stress. Plant Soil 2010, 340, 443–452. [Google Scholar] [CrossRef]
- Shu, S.; Guo, S.R.; Sun, J.; Yuan, L.Y. Effects of Salt Stress on the Structure and Function of the Photosynthetic Apparatus in Cucumis Sativus and its Protection by Exogenous Putrescine. Physiol. Plant 2012, 146, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Song, C.P.; Wang, B.; Zhou, J.; Kangasjarvi, J.; Zhu, J.K.; Gong, Z. ROS Signaling and Stomatal Movement in Plant Responses to Drought Stress and Pathogen Attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef] [PubMed]
- Cavusoglu, K.; Kiliç, S.; Kabar, K. Effects of some plant growth regulators on stem anatomy of radish seedlings grown under saline (NaCl) conditions. Plant Soil Environ. 2008, 54, 428–433. [Google Scholar] [CrossRef]
- Li, P.; Zhu, Y.; Song, X.; Song, F. Negative effects of long-term moderate salinity and short-term drought stress on the photosynthetic performance of Hybrid Pennisetum. Plant Physiol. Biochem. 2020, 155, 93–104. [Google Scholar] [CrossRef]
- Zhang, H.-F.; Liu, S.-Y.; Ma, J.-H.; Wang, X.-K.; Haq, S.U.; Meng, Y.-C.; Zhang, Y.-M.; Chen, R.-G. CaDHN4, a Salt and Cold Stress-Responsive Dehydrin Gene from Pepper Decreases Abscisic Acid Sensitivity in Arabidopsis. Int. J. Mol. Sci. 2019, 21, 26. [Google Scholar] [CrossRef]
- Du, Y.-T.; Zhao, M.-J.; Wang, C.-T.; Gao, Y.; Wang, Y.-X.; Liu, Y.-W.; Chen, M.; Chen, J.; Zhou, Y.-B.; Xu, Z.-S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 1–18. [Google Scholar] [CrossRef]
- Turan, S.; Tripathy, B.C. Salt-stress induced modulation of chlorophyll biosynthesis during de-etiolation of rice seedlings. Physiol. Plant. 2014, 153, 477–491. [Google Scholar] [CrossRef]
- Barhoumi, Z.; Djebali, W.; Chaïbi, W.; Abdelly, C.; Smaoui, A. Salt impact on photosynthesis and leaf ultrastructure of Aeluropus littoralis. J. Plant Res. 2007, 120, 529–537. [Google Scholar] [CrossRef]
- Bejaoui, F.; Salas, J.J.; Nouairi, I.; Smaoui, A.; Abdelly, C.; Martínez-Force, E.; Ben Youssef, N. Changes in chloroplast lipid contents and chloroplast ultrastructure in Sulla carnosa and Sulla coronaria leaves under salt stress. J. Plant Physiol. 2016, 198, 32–38. [Google Scholar] [CrossRef]
- Goussi, R.; Manaa, A.; Derbali, W.; Cantamessa, S.; Abdelly, C.; Barbato, R. Comparative analysis of salt stress, duration and intensity, on the chloroplast ultrastructure and photosynthetic apparatus in Thellungiella salsuginea. J. Photochem. Photobiol. B Biol. 2018, 183, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Zheng, Z.; Huan, L.; Wang, G. G6PDH activity highlights the operation of the cyclic electron flow around PSI in Physcomitrella patens during salt stress. Sci. Rep. 2016, 6, 21245. [Google Scholar] [CrossRef]
- Ji, X.; Cheng, J.; Gong, D.; Zhao, X.; Qi, Y.; Su, Y.; Ma, W. The effect of NaCl stress on photosynthetic efficiency and lipid production in freshwater microalga—Scenedesmus obliquus XJ002. Sci. Total Environ. 2018, 633, 593–599. [Google Scholar] [CrossRef]
- Ioannidis, N.E.; Sfichi, L.; Kotzabasis, K. Putrescine stimulates chemiosmotic ATP synthesis. Biochim. Biophys. Acta 2006, 1757, 821–828. [Google Scholar] [CrossRef][Green Version]
- Shu, S.; Chen, L.; Lu, W.; Sun, J.; Guo, S.; Yuan, Y.; Li, J. Effects of exogenous spermidine on photosynthetic capacity and expression of Calvin cycle genes in salt-stressed cucumber seedlings. J. Plant Res. 2014, 127, 763–773. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2008, 103, 551–560. [Google Scholar] [CrossRef]
- Silveira, J.A.; Carvalho, F.E. Proteomics, photosynthesis and salt resistance in crops: An integrative view. J. Proteom. 2016, 143, 24–35. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Plant salt tolerance: Adaptations in halophytes. Ann. Bot. 2015, 115, 327–331. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, L.J.; Zhao, Y.J.; Tang, N.Y.; Qu, R.J.; Tang, X.Q.; Wang, K.C. Changes of Ion Absorption, Distribution and Essential Oil Components of Flowering Schizonepeta Tenuifolia under Salt Stress. Zhongguo Zhong Yao Za Zhi 2018, 43, 4410–4418. [Google Scholar] [CrossRef] [PubMed]
- Ottow, E.A.; Brinker, M.; Teichmann, T.; Fritz, E.; Kaiser, W.; Brosche, M.; Kangasjarvi, J.; Jiang, X.; Polle, A. Populus Euphratica Displays Apoplastic Sodium Accumulation, Osmotic Adjustment by Decreases in Calcium and Soluble Car-bohydrates, and Develops Leaf Succulence under Salt Stress. Plant Physiol. 2005, 139, 1762–1772. [Google Scholar] [CrossRef]
- Song, J.; Ding, X.; Feng, G.; Zhang, F. Nutritional and osmotic roles of nitrate in a euhalophyte and a xerophyte in saline conditions. New Phytol. 2006, 171, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.A.; Molla, K.A.; Henry, R.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhang, K.; Sun, Y.; Cui, H.; Cao, S.; Yan, L.; Xu, M. Growth, Physiology, and Tran-scriptional Analysis of Two Contrasting Carex Rigescens Genotypes under Salt Stress Reveals Salt-Tolerance Mechanisms. J. Plant Physiol. 2018, 229, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Leshem, Y.; Melamed-Book, N.; Cagnac, O.; Ronen, G.; Nishri, Y.; Solomon, M.; Cohen, G.; Levine, A. Suppression of Arabidopsis vesicle-SNARE expression inhibited fusion of H2O2-containing vesicles with tonoplast and increased salt tolerance. Proc. Natl. Acad. Sci. USA 2006, 103, 18008–18013. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Wang, X.; Ma, B.; Du, C.; Zheng, L.; Wang, Y. Expression of a Na(+)/H(+) Antiporter Rtnhx1 from a Recreto-halophyte Reaumuria Trigyna Improved Salt Tolerance of Transgenic Arabidopsis Thaliana. J. Plant Physiol. 2017, 218, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Debouba, M.; Maâroufi-Dghimi, H.; Suzuki, A.; Ghorbel, M.H.; Gouia, H. Changes in Growth and Activity of Enzymes Involved in Nitrate Reduction and Ammonium Assimilation in Tomato Seedlings in Response to NaCl Stress. Ann. Bot. 2007, 99, 1143–1151. [Google Scholar] [CrossRef]
- Shahbaz, M.; Ashraf, M.; Akram, N.A.; Hanif, A.; Hameed, S.; Joham, S.; Rehman, R. Salt-induced modulation in growth, photosynthetic capacity, proline content and ion accumulation in sunflower (Helianthus annuus L.). Acta Physiol. Plant. 2011, 33, 1113–1122. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. A Halotolerant Bacterium Bacillus Licheniformis Hsw-16 Augments Induced Systemic Tolerance to Salt Stress in Wheat Plant (Triticum Aestivum). Front. Plant Sci. 2016, 7, 1890. [Google Scholar] [CrossRef]
- Umnajkitikorn, K.; Faiyue, B. Enhancing Antioxidant Properties of Germinated Thai rice (Oryza sativa L.) cv. Kum Doi Saket with Salinity. Rice Res. Open Access 2016, 4. [Google Scholar] [CrossRef]
- El-Katony, T.; El-Bastawisy, Z.M.; El-Ghareeb, S.S. Timing of salicylic acid application affects the response of maize (Zea mays L.) hybrids to salinity stress. Heliyon 2019, 5, e01547. [Google Scholar] [CrossRef]
- Lv, S.; Jiang, P.; Chen, X.; Fan, P.; Wang, X.; Li, Y. Multiple compartmentalization of sodium conferred salt tolerance in Salicornia europaea. Plant Physiol. Biochem. 2012, 51, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Zhao, Y.; Jin, D.; Zhang, L.; Bi, X.; Chen, H.; Xu, Q.; Ma, C.; Li, G. Salicylic acid-altering Arabidopsis mutants response to salt stress. Plant Soil 2011, 354, 81–95. [Google Scholar] [CrossRef]
- Soltabayeva, A.; Ongaltay, A.; Omondi, J.; Srivastava, S. Morphological, Physiological and Molecular Markers for Salt-Stressed Plants. Plants 2021, 10, 243. [Google Scholar] [CrossRef]
- Faseela, P.; Sinisha, A.K.; Brestic, M.; Puthur, J. Special issue in honour of Prof. Reto J. Strasser—Chlorophyll a fluorescence parameters as indicators of a particular abiotic stress in rice. Photosynthetica 2020, 58, 293–300. [Google Scholar] [CrossRef]
- Sun, Z.W.; Ren, L.K.; Fan, J.W.; Li, Q.; Wang, K.J.; Guo, M.M.; Wang, L.; Li, J.; Zhang, G.X.; Yang, Z.Y.; et al. Salt Response of Photosynthetic Electron Transport System in Wheat Cultivars with Contrasting Tolerance. Plant Soil Environ. 2016, 62, 515–521. [Google Scholar] [CrossRef]
- Choi, J.Y.; Seo, Y.S.; Kim, S.J.; Kim, W.T.; Shin, J.S. Constitutive Expression of Caxth3, a Hot Pepper Xyloglucan Endotransglucosylase/Hydrolase, Enhanced Tolerance to Salt and Drought Stresses without Phenotypic Defects in Tomato Plants (Solanum Lycopersicum Cv. Dotaerang). Plant Cell Rep. 2011, 30, 867–877. [Google Scholar] [CrossRef]
- Estrada, B.; Aroca, R.; Barea, J.M.; Ruiz-Lozano, J.M. Native arbuscular mycorrhizal fungi isolated from a saline habitat improved maize antioxidant systems and plant tolerance to salinity. Plant Sci. 2013, 201-202, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Barros, C.V.S.D.; Melo, Y.L.; Souza, M.D.F.; Silva, D.V.; De Macedo, C.E.C. Sensitivity and biochemical mechanisms of sunflower genotypes exposed to saline and water stress. Acta Physiol. Plant. 2019, 41. [Google Scholar] [CrossRef]
- El-Hendawy, S.; Al-Suhaibani, N.; Hassan, W.; Tahir, M.; Schmidhalter, U. Hyperspectral reflectance sensing to assess the growth and photosynthetic properties of wheat cultivars exposed to different irrigation rates in an irrigated arid region. PLoS ONE 2017, 12, e0183262. [Google Scholar] [CrossRef]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef]
- Schobert, B.; Tschesche, H. Unusual solution properties of proline and its interaction with proteins. Biochim. Biophys. Acta BBA Gen. Subj. 1978, 541, 270–277. [Google Scholar] [CrossRef]
- Arakawa, T.; Timasheff, S. The stabilization of proteins by osmolytes. Biophys. J. 1985, 47, 411–414. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline Mechanisms of Stress Survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef] [PubMed]
- Per, T.S.; Khan, N.A.; Reddy, P.S.; Masood, A.; Hasanuzzaman, M.; Khan, M.I.R.; Anjum, N.A. Approaches in modulating proline metabolism in plants for salt and drought stress tolerance: Phytohormones, mineral nutrients and transgenics. Plant Physiol. Biochem. 2017, 115, 126–140. [Google Scholar] [CrossRef]
- Wani, A.S.; Ahmad, A.; Hayat, S.; Tahir, I. Epibrassinolide and Proline Alleviate the Photosynthetic and Yield Inhi-bition under Salt Stress by Acting on Antioxidant System in Mustard. Plant Physiol. Biochem. 2019, 135, 385–394. [Google Scholar] [CrossRef]
- Gao, Y.; Li, M.; Zhang, X.; Yang, Q.; Huang, B. Up-regulation of lipid metabolism and glycine betaine synthesis are associated with choline-induced salt tolerance in halophytic seashore paspalum. Plant Cell Environ. 2020, 43, 159–173. [Google Scholar] [CrossRef]
- Chen, F.; Fang, P.; Zeng, W.; Ding, Y.; Zhuang, Z.; Peng, Y. Comparing transcriptome expression profiles to reveal the mechanisms of salt tolerance and exogenous glycine betaine mitigation in maize seedlings. PLoS ONE 2020, 15, e0233616. [Google Scholar] [CrossRef]
- Byerrum, R.U.; Sato, C.S.; Ball, C.D. Utilization of Betaine as a Methyl Group Donor in Tobacco. Plant Physiol. 1956, 31, 374–377. [Google Scholar] [CrossRef]
- Peng, J.; Liu, J.; Zhang, L.; Luo, J.; Dong, H.; Ma, Y.; Zhao, X.; Chen, B.; Sui, N.; Zhou, Z.; et al. Effects of Soil Salinity on Sucrose Metabolism in Cotton Leaves. PLoS ONE 2016, 11, e0156241. [Google Scholar] [CrossRef]
- Al Hassan, M.; Morosan, M.; López-Gresa, M.D.P.; Prohens, J.; Vicente, O.; Boscaiu, M. Salinity-Induced Variation in Biochemical Markers Provides Insight into the Mechanisms of Salt Tolerance in Common (Phaseolus vulgaris) and Runner (P. coccineus) Beans. Int. J. Mol. Sci. 2016, 17, 1582. [Google Scholar] [CrossRef]
- Li, Q.; Yang, A.; Zhang, W.-H. Comparative studies on tolerance of rice genotypes differing in their tolerance to moderate salt stress. BMC Plant Biol. 2017, 17, 1–13. [Google Scholar] [CrossRef]
- Livingston, D.P., 3rd; Hincha, D.K.; Heyer, A.G. Fructan and Its Relationship to Abiotic Stress Tolerance in Plants. Cell Mol. Life Sci. 2009, 66, 2007–2023. [Google Scholar] [CrossRef]
- Ruan, Y.-L. Sucrose Metabolism: Gateway to Diverse Carbon Use and Sugar Signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef] [PubMed]
- Conde, A.; Regalado, A.; Rodrigues, D.; Costa, J.M.; Blumwald, E.; Chaves, M.M.; Gerós, H. Polyols in grape berry: Transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 2014, 66, 889–906. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Shabala, L. Ion transport and osmotic adjustment in plants and bacteria. Biomol. Concepts 2011, 2, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Bhaduri, D.; Meena, H.N.; Kalariya, K. External potassium (K+) application improves salinity tolerance by promoting Na+-exclusion, K+-accumulation and osmotic adjustment in contrasting peanut cultivars. Plant Physiol. Biochem. 2016, 103, 143–153. [Google Scholar] [CrossRef]
- Keisham, M.; Mukherjee, S.; Bhatla, S.C. Mechanisms of Sodium Transport in Plants—Progresses and Challenges. Int. J. Mol. Sci. 2018, 19, 647. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Qiu, N.; Wang, P.; Zhang, W.; Yang, X.; Chen, M.; Wang, B.; Sun, J. Na+ compartmentation strategy of Chinese cabbage in response to salt stress. Plant Physiol. Biochem. 2019, 140, 151–157. [Google Scholar] [CrossRef]
- Rodriguez, H.; González, J.; Roberts, K.M.; Jordan, W.R.; Drew, M.C. Growth, Water Relations, and Accumulation of Organic and Inorganic Solutes in Roots of Maize Seedlings During Salt Stress. Plant Physiol. 1997, 113, 881–893. [Google Scholar] [CrossRef]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Bose, J.; Rodrigo-Moreno, A.; Shabala, S. ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 2014, 65, 1241–1257. [Google Scholar] [CrossRef]
- Kim, Y.H.; Khan, A.L.; Waqas, M.; Lee, I.J. Silicon Regulates Antioxidant Activities of Crop Plants under Abiotic-Induced Oxidative Stress: A Review. Front. Plant Sci. 2017, 8, 510. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Li, Q.; Park, S.-C.; Wang, X.; Liu, Y.-J.; Zhang, Y.-G.; Tang, W.; Kou, M.; Ma, D.-F. Overexpression of CuZnSOD and APX enhance salt stress tolerance in sweet potato. Plant Physiol. Biochem. 2016, 109, 20–27. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Parvin, K.; Bhuiyan, T.F.; Anee, T.I.; Nahar, K.; Hossen, S.; Zulfiqar, F.; Alam, M.; Fujita, M. Regulation of ROS Metabolism in Plants under Environmental Stress: A Review of Recent Experimental Evidence. Int. J. Mol. Sci. 2020, 21, 8695. [Google Scholar] [CrossRef]
- He, Z.; He, C.; Zhang, Z.; Zou, Z.; Wang, H. Changes of antioxidative enzymes and cell membrane osmosis in tomato colonized by arbuscular mycorrhizae under NaCl stress. Colloids Surf. B Biointerfaces 2007, 59, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Willekens, H.; Inzé, D.; Van Montagu, M.; Van Camp, W. Catalases in plants. Mol. Breed. 1995, 1, 207–228. [Google Scholar] [CrossRef]
- Gondim, F.A.; Gomes-Filho, E.; Hélio Costa, J.; Mendes Alencar, N.L.; Tarquinio Prisco, J. Catalase Plays a Key Role in Salt Stress Acclimation Induced by Hydrogen Peroxide Pretreatment in Maize. Plant Phys. Biochem. 2012, 56, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Jespersen, H.M.; Kjærsgård, I.V.H.; Østergaard, L.; Welinder, K.G. From sequence analysis of three novel ascorbate peroxidases from Arabidopsis thaliana to structure, function and evolution of seven types of ascorbate peroxidase. Biochem. J. 1997, 326, 305–310. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.; Gill, T.; Zahoor, I.; Ahuja, P.S.; Sreenivasulu, Y.; Kumar, S.; Singh, A.K. Ectopic expression of SOD and APX genes in Arabidopsis alters metabolic pools and genes related to secondary cell wall cellulose biosynthesis and improve salt tolerance. Mol. Biol. Rep. 2019, 46, 1985–2002. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Al Mahmud, J.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Edas, P.; Nutan, K.K.; Singla-Pareek, S.L.; Epareek, A. Oxidative environment and redox homeostasis in plants: Dissecting out significant contribution of major cellular organelles. Front. Environ. Sci. 2015, 2. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Hossain, S.; Al Mahmud, J.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Coordinated Actions of Glyoxalase and Antioxidant Defense Systems in Conferring Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2017, 18, 200. [Google Scholar] [CrossRef]
- Bela, K.; Horváth, E.; Gallé, Á.; Szabados, L.; Tari, I.; Csiszár, J. Plant glutathione peroxidases: Emerging role of the antioxidant enzymes in plant development and stress responses. J. Plant Physiol. 2015, 176, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Nianiou-Obeidat, I.; Madesis, P.; Kissoudis, C.; Voulgari, G.; Chronopoulou, E.; Tsaftaris, A.; Labrou, N.E. Plant Glutathione Transferase-Mediated Stress Tolerance: Functions and Biotechnological Applications. Plant Cell Rep. 2017, 36, 791–805. [Google Scholar] [CrossRef]
- Gaafar, R.M.; Seyam, M.M. Ascorbate–glutathione cycle confers salt tolerance in Egyptian lentil cultivars. Physiol. Mol. Biol. Plants 2018, 24, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Müller-Moulé, P.; Conklin, P.L.; Niyogi, K.K. Ascorbate Deficiency Can Limit Violaxanthin De-Epoxidase Activity in Vivo. Plant Physiol. 2002, 128, 970–977. [Google Scholar] [CrossRef]
- McCarrell, E.M.; Gould, S.W.J.; Fielder, M.D.; Kelly, A.F.; El Sankary, W.; Naughton, D.P. Antimicrobial activities of pomegranate rind extracts: Enhancement by addition of metal salts and vitamin C. BMC Complement. Altern. Med. 2008, 8, 64–67. [Google Scholar] [CrossRef]
- Singh, N.; Bhardwaj, R.D. Ascorbic acid alleviates water deficit induced growth inhibition in wheat seedlings by modulating levels of endogenous antioxidants. Biologia 2016, 71. [Google Scholar] [CrossRef]
- Noctor, G.; Foyer, C.H. Ascorbate and Glutathione: Keeping Active Oxygen Under Control. Annu. Rev. Plant Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef]
- Whitbread, A.K.; Masoumi, A.; Tetlow, N.; Schmuck, E.; Coggan, M.; Board, P.G. Characterization of the Omega Class of Glutathione Transferases. Methods Enzymol. 2005, 401, 78–99. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, S.; Zhou, F.; Ding, C. Exogenous Ascorbic Acid and Glutathione Alleviate Oxidative Stress Induced by Salt Stress in the Chloroplasts of Oryza sativa L. Z. Nat. C 2014, 69, 226–236. [Google Scholar] [CrossRef]
- Havaux, M.; Niyogi, K.K. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc. Natl. Acad. Sci. USA 1999, 96, 8762–8767. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. ROS and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.K. ROS Homeostasis in Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2020, 21, 5208. [Google Scholar] [CrossRef]
- Cheeseman, J.M. Mechanisms of Salinity Tolerance in Plants. Plant Physiol. 1988, 87, 547–550. [Google Scholar] [CrossRef] [PubMed]
- Michelet, B.; Boutry, M. The Plasma Membrane H+-Atpase (a Highly Regulated Enzyme with Multiple Physiological Functions). Plant Physiol. 1995, 108, 1. [Google Scholar] [CrossRef]
- Allakhverdiev, S.; Nishiyama, Y.; Suzuki, I.; Tasaka, Y.; Murata, N. Genetic engineering of the unsaturation of fatty acids in membrane lipids alters the tolerance of Synechocystis to salt stress. Proc. Natl. Acad. Sci. USA 1999, 96, 5862–5867. [Google Scholar] [CrossRef] [PubMed]
- Schuler, M.L.; Mantegazza, O.; Weber, A.P. Engineering C4photosynthesis into C3chassis in the synthetic biology age. Plant J. 2016, 87, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Frommhagen, M.; Haar, M.K.; van Erven, G.; Bakx, E.J.; van Berkel, W.J.H.; Kabel, M.A. Mass Spectrometric Fragmentation Patterns Discriminate C1- and C4-Oxidised Cello-Oligosaccharides from Their Non-Oxidised and Reduced Forms. Carbohydr. Polym. 2020, 234, 115917. [Google Scholar] [CrossRef]
- Cushman, J.C.; Meyer, G.; Michalowski, C.B.; Schmitt, J.M.; Bohnert, H.J. Salt Stress Leads to Differential Expression of Two Isogenes of Phosphoenolpyruvate Carboxylase During Crassulacean Acid Metabolism Induction in the Common Ice Plant. Plant Cell 1989, 1, 715–725. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Bose, J.; Hedrich, R. Salt bladders: Do they matter? Trends Plant Sci. 2014, 19, 687–691. [Google Scholar] [CrossRef]
- Kiani-Pouya, A.; Roessner, U.; Jayasinghe, N.S.; Lutz, A.; Rupasinghe, T.; Bazihizina, N.; Bohm, J.; Alharbi, S.; Hedrich, R.; Shabala, S. Epidermal bladder cells confer salinity stress tolerance in the halophyte quinoa and Atriplex species. Plant Cell Environ. 2017, 40, 1900–1915. [Google Scholar] [CrossRef] [PubMed]
- Barkla, B.J.; Vera-Estrella, R.; Pantoja, O. Protein Profiling of Epidermal Bladder Cells from the Halophyte Mesem-bryanthemum Crystallinum. Proteomics 2012, 12, 2862–2865. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R. Single cell-type comparative metabolomics of epidermal bladder cells from the halophyte Mesembryanthemum crystallinum. Front. Plant Sci. 2015, 6, 435. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R.; Raymond, C. Single-Cell-Type Quantitative Proteomic and Ionomic Analysis of Epidermal Bladder Cells from the Halophyte Model Plant Mesembryanthemum Crystallinum to Identify Salt-Responsive Proteins. BMC Plant Biol. 2016, 16, 110. [Google Scholar] [CrossRef]
- White, P.J.; Broadley, M.R. Calcium in Plants. Ann. Bot. 2003, 92, 487–511. [Google Scholar] [CrossRef]
- Ullah, A.; Dutta, D.; Fliegel, L. Expression and characterization of the SOS1 Arabidopsis salt tolerance protein. Mol. Cell. Biochem. 2016, 415, 133–143. [Google Scholar] [CrossRef]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.K. The Putative Plasma Membrane Na (+)/H(+) Antiporter Sos1 Controls Long-Distance Na(+) Transport in Plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ishitani, M.; Halfter, U.; Kim, C.S.; Zhu, J.K. The Arabidopsis Thaliana Sos2 Gene Encodes a Protein Kinase That Is Required for Salt Tolerance. Proc. Natl. Acad. Sci. USA 2000, 97, 3730–3734. [Google Scholar] [CrossRef] [PubMed]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.-S.; Shi, W.; Zhu, J.-K. SOS3 Function in Plant Salt Tolerance Requires N-Myristoylation and Calcium Binding. Plant Cell 2000, 12, 1667. [Google Scholar] [CrossRef] [PubMed]
- Halfter, U.; Ishitani, M.; Zhu, J.K. The Arabidopsis Sos2 Protein Kinase Physically Interacts with and Is Activated by the Calcium-Binding Protein Sos3. Proc. Natl. Acad. Sci. USA 2000, 97, 3735–3740. [Google Scholar] [CrossRef] [PubMed]
- Quintero, F.J.; Ohta, M.; Shi, H.; Zhu, J.-K.; Pardo, J.M. Reconstitution in yeast of the Arabidopsis SOS signaling pathway for Na+ homeostasis. Proc. Natl. Acad. Sci. USA 2002, 99, 9061–9066. [Google Scholar] [CrossRef]
- Sottosanto, J.B.; Gelli, A.; Blumwald, E. DNA Array Analyses of Arabidopsis Thaliana Lacking a Vacuolar Na+/H+ Antiporter: Impact of Atnhx1 on Gene Expression. Plant J. 2004, 40, 752–771. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhu, J.-K. Regulation of expression of the vacuolar Na+/H+ antiporter gene AtNHX1 by salt stress and abscisic acid. Plant Mol. Biol. 2002, 50, 543–550. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Palmgren, M.G.; Schumacher, K. Plant proton pumps. FEBS Lett. 2007, 581, 2204–2214. [Google Scholar] [CrossRef]
- Qiu, Q.-S.; Guo, Y.; Quintero, F.J.; Pardo, J.M.; Schumaker, K.S.; Zhu, J.-K. Regulation of Vacuolar Na+/H+ Exchange in Arabidopsis thaliana by the Salt-Overly-Sensitive (SOS) Pathway. J. Biol. Chem. 2004, 279, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.; Bressan, R.A.; Song, C.; Zhu, J.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Jiang, F.; Hartung, W. Long-distance signalling of abscisic acid (ABA): The factors regulating the intensity of the ABA signal. J. Exp. Bot. 2008, 59, 37–43. [Google Scholar] [CrossRef]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The Abi1 and Abi2 Protein Phosphatases 2c Act in a Negative Feedback Regulatory Loop of the Abscisic Acid Signalling Pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arabidopsis Book 2013, 11, e0166. [Google Scholar] [CrossRef]
- Hamilton, D.W.A.; Hills, A.; Köhler, B.; Blatt, M.R. Ca2+ channels at the plasma membrane of stomatal guard cells are activated by hyperpolarization and abscisic acid. Proc. Natl. Acad. Sci. USA 2000, 97, 4967–4972. [Google Scholar] [CrossRef]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A protein kinase-phosphatase pair interacts with an ion channel to regulate ABA signaling in plant guard cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef]
- Dammann, C.; Ichida, A.; Hong, B.; Romanowsky, S.M.; Hrabak, E.M.; Harmon, A.C.; Pickard, B.G.; Harper, J.F. Subcellular Targeting of Nine Calcium-Dependent Protein Kinase Isoforms from Arabidopsis. Plant Physiol. 2003, 132, 1840–1848. [Google Scholar] [CrossRef] [PubMed]
- Harmon, A.C.; Gribskov, M.; Harper, J.F. Cdpks–A Kinase for Every Ca2+ Signal? Trends Plant Sci. 2000, 5, 154–159. [Google Scholar] [CrossRef]
- Asano, T.; Hayashi, N.; Kikuchi, S.; Ohsugi, R. CDPK-mediated abiotic stress signaling. Plant Signal. Behav. 2012, 7, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Hirt, H. Nuclear Signaling of Plant MAPKs. Front. Plant Sci. 2018, 9, 469. [Google Scholar] [CrossRef] [PubMed]
- Jalmi, S.K.; Sinha, A.K. ROS mediated MAPK signaling in abiotic and biotic stress- striking similarities and differences. Front. Plant Sci. 2015, 6, 769. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; Chen, K.; Wang, S.S.; Gong, M. Osmoregulation as a key factor in drought hardening-induced drought tolerance in Jatropha curcas. Biol. Plant. 2015, 59, 529–536. [Google Scholar] [CrossRef]
- Chen, J.B.; Yang, J.W.; Zhang, Z.Y.; Feng, X.F.; Wang, S.M. Two P5CS genes from common bean exhibiting different tolerance to salt stress in transgenic Arabidopsis. J. Genet. 2013, 92, 461–469. [Google Scholar] [CrossRef]
- Hanson, A.D.; Wyse, R. Biosynthesis, Translocation, and Accumulation of Betaine in Sugar Beet and Its Progenitors in Relation to Salinity. Plant Physiol. 1982, 70, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, Y.; Zeng, S.; Patra, B.; Yuan, L.; Wang, Y. Isolation and characterization of a salt stress-responsive betaine aldehyde dehydrogenase in Lycium ruthenicum Murr. Physiol. Plant. 2018, 163, 73–87. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Lai, L.-D.; Tong, S.-M.; Li, Q.-L. Constitutive and salt-inducible expression of SlBADH gene in transgenic tomato (Solanum lycopersicum L. cv. Micro-Tom) enhances salt tolerance. Biochem. Biophys. Res. Commun. 2013, 432, 262–267. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Broyart, C.; Rabanal, F.A.; Jonak, C. The Redox-Sensitive Chloroplast Trehalose-6-Phosphate Phosphatase AtTPPD Regulates Salt Stress Tolerance. Antioxidants Redox Signal. 2014, 21, 1289–1304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Y.; Tong, J.; He, X.; Xu, Z.; Xu, L.; Wei, P.; Huang, Y.; Ebrestic, M.; Ema, H.; Eshao, H.-B. A Novel Soybean Intrinsic Protein Gene, GmTIP2;3, Involved in Responding to Osmotic Stress. Front. Plant Sci. 2016, 6, 1237. [Google Scholar] [CrossRef] [PubMed]
- Trouverie, J.; Chateau-Joubert, S.; Jacquemot, M.-P.; Prioul, J.-L. Regulation of vacuolar invertase by abscisic acid or glucose in leaves and roots from maize plantlets. Planta 2004, 219, 894–905. [Google Scholar] [CrossRef]
- Tada, Y.; Kashimura, T. Proteomic Analysis of Salt-Responsive Proteins in the Mangrove Plant, Bruguiera gymnorhiza. Plant Cell Physiol. 2009, 50, 439–446. [Google Scholar] [CrossRef]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Babgohari, M.Z.; Ebrahimie, E.; Niazi, A. In silico analysis of high affinity potassium transporter (HKT) isoforms in different plants. Aquat. Biosyst. 2014, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yin, S.; Huang, L.; Wu, X.; Zeng, J.; Liu, X.; Qiu, L.; Munns, R.; Chen, Z.-H.; Zhang, G. A Sodium Transporter Hvhkt1; 1 Confers Salt Tolerance in Barley Via Regulating Tissue and Cell Ion Homeostasis. Plant Cell Phys. 2018, 59, 1976–1989. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Kuang, L.; Wu, L.; Shen, Q.; Han, Y.; Jiang, L.; Wu, D.; Zhang, G. The HKT Transporter HvHKT1;5 Negatively Regulates Salt Tolerance. Plant Physiol. 2020, 182, 584–596. [Google Scholar] [CrossRef]
- Kader, A.; Seidel, T.; Golldack, D.; Lindberg, S. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J. Exp. Bot. 2006, 57, 4257–4268. [Google Scholar] [CrossRef]
- Wang, S.-M.; Zhang, J.; Flowers, T.J. Low-Affinity Na+ Uptake in the Halophyte Suaeda maritima. Plant Physiol. 2007, 145, 559–571. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Rao, R.; Sherman, A.; Grisafi, P.; Alper, S.L.; Fink, G.R. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc. Natl. Acad. Sci. USA 1999, 96, 1480–1485. [Google Scholar] [CrossRef] [PubMed]
- Adabnejad, H.; Kavousi, H.R.; Hamidi, H.; Tavassolian, I. Assessment of the Vacuolar Na+/H+ Antiporter (Nhx1) Transcriptional Changes in Leptochloa Fusca L. in Response to Salt and Cadmium Stresses. Mol. Biol. Res. Commun. 2015, 4, 133–142. [Google Scholar]
- Yang, Y.; Guo, Z.; Liu, Q.; Tang, J.; Huang, S.; Dhankher, O.P.; Yuan, H. Growth, physiological adaptation, and NHX gene expression analysis of Iris halophila under salt stress. Environ. Sci. Pollut. Res. 2018, 25, 25207–25216. [Google Scholar] [CrossRef] [PubMed]
- Guan, B.; Hu, Y.; Zeng, Y.; Wang, Y.; Zhang, F. Molecular characterization and functional analysis of a vacuolar Na+/H+ antiporter gene (HcNHX1) from Halostachys caspica. Mol. Biol. Rep. 2011, 38, 1889–1899. [Google Scholar] [CrossRef]
- Asif, M.A.; Zafar, Y.; Iqbal, J.; Iqbal, M.M.; Rashid, U.; Ali, G.M.; Arif, A.; Nazir, F. Enhanced Expression of AtNHX1, in Transgenic Groundnut (Arachis hypogaea L.) Improves Salt and Drought Tolerence. Mol. Biotechnol. 2011, 49, 250–256. [Google Scholar] [CrossRef]
- Shi, L.-Y.; Li, H.-Q.; Pan, X.-P.; Wu, G.-J.; Li, M.-R. Improvement of Torenia fournieri salinity tolerance by expression of Arabidopsis AtNHX5. Funct. Plant Biol. 2008, 35, 185–192. [Google Scholar] [CrossRef]
- Verma, D.; Singla-Pareek, S.L.; Rajagopal, D.; Reddy, M.K.; Sopory, S.K. Functional validation of a novel isoform of Na+/H+ antiporter from Pennisetum glaucum for enhancing salinity tolerance in rice. J. Biosci. 2007, 32, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Alavilli, H.; Lee, B.-H.; Panda, S.K.; Sahoo, L. Cloning and Functional Characterization of a Vacuolar Na+/H+ Antiporter Gene from Mungbean (VrNHX1) and Its Ectopic Expression Enhanced Salt Tolerance in Arabidopsis thaliana. PLoS ONE 2014, 9, e106678. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, R.J.; Li, B.; Wang, H.H.; Jin, Y.L.; Jiang, C.M.; Bao, Y.; Su, H.Y.; Zhao, N.; Ma, X.J.; et al. Overexpression of a Populus trichocarpa H+-pyrophosphatase gene PtVP1.1 confers salt tolerance on transgenic poplar. Tree Physiol. 2015, 35, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Brini, F.X.; Hanin, M.; Mezghani, I.; Berkowitz, G.A.; Masmoudi, K. Overexpression of wheat Na+/H+ antiporter TNHX1 and H+-pyrophosphatase TVP1 improve salt- and drought-stress tolerance in Arabidopsis thaliana plants. J. Exp. Bot. 2007, 58, 301–308. [Google Scholar] [CrossRef]
- Bhaskaran, S.; Savithramma, D.L. Co-Expression of Pennisetum Glaucum Vacuolar Na (+)/H (+) Antiporter and Arabidopsis H(+)-Pyrophosphatase Enhances Salt Tolerance in Transgenic Tomato. J. Exp. Bot. 2011, 62, 5561–5570. [Google Scholar] [CrossRef]
- Verma, D.; Lakhanpal, N.; Singh, K. Genome-wide identification and characterization of abiotic-stress responsive SOD (superoxide dismutase) gene family in Brassica juncea and B. rapa. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Wu, C.; Ding, X.; Ding, Z.; Tie, W.; Yan, Y.; Wang, Y.; Yang, H.; Hu, W. The Class III Peroxidase (POD) Gene Family in Cassava: Identification, Phylogeny, Duplication, and Expression. Int. J. Mol. Sci. 2019, 20, 2730. [Google Scholar] [CrossRef]
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The Catalase Gene Family in Cotton: Genome-Wide Characterization and Bioinformatics Analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed]
- Tao, C.; Jin, X.; Zhu, L.; Xie, Q.; Wang, X.; Li, H. Genome-wide investigation and expression profiling of APX gene family in Gossypium hirsutum provide new insights in redox homeostasis maintenance during different fiber development stages. Mol. Genet. Genom. 2018, 293, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Noctor, G.; Mhamdi, A.; Chaouch, S.; Han, Y.; Neukermans, J.; Marquez-Garcia, B.; Queval, G.; Foyer, C.H. Glutathione in plants: An integrated overview. Plant Cell Environ. 2012, 35, 454–484. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [PubMed]
- Horvath, E.; Bela, K.; Holinka, B.; Riyazuddin, R.; Galle, A.; Hajnal, A.; Hurton, A.; Feher, A.; Csiszar, J. The Arabidopsis Glutathione Transferases, Atgstf8 and Atgstu19 Are Involved in the Maintenance of Root Redox Homeostasis Affecting Meristem Size and Salt Stress Sensitivity. Plant Sci. 2019, 283, 366–374. [Google Scholar] [CrossRef]
- Na Sun, L.; Wang, F.; Wang, J.W.; Gao, W.R.; Song, X.S.; Sun, L.J. Overexpression of the ChVDE gene, encoding a violaxanthin de-epoxidase, improves tolerance to drought and salt stress in transgenic Arabidopsis. 3 Biotech 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Guo, K.-M.; Babourina, O.; Rengel, Z. Na+/H+antiporter activity of theSOS1gene: Lifetime imaging analysis and electrophysiological studies on Arabidopsis seedlings. Physiol. Plant. 2009, 137, 155–165. [Google Scholar] [CrossRef]
- Shi, H.; Zhu, J.-K. SOS4, A Pyridoxal Kinase Gene, Is Required for Root Hair Development in Arabidopsis. Plant Physiol. 2002, 129, 585–593. [Google Scholar] [CrossRef]
- Shi, H.; Kim, Y.; Guo, Y.; Stevenson, B.; Zhu, J.-K. The Arabidopsis SOS5 Locus Encodes a Putative Cell Surface Adhesion Protein and Is Required for Normal Cell Expansion. Plant Cell 2003, 15, 19–32. [Google Scholar] [CrossRef]
- Zhu, J.; Lee, B.H.; Dellinger, M.; Cui, X.; Zhang, C.; Wu, S.; Nothnagel, E.A.; Zhu, J.K. A Cellulose Synthase-Like Protein Is Required for Osmotic Stress Tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [CrossRef]
- Cheng, C.; Zhong, Y.; Wang, Q.; Cai, Z.; Wang, D.; Li, C. Genome-wide identification and gene expression analysis of SOS family genes in tuber mustard (Brassica juncea var. tumida). PLoS ONE 2019, 14, e0224672. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; William, D.; Sandhu, D. Isolation and characterization of Salt Overly Sensitive family genes in spinach. Physiol. Plant. 2021, 171, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Expression and Signal Transduction in Water-Stress Response. Plant Physiol. 1997, 115, 327. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, H.; Liu, G.; Ma, W.; Li, C.; Huo, H.; He, J.; Liu, L. PpSARK Regulates Moss Senescence and Salt Tolerance through ABA Related Pathway. Int. J. Mol. Sci. 2018, 19, 2609. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Huang, J.; Xia, J. OsNAC45 is Involved in ABA Response and Salt Tolerance in Rice. Rice 2020, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA–mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e1007662. [Google Scholar] [CrossRef]
- Shi, S.; Li, S.; Asim, M.; Mao, J.; Xu, D.; Ullah, Z.; Liu, G.; Wang, Q.; Liu, H. The Arabidopsis Calcium-Dependent Protein Kinases (CDPKs) and Their Roles in Plant Growth Regulation and Abiotic Stress Responses. Int. J. Mol. Sci. 2018, 19, 1900. [Google Scholar] [CrossRef]
- Zhao, R.; Sun, H.; Zhao, N.; Jing, X.; Shen, X.; Chen, S. The Arabidopsis Ca2+-dependent protein kinase CPK27 is required for plant response to salt-stress. Gene 2015, 563, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Y.; Deng, C.; Deng, S.; Li, N.; Zhao, C.; Zhao, R.; Liang, S.; Chen, S. The Arabidopsis Ca2+-Dependent Protein Kinase CPK12 Is Involved in Plant Response to Salt Stress. Int. J. Mol. Sci. 2018, 19, 4062. [Google Scholar] [CrossRef]
- Xu, J.; Tian, Y.-S.; Peng, R.-H.; Xiong, A.-S.; Zhu, B.; Jin, X.-F.; Gao, F.; Fu, X.-Y.; Hou, X.-L.; Yao, Q.-H. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta 2010, 231, 1251–1260. [Google Scholar] [CrossRef]
- Zou, J.J.; Li, X.D.; Ratnasekera, D.; Wang, C.; Liu, W.X.; Song, L.F.; Zhang, W.Z.; Wu, W.H. Arabidopsis Calcium-Dependent Protein Kinase8 and Catalase3 Function in Abscisic acid-mediated Signaling and H2o2 Homeostasis in Stomatal Guard Cells under Drought Stress. Plant Cell 2015, 27, 1445–1460. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 Pathway Mediates Cold and Salt Stress Signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Yang, C.; Wang, R.; Gou, L.; Si, Y.; Guan, Q. Overexpression of Populus trichocarpa Mitogen-Activated Protein Kinase Kinase4 Enhances Salt Tolerance in Tobacco. Int. J. Mol. Sci. 2017, 18, 2090. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Liang, Y.-H.; Zhang, J.-Y.; Cheng, Z.-M. Cloning, molecular and functional characterization by overexpression in Arabidopsis of MAPKK genes from grapevine (Vitis vinifera). BMC Plant Biol. 2020, 20, 194. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Pan, J.; Zhang, M.; Xing, X.; Zhou, Y.; Liu, Y.; Li, D.; Li, D. ZmMKK4, a novel group C mitogen-activated protein kinase kinase in maize (Zea mays), confers salt and cold tolerance in transgenic Arabidopsis. Plant Cell Environ. 2011, 34, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jiang, S.; Pan, J.; Kong, X.; Zhou, Y.; Liu, Y.; Li, D. The overexpression of a maize mitogen-activated protein kinase gene (ZmMPK5) confers salt stress tolerance and induces defence responses in tobacco. Plant Biol. 2014, 16, 558–570. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Zhu, M.; Jia, H.; Liu, D.; Hao, L.; Guo, X. A cotton Raf-like MAP3K gene, GhMAP3K40, mediates reduced tolerance to biotic and abiotic stress in Nicotiana benthamiana by negatively regulating growth and development. Plant Sci. 2015, 240, 10–24. [Google Scholar] [CrossRef]
- Zhang, L.; Xi, D.; Li, S.; Gao, Z.; Zhao, S.; Shi, J.; Wu, C.; Guo, X. A cotton group C MAP kinase gene, GhMPK2, positively regulates salt and drought tolerance in tobacco. Plant Mol. Biol. 2011, 77, 17–31. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, T.-F.; Ma, J.; Chen, J.; Zhou, Y.-B.; Chen, M.; Ma, Y.-Z.; Wei, W.-L.; Xu, Z.-S. The Soybean bZIP Transcription Factor Gene GmbZIP2 Confers Drought and Salt Resistances in Transgenic Plants. Int. J. Mol. Sci. 2020, 21, 670. [Google Scholar] [CrossRef]
- Rajappa, S.; Krishnamurthy, P.; Kumar, P.P. Regulation of AtKUP2 Expression by bHLH and WRKY Transcription Factors Helps to Confer Increased Salt Tolerance to Arabidopsis thaliana Plants. Front. Plant Sci. 2020, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2r3-Myb Transcription Factor, Atmyb20, Negatively Regulates Type 2c Ser-ine/Threonine Protein Phosphatases to Enhance Salt Tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Yu, L.; Zhang, K.; Zhao, Y.; Guo, Y.; Gao, C. A ThDREB gene from Tamarix hispida improved the salt and drought tolerance of transgenic tobacco and T. hispida. Plant Physiol. Biochem. 2017, 113, 187–197. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Shi, Q.; Ren, Z. SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 2020, 291, 110356. [Google Scholar] [CrossRef]
- Ju, Y.-L.; Yue, X.-F.; Min, Z.; Wang, X.-H.; Fang, Y.-L.; Zhang, J.-X. VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2020, 146, 98–111. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, S.; Zhou, M.; Yuan, N.; Li, Z.; Hu, Q.; Bethea, F.G., Jr.; Liu, H.; Li, S.; Luo, H. Transgenic Creeping Bentgrass Overexpressing Osa-Mir393a Exhibits Altered Plant Development and Improved Multiple Stress Tolerance. Plant Biotechnol. J. 2019, 17, 233–251. [Google Scholar] [CrossRef]
- Bai, Q.; Wang, X.; Chen, X.; Shi, G.; Liu, Z.; Guo, C.; Xiao, K. Wheat miRNA TaemiR408 Acts as an Essential Mediator in Plant Tolerance to Pi Deprivation and Salt Stress via Modulating Stress-Associated Physiological Processes. Front. Plant Sci. 2018, 9, 499. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, L.; Cui, Y.-N.; Wang, S.-M.; Bao, A.-K. Identification of candidate genes related to salt tolerance of the secretohalophyte Atriplex canescens by transcriptomic analysis. BMC Plant Biol. 2019, 19, 213. [Google Scholar] [CrossRef]
- İbrahimova, U.; Kumari, P.; Yadav, S.; Rastogi, A.; Antala, M.; Suleymanova, Z.; Zivcak, M.; Tahjib-Ul-Arif, M.; Hussain, S.; Abdelhamid, M.; et al. Progress in Understanding Salt Stress Response in Plants Using Biotechnological Tools. J. Biotechnol. 2021, 329, 180–191. [Google Scholar] [CrossRef]
- Sanoubar, R.; Cellini, A.; Gianfranco, G.; Spinelli, F. Osmoprotectants and Antioxidative Enzymes as Screening Tools for Salinity Tolerance in Radish (Raphanus sativus). Hortic. Plant J. 2020, 6, 14–24. [Google Scholar] [CrossRef]
- Jiang, C.; Cui, Q.; Feng, K.; Xu, D.; Li, C.; Zheng, Q. Melatonin improves antioxidant capacity and ion homeostasis and enhances salt tolerance in maize seedlings. Acta Physiol. Plant. 2016, 38, 1–9. [Google Scholar] [CrossRef]
- Azad, K.; Kaminskyj, S. A fungal endophyte strategy for mitigating the effect of salt and drought stress on plant growth. Symbiosis 2015, 68, 73–78. [Google Scholar] [CrossRef]
- Bargaz, A.; Nassar, R.M.A.; Rady, M.M.; Gaballah, M.S.; Thompson, S.M.; Brestic, M.; Schmidhalter, U.; Abdelhamid, M.T. Improved Salinity Tolerance by Phosphorus Fertilizer in Two Phaseolus Vulgaris Recombinant Inbred Lines Contrasting in Their P-Efficiency. J. Agron. Crop Sci. 2016, 202, 497–507. [Google Scholar] [CrossRef]
- Bashir, M.A.; Silvestri, C.; Ahmad, T.; Hafiz, I.A.; Abbasi, N.A.; Manzoor, A.; Cristofori, V.; Rugini, E. Osmotin: A Cationic Protein Leads to Improve Biotic and Abiotic Stress Tolerance in Plants. Plants 2020, 9, 992. [Google Scholar] [CrossRef] [PubMed]
- Hakim; Ullah, A.; Hussain, A.; Shaban, M.; Khan, A.H.; Alariqi, M.; Gul, S.; Jun, Z.; Lin, S.; Li, J.; et al. Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 2018, 123, 149–159. [Google Scholar] [CrossRef]
- Wan, Q.; Hongbo, S.; Zhaolong, X.; Jia, L.; Dayong, Z.; Yihong, H. Salinity Tolerance Mechanism of Osmotin and Osmot-in-Like Proteins: A Promising Candidate for Enhancing Plant Salt Tolerance. Curr. Genom. 2017, 18, 553–556. [Google Scholar] [CrossRef]
- Subramanyam, K.; Sailaja, K.V.; Subramanyam, K.; Rao, D.M.; Lakshmidevi, K. Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.). Plant Cell Tissue Organ Cult. PCTOC 2010, 105, 181–192. [Google Scholar] [CrossRef]
- Subramanyam, K.; Arun, M.; Mariashibu, T.S.; Theboral, J.; Rajesh, M.; Singh, N.K.; Manickavasagam, M.; Ganapathi, A. Overexpression of tobacco osmotin (Tbosm) in soybean conferred resistance to salinity stress and fungal infections. Planta 2012, 236, 1909–1925. [Google Scholar] [CrossRef] [PubMed]
- Bashir, M.; Silvestri, C.; Coppa, E.; Brunori, E.; Cristofori, V.; Rugini, E.; Ahmad, T.; Hafiz, I.; Abbasi, N.; Shah, M.N.; et al. Response of Olive Shoots to Salinity Stress Suggests the Involvement of Sulfur Metabolism. Plants 2021, 10, 350. [Google Scholar] [CrossRef]
- Rugini, E.; Cristofori, V.; Silvestri, C. Genetic improvement of olive (Olea europaea L.) by conventional and in vitro biotechnology methods. Biotechnol. Adv. 2016, 34, 687–696. [Google Scholar] [CrossRef]


| Abbreviation | Full Name | Abbreviation | Full Name |
|---|---|---|---|
| ROS | reactive oxygen species | ABA | abscisic acid |
| OST1 | ABA-activated SnRK2 protein kinase open stomata1 | CDPK | calcium-dependent protein kinase |
| MAPK | mitogen-activated protein kinase | QA/B | plastoquinone A/B |
| NADPH | reductive coenzyme | RuBP | ribulose-1, 5-bisphosphate |
| MDA | malondialdehyde | PPI | pyrophosphoric acid |
| AsA | ascorbic acid | MDHA | monodehydroascorbic acid |
| DHA | dehydroascorbic acid | GSH | glutathione |
| GSSG | oxidized glutathione | CAR | carotenoids |
| Ve | α-tocopherol | ABRE | ABA response element |
| P5CS | 1 pyrroline—5—carboxylic acid synthetase | NSCC | non-selective cation channel |
| GDH | glutamate dehydrogenase | CAX | Ca2+/H+ antiporter |
| OAT | ornithine aminotransferase | VP | vacuolar H+ phosphorylase |
| ProDH | proline dehydrogenase | SOD | superoxide dismutase |
| BADH | betaine aldehyde dehydrogenase | POD | peroxidase |
| CMO | choline monooxygenase | APX | ascorbic peroxidase |
| TPS | trehalose phosphate synthase | CAT | catalase |
| FBP | 1, 6-diphosphate fructose | GCL | glutamate cysteine ligase |
| TPP | trehalose phosphate phosphatase | MDHAR | monodehydroascorbic acid reductase |
| SPS | phosphate sucrose synthase | GS | glutathione synthase |
| CWIN | cell wall invertase | AspX | ascorbate peroxidase |
| VIN | vacuolar invertase | DHAR | GSH-dependent dehydroascorbic acid reductase |
| CIN | cytoplasmic invertase | GR | glutathione reductase |
| HKT | high-affinity K+ transporter | GST | glutathione S-transferase |
| NHX | Na+/H+ antiporter | SOS1 | salt overly sensitive 1 |
| KT | K+ transporter | SOS2 | salt overly sensitive 2 |
| SKOR | stelar K+ outward rectifier | SOS3 | salt overly sensitive 3 |
| GORK | guard cell outward rectifying K+ channel | ACA | Ca2+-ATPase isomer |
| G6PDH | glucose-6-phosphate dehydrogenase | SuSy | sucrose synthase |
| CAM | calmodulin | Orn | ornithine |
| Glu | glutamate | PEPCase | phosphoenolpyruvate carboxylase |
| CAM | crassulacean acid metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, S.; Wang, Y.; Yan, Y.; Liu, Y.; Wang, J.; Chen, S. A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae 2021, 7, 132. https://doi.org/10.3390/horticulturae7060132
Hao S, Wang Y, Yan Y, Liu Y, Wang J, Chen S. A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae. 2021; 7(6):132. https://doi.org/10.3390/horticulturae7060132
Chicago/Turabian StyleHao, Shanhu, Yiran Wang, Yunxiu Yan, Yuhang Liu, Jingyao Wang, and Su Chen. 2021. "A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance" Horticulturae 7, no. 6: 132. https://doi.org/10.3390/horticulturae7060132
APA StyleHao, S., Wang, Y., Yan, Y., Liu, Y., Wang, J., & Chen, S. (2021). A Review on Plant Responses to Salt Stress and Their Mechanisms of Salt Resistance. Horticulturae, 7(6), 132. https://doi.org/10.3390/horticulturae7060132






