Evaluation of Anthracnose Resistance in Pepper (Capsicum spp.) Genetic Resources
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Inoculum Preparation
2.3. Inoculation Method
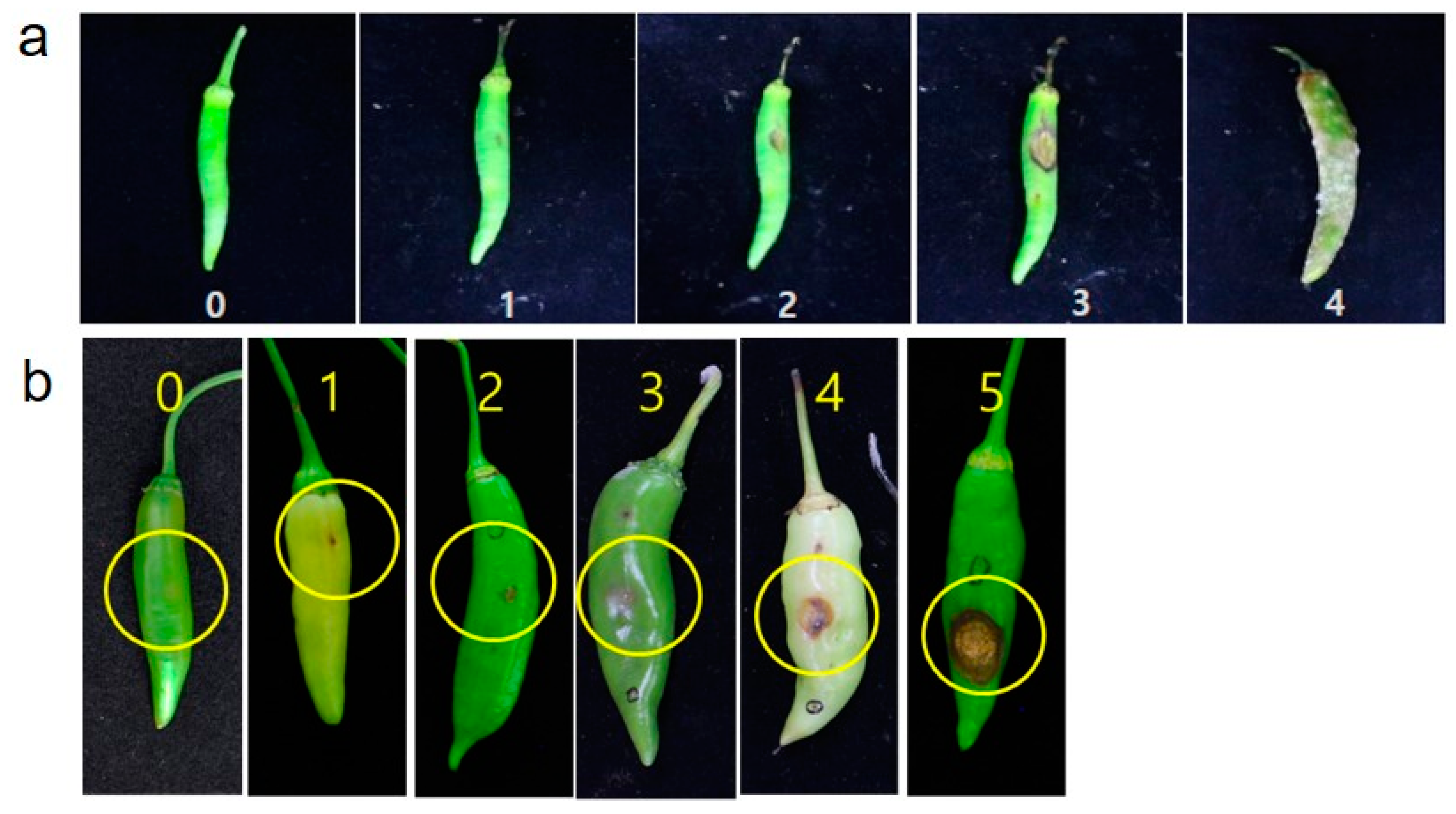
2.4. Disease Evaluation
2.5. DNA Extraction and HRM Maker Analysis
2.6. Fruit Characterization and Statistical Analyses
3. Results
3.1. Pepper Germplasm against C. acutatum with Non-Wound Inoculation
3.2. Pepper Germplasm against C. acutatum with Wound Inoculation
3.3. Maker Validation
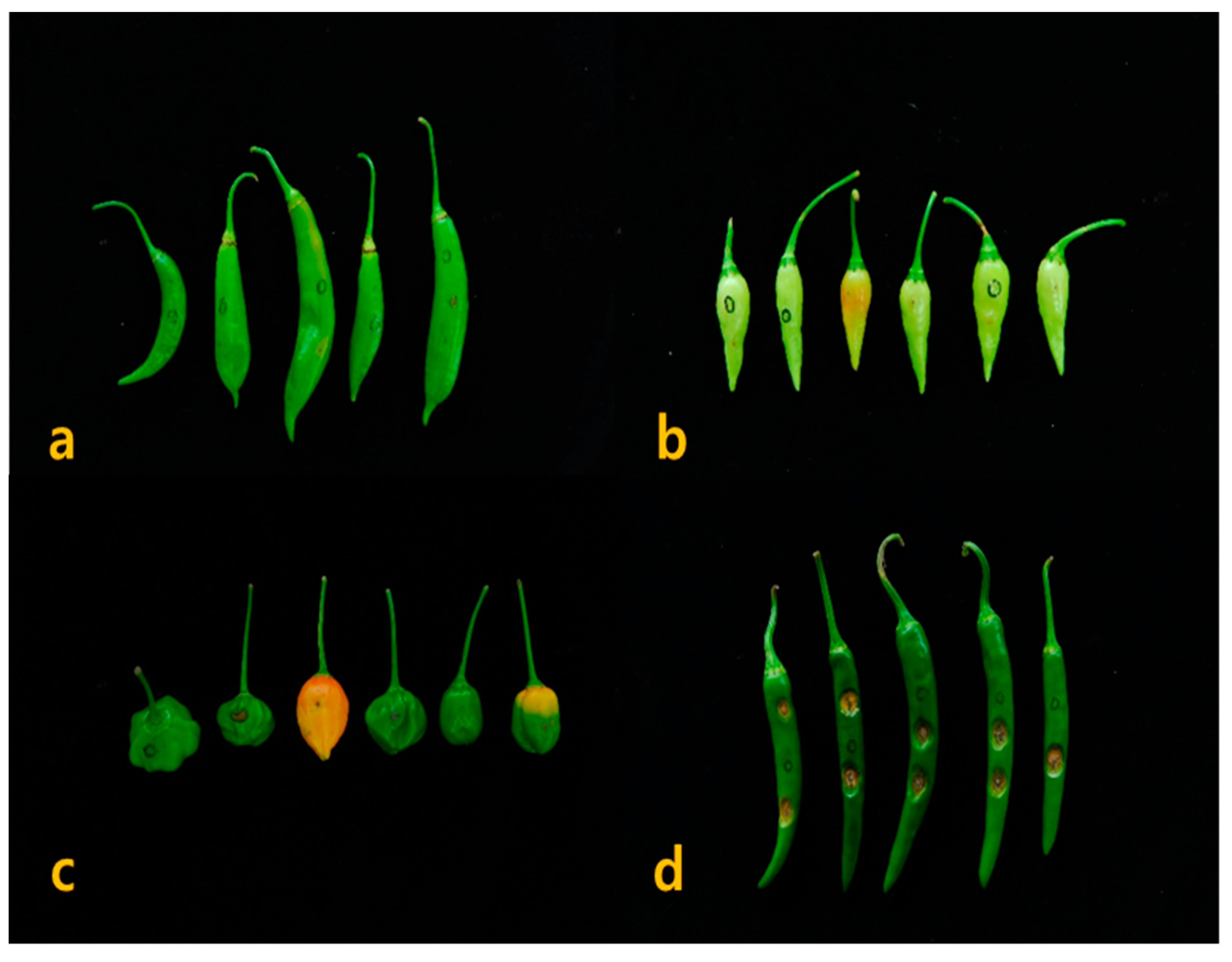
3.4. Fruit Characteristics
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Appendix A. Abbreviations of Country Names. | |
| Abbreviation | Nation |
| 9 | Europe |
| 27 | South America |
| AFG | Islamic State of Afghanistan |
| ARG | Argentine Republic |
| ARM | Republic of Armenia |
| AUS | Australia |
| AUT | Republic of Austria |
| AZE | Azerbaijani Republic |
| BEL | Kindgom of Belgium |
| BFA | Burkina Faso |
| BGD | People’s Republic of Bangladesh |
| BGR | Republic of BμL garia |
| BHS | Commonwealth of the Bahamas |
| BLR | Republic of Belarus |
| BLZ | Belize |
| BOL | Republic of Bolivia |
| BRA | Federative Republic of Brazil |
| BTN | Kingdom of Bhutan |
| BWA | Republic of Botswana |
| CAN | Canada |
| CHE | Swiss Confederation |
| CHL | Republic of Chile |
| CHN | People’s Republic of China |
| COL | Republic of Colombia |
| CRI | Republic of Costa Rica |
| CSK | Czechoslovakia |
| CUB | Republic of Cuba |
| CZE | Czech Republic |
| DEU | Federal Republic of Germany |
| DNK | Kingdom of Denmark |
| DZA | People’s Democratic Republic of Algeria |
| ECU | Republic of Ecuador |
| EGY | Arab Republic of Egypt |
| ESP | Kingdom of Spain |
| ETH | Ethiopia |
| FJI | Republic of Fiji |
| FRA | French Republic |
| GAB | Gabonese Republic |
| GBR | United Kingdom of Great Britain and Northern Ireland |
| GEO | Republic of Georgia |
| GIN | The Republic of Guinea |
| GRC | The Hellenic Republic |
| GRD | Grenada |
| GTM | Republic of Guatemala |
| GUY | Cooperative Republic of Guyana |
| HND | Republic of Honduras |
| HUN | Republic of Hungary |
| IDN | Republic of Indonesia |
| IND | Republic of India |
| IRN | Islamic Republic of Iran |
| IRQ | The Republic of Iraq |
| ISR | State of Israel |
| ITA | The Italian Republic |
| JAM | Jamaica |
| JPN | Japan |
| KAZ | Republic of Kazakhstan |
| KEN | The Republic of Kenya |
| KGZ | Kyrgyz Republic |
| KHM | Cambodia |
| KOR | Republic of Korea |
| LAO | Lao People’s Democratic Republic |
| LBY | Socialist People’s Libyan Arab Jamahiriya |
| LKA | Democratic Socialist Republic of Sri Lanka |
| MAR | Kingdom of Morocco |
| MDA | Republic of Moldova |
| MDV | Republic of Maldives |
| MEX | United Mexican States |
| MMR | Union of Myanmar |
| MNG | Mongolia |
| MWI | Republic of Malawi |
| MYS | Malaysia |
| NGA | Federal Republic of Nigeria |
| NIC | Republic of Nicaragua |
| NLD | Kingdom of the Netherlands |
| NPL | Kingdom of Nepal |
| PAK | Islamic Republic of Pakistan |
| PAN | Republic of Panama |
| PER | Republic of Peru |
| PHL | Republic of the Philippines |
| PNG | Papua New Guinea |
| PRI | Puerto Rico |
| PRK | Democratic People’s Republic of Korea |
| PRT | Portuguese Republic |
| PRY | Republic of Paraguay |
| ROM | Romania |
| RUS | Russian Federation |
| SDN | Republic of the Sudan |
| SEN | Republic of Senegal |
| SLV | Republic of El Salvador |
| SRB | Republic of Serbia |
| SUN | Union of Soviet Socialist Republics |
| SUR | Republic of Suriname |
| SVK | Slovak Republic |
| SYR | Syrian Arab Republic |
| THA | Kingdom of Thailand |
| TJK | Republic of Tajikistan |
| TKM | Turkmenistan |
| TUN | Republic of Tunisia |
| TUR | Republic of Turkey |
| TWN | Taiwan Province of China |
| TZA | United Republic of Tanzania |
| UGA | Republic of Uganda |
| UKR | Ukraine |
| UNK | Unknown |
| URY | Eastern Republic of Uruguay |
| USA | United States of America |
| UZB | Republic of Uzbekistan |
| VEN | Republic of Venezuela |
| VIR | Virgin Islands of the United States |
| VNM | Socialist Republic of Viet Nam |
| YEM | Republic of Yemen |
| YUG | Federal Republic of Yugoslavia |
| ZAR | Republic of Zaire |
| ZMB | Republic of Zambia |
References
- Eshbaugh, W.H. The genus Capsicum (Solanaceae) in Africa. Bothalia 1983, 14, 4. [Google Scholar] [CrossRef]
- Krishna De, A. Capsicum: The Genus Capsicum. Medicinal and Aromatic Plants—Industrial Profile; Taylor & Francis: London, UK; New York, NY, USA, 2003; Volume 33, p. 275. [Google Scholar]
- Davenport, L.J. Genera Solanacearum: The Genera of Solanaceae Illustrated, Arranged According to a New System by Armando T. Hunziker. Syst. Bot. 2004, 29, 221–222. [Google Scholar] [CrossRef]
- Gonzalez-Perez, S.; Garces-Claver, A.; Mallor, C.; Saenz de Miera, L.E.; Fayos, O.; Pomar, F.; Merino, F.; Silvar, C. New insights into Capsicum spp. relatedness and the diversification process of Capsicum annuum in Spain. PLoS ONE 2014, 9, e116276. [Google Scholar] [CrossRef] [Green Version]
- Pozzobon, M.T.; Schifino-Wittmann, M.T.; De Bem Bianchetti, L. Chromosome numbers in wild and semidomesticated Brazilian Capsicum L. (Solanaceae) species: Do x = 12 and x = 13 represent two evolutionary lines? Bot. J. Linn. Soc. 2006, 151, 259–269. [Google Scholar] [CrossRef] [Green Version]
- Moscone, E.A.; Scaldaferro, M.A.; Grabiele, M.; Cecchini, N.M.; Sánchez García, Y.; Jarret, R.; Daviña, J.R.; Ducasse, D.A.; Barboza, G.E.; Ehrendorfer, F. The Evolution of Chili Peppers (Capsicum-Solanaceae): A Cytogenetic Perspective. Act. Hortic. 2007, 137–169. [Google Scholar] [CrossRef]
- Hernandez-Verdugo, S.; Guevara-Gonzalez, R.G.; Rivera-Bustamante, R.F.; Oyama, K. Screening wild plants of Capsicum annuum for resistance to pepper huasteco virus (PHV): Presence of viral DNA and differentiation among populations. Euphytica 2001, 122, 31–36. [Google Scholar] [CrossRef]
- Hammer, K.; Arrowsmith, N.; Gladis, T. Agrobiodiversity with emphasis on plant genetic resources. Naturwissenschaften 2003, 90, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Than, P.P.; Prihastuti, H.; Phoulivong, S.; Taylor, P.W.J.; Hyde, K.D. Chilli anthracnose disease caused by Colletotrichum species. J. Zhejiang Univ. Sci. B 2008, 9, 764. [Google Scholar] [CrossRef] [Green Version]
- Kang, B.K.; Kim, J.H.; Lee, K.H.; Lim, S.C.; Ji, J.J.; Lee, J.W.; Kim, H.T. Effects of Temperature and Moisture on the Survival of Colletotrichum acutatum, the Causal Agent of Pepper Anthracnose in Soil and Pepper Fruit Debris. Plant Pathol. J. 2009, 25, 128–135. [Google Scholar] [CrossRef] [Green Version]
- Manandhar, J.B.; Hartman, G.L. Anthracnose development on pepper fruits inoculated with Colletotrichum gloeosporioides. Plant Dis. 1995, 79, 380–383. [Google Scholar] [CrossRef]
- Srikhong, P.; Lertmongkonthum, K.; Sowanpreecha, R.; Rerngsamran, P. Bacillus sp. strain M10 as a potential biocontrol agent protecting chili pepper and tomato fruits from anthracnose disease caused by Colletotrichum capsici. BioControl 2018, 63, 833–842. [Google Scholar] [CrossRef]
- Park, S.-K.; Kim, S.H.; Park, H.; Yoon, J. Capsicum Germplasm Resistant to Pepper Anthracnose Differentially Interact with Colletotrichum Isolates. Hortic. Environ. Biotechnol. 2009, 50, 17–23. [Google Scholar]
- Lee, J.; Hong, J.-H.; Do, J.W.; Yoon, J.B. Identification of QTLs for resistance to anthracnose to two Colletotrichum species in pepper. J. Crop Sci. Biotechnol. 2010, 13, 227–233. [Google Scholar] [CrossRef]
- Lee, J.; Do, J.W.; Yoon, J.B. Development of STS markers linked to the major QTLs for resistance to the pepper anthracnose caused by Colletotrichum acutatum and C. capsici. Hortic. Environ. Biotechnol. 2011, 52, 596–601. [Google Scholar] [CrossRef]
- Yoon, J.; Park, H. Trispecies Bridge Crosses, (Capsicum annuum× C. chinense)× C. baccatum, as an Alternative for Introgression of AnthracnoseResistance from C. baccatum into C. annuum. Hortic. Environ. Biotechnol. 2005, 46, 5–9. [Google Scholar]
- AVRDC (Ed.) Host resistance to pepper anthracnose. In AVRDC Report; AVRDC-The World Vegetable Centre: Shanhua, Taiwan, 2003; pp. 29–30. [Google Scholar]
- Pakdeevaraporn, P.; Wasee, S.; Taylor, P.W.J.; Mongkolporn, O. Inheritance of resistance to anthracnose caused by Colletotrichum capsici in Capsicum. Plant Breed. 2005, 124, 206–208. [Google Scholar] [CrossRef]
- Ridzuan, R.; Rafii, M.Y.; Ismail, S.I.; Mohammad Yusoff, M.; Miah, G.; Usman, M. Breeding for Anthracnose Disease Resistance in Chili: Progress and Prospects. Int. J. Mol. Sci. 2018, 19, 3122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.B.; Do, J.W.; Yang, D.C.; Park, H.G. Interspecific Cross Compatibility among Five Domesticated Species of Capsicum Genus. Hortic. Environ. Biotechnol. 2004, 45, 324–329. [Google Scholar]
- Kim, S.-G.; Ro, N.-Y.; Hur, O.-S.; Ko, H.-C.; Gwag, J.-G.; Huh, Y.-C. Evaluation of Resistance to Colletotrichum acutatum in Pepper Genetic Resources. Res. Plant Dis. 2012, 18, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Su, K.; Ki-taek, K.; DongHwi, K.; EunYoung, Y.; MyeongCheoul, C.; Jamal, A.; Young, C.; DoHam, P.; DaeGeun, O.; JuKwang, H. Identification of quantitative trait loci associated with anthracnose resistance in chili pepper (Capsicum spp.). Korean J. Hortic. Sci. Technol. 2010, 28, 1014–1024. [Google Scholar]
- Kim, K.H.; Yoon, J.B.; Park, H.G.; Park, E.W.; Kim, Y.H. Structural Modifications and Programmed Cell Death of Chili Pepper Fruit Related to Resistance Responses to Colletotrichum gloeosporioides Infection. Phytopathology 2004, 94, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Oh, B.J.; Kim, K.D.; Kim, Y.S. Effect of Cuticular Wax Layers of Green and Red Pepper Fruits on Infection by Colletotrichum gloeosporioides. J. Phytopathol. 1999, 147, 547–552. [Google Scholar] [CrossRef]
- Suwor, P.; Thummabenjapone, P.; Sanitchon, J.; Kumar, S.; Techawongstien, S. Phenotypic and genotypic responses of chili (Capsicum annuum L.) progressive lines with different resistant genes against anthracnose pathogen (Colletotrichum spp.). Eur. J. Plant Pathol. 2015, 143, 725–736. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, Y.; Zhang, Z.; Cao, Y.; Yu, H.; Ma, W.; Zhang, B.; Wang, R.; Gao, J.; Wang, L. Fine mapping of the major anthracnose resistance QTL AnRGO5 in Capsicum chinense ‘PBC932’. BMC Plant Biol. 2020, 20, 189. [Google Scholar] [CrossRef] [PubMed]
- Mahasuk, P.; Khumpeng, N.; Wasee, S.; Taylor, P.W.J.; Mongkolporn, O. Inheritance of resistance to anthracnose (Colletotrichum capsici) at seedling and fruiting stages in chili pepper (Capsicum spp.). Plant Breed. 2009, 128, 701–706. [Google Scholar] [CrossRef]
- Mahasuk, P.; Taylor, P.W.J.; Mongkolporn, O. Identification of Two New Genes Conferring Resistance to Colletotrichum acutatum in Capsicum baccatum. Phytopathology® 2009, 99, 1100–1104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahasuk, P.; Struss, D.; Mongkolporn, O. QTLs for resistance to anthracnose identified in two Capsicum sources. Mol. Breed. 2016, 36, 10. [Google Scholar] [CrossRef]
- Kim, S.H.; Yoon, J.B.; Park, H.G. Inheritance of anthracnose resistance in a new genetic resource, Capsicum baccatum PI594137. J. Crop Sci. Biotechnol. 2008, 11, 13–16. [Google Scholar]
- Kim, H.; Yoon, J.B.; Lee, J. Development of Fluidigm SNP Type Genotyping Assays for Marker-assisted Breeding of Chili Pepper (Capsicum annuum L.). Hortic. Sci. Technol. 2017, 35, 465–479. [Google Scholar] [CrossRef]
- Lee, Y.R.; Kim, J.; Lee, S.Y.; Lee, J. Diallelic SNP marker development and genetic linkage map construction in octoploid strawberry (Fragaria × ananassa) through next-generation resequencing and high-resolution melting analysis. Hortic. Environ. Biotechnol. 2020, 61, 371–383. [Google Scholar] [CrossRef]
- RDA. Pepper (Capsicum spp.). Rural Development Administration, Korea, 2012, pp. 1–53. Available online: http://www.rda.go.kr/foreign/ten/index.jsp (accessed on 18 August 2021).
- Kim, J.-T.; Kim, J.; Park, S.; Choi, W. Characterization of Colletotrichum Isolates Causing Anthracnose of Pepper in Korea. Plant Pathol. J. 2008, 24, 17–23. [Google Scholar] [CrossRef]
- Montri, P.; Taylor, P.W.J.; Mongkolporn, O. Pathotypes of Colletotrichum capsici, the Causal Agent of Chili Anthracnose, in Thailand. Plant Dis. 2009, 93, 17–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-G. Structural Changes in the Development of Phytophthora Blight and Anthracnose on Chili Pepper (Capsicum annuum L.). Ph.D. Thesis, Seoul National University, Seoul, Korea, 2009. [Google Scholar]
- Ziv, C.; Zhao, Z.; Gao, Y.G.; Xia, Y. Multifunctional Roles of Plant Cuticle During Plant-Pathogen Interactions. Front. Plant Sci. 2018, 9, 1088. [Google Scholar] [CrossRef]
- Underwood, W. The Plant Cell Wall: A Dynamic Barrier Against Pathogen Invasion. Front. Plant Sci. 2012, 3, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malinovsky, F.G.; Fangel, J.U.; Willats, W.G.T. The role of the cell wall in plant immunity. Front. Plant Sci. 2014, 5, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkan, N.; Friedlander, G.; Ment, D.; Prusky, D.; Fluhr, R. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol. 2015, 205, 801–815. [Google Scholar] [CrossRef]
- Marques, J.P.R.; Amorim, L.; Spósito, M.B.; Appezzato-da-Glória, B. Ultrastructural changes in the epidermis of petals of the sweet orange infected by Colletotrichum acutatum. Protoplasma 2016, 253, 1233–1242. [Google Scholar] [CrossRef] [PubMed]
- Ziv, C.; Kumar, D.; Sela, N.; Itkin, M.; Malitsky, S.; Schaffer, A.A.; Prusky, D.B. Sugar-regulated susceptibility of tomato fruit to Colletotrichum and Penicillium requires differential mechanisms of pathogenicity and fruit responses. Environ. Microbiol. 2020, 22, 2870–2891. [Google Scholar] [CrossRef] [PubMed]
- Reymond, P.; Farmer, E.E. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1998, 1, 404–411. [Google Scholar] [CrossRef]
- Lin, S.W.; Gniffke, P.A.; Wang, T.C. Inheritance of Resistance to Pepper Anthracnose Caused by Colletotrichum acutatum. Acta. Hortic. 2007, 329–334. [Google Scholar] [CrossRef]
- Jo, I.-H.; Sung, J.; Hong, C.-E.; Raveendar, S.; Bang, K.-H.; Chung, J.-W. Development of cleaved amplified polymorphic sequence (CAPS) and high-resolution melting (HRM) markers from the chloroplast genome of Glycyrrhiza species. 3 Biotech 2018, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Geleta, L.F.; Labuschagne, M.T.; Viljoen, C.D. Genetic Variability in Pepper (Capsicum annuum L.) Estimated by Morphological Data and Amplified Fragment Length Polymorphism Markers. Biodivers. Conserv. 2005, 14, 2361–2375. [Google Scholar] [CrossRef]
- Greene, S.L.; Gritsenko, M.; Vandemark, G. Relating Morphologic and RAPD Marker Varlation to Collection Site Environment in wild Populations of Red Clover (Trifolium Pratense L.). Genet. Resour. Crop Evol. 2004, 51, 643–653. [Google Scholar] [CrossRef]
- Luitel, B.P.; Ro, N.-Y.; Ko, H.-C.; Sung, J.-S.; Rhee, J.-H.; Hur, O.-S. Phenotypic Variation in a Germplasm Collection of Pepper (Capsicum chinense Jacq.) from Korea. J. Crop Sci. Biotechnol. 2018, 21, 499–506. [Google Scholar] [CrossRef]



| Continent | No. of Accession | Country * |
|---|---|---|
| South America | 885 | ARG, BOL, BRA, CHL, COL, ECU, GUY, PER, PRI, PRY, SUR, URY, VEN |
| North America | 585 | BHS, BLZ, CAN, CRI, CUB, GRD, GTM, HND, JAM, MEX, NIC, PAN, SLV, USA, VIR |
| Asia | 1185 | AFG, ARM, AZE, BGD, BTN, CHN, GEO, IDN, IND, IRN, IRQ, ISR, JPN, KAZ, KGZ, KHM, KOR, LAO, LKA, MDV, MMR, MNG, MYS, NPL, PAK, PHL, PRK, SYR, THA, TJK, TKM, TUR, TWN, UZB, VNM, YEM |
| Africa | 61 | MAR, BFA, BWA, DZA, EGY, ETH, GAB, GIN, KEN, LBY, MWI, NGA, SDN, SEN, TUN, TZA, UGA, ZAR, ZMB |
| Oceania | 9 | AUS, FJI, PNG |
| Europe | 724 | SUN, AUT, BEL, BGR, BLR, CHE, CSK, CZE, DEU, DNK, ESP, FRA, GBR, GRC, HUN, ITA, MDA, NLD, PRT, ROM, RUS, SRB, SVK, UKR, YUG |
| Unknown | 289 | |
| Total | 3738 |
| Species | Disease Rating Scale | Total | |||
|---|---|---|---|---|---|
| 0–1 (R) | 1–2 (MR) | 2–3 (S) | 3–4 (HS) | ||
| C. annuum | 2 | 3 | 8 | 684 | 697 |
| C. annuum var annuum | 49 | 27 | 26 | 1213 | 1315 |
| C. annuum var glabriusculum | 1 | - | - | 3 | 4 |
| C. baccatum | 18 | 12 | 6 | 55 | 91 |
| C. baccatum var baccatum | 9 | 8 | 15 | 136 | 168 |
| C. baccatum var pendulum | 5 | 15 | 6 | 97 | 123 |
| C. baccatum var praetermissum | - | - | - | 4 | 4 |
| C. chacoense | 3 | 2 | - | 12 | 17 |
| C. chinense | 84 | 43 | 40 | 512 | 679 |
| C. eximium | - | - | - | 1 | 1 |
| C. frutescens | 86 | 35 | 36 | 308 | 465 |
| C. galapagoense | - | - | - | 2 | 2 |
| C. pubescens | - | 2 | 2 | 25 | 29 |
| C. tovarii | 1 | - | - | - | 1 |
| C. sp. | 3 | - | 1 | 138 | 142 |
| Total | 261 | 147 | 140 | 3190 | 3738 |
| Species | Disease Incidence (%) | Total * | |||
|---|---|---|---|---|---|
| 0–25 | 25–50 | 50–75 | 75–100 | ||
| C. annuum | - | - | 1 | 2 | 3 ab |
| C. annuum var. annuum | - | - | - | 5 | 5 ab |
| C. baccatum | 3 | 3 | 9 | 9 | 24 b |
| C. baccatum var. baccatum | 2 | 5 | 12 | 10 | 29 ab |
| C. baccatum var. pendulum | 2 | 6 | 5 | 9 | 22 b |
| C. chacoense | - | - | - | 5 | 5 a |
| C. chinense | 4 | 5 | 20 | 79 | 108 ab |
| C. frutescens | 1 | 1 | 7 | 8 | 17 ab |
| C. pubescens | - | - | 1 | - | 1 ab |
| C. sp. | - | - | 1 | - | 1- |
| Total | 12 | 20 | 56 | 127 | 215 |
| Acc. No. | Species | Incidence (%) | Lesion (mm) | Disease Rating Scale | CA12g19240 | CaR12.2M1-CAPS |
|---|---|---|---|---|---|---|
| 158502 | C. chinense | 14.3 | 1.4 ± 0.5 | 1 | S/- | S/Ud |
| 158769 | C. baccatum var. pendulum | 22.2 | 2.0 ± 0.7 | 1 | R | R |
| 218958 | C. baccatum var. baccatum | 16.7 | 1.6 ± 0.6 | 1 | R | R |
| 229147 | C. baccatum var. baccatum | 20 | 2.0 ± 1.2 | 1 | R | R |
| 229200 | C. chinense | 11.1 | 0.9 ± 0.3 | 1 | S | S |
| 240869 | C. baccatum | 15.8 | 1.5 ± 0.7 | 1 | R | R |
| 258953 | C. baccatum | 22.8 | 3.9 ± 1.3 | 2 | R | R |
| 270479 | C. chinense | 10 | 0.3 ± 0.2 | 1 | S | S |
| 276470 | C. frutescens | 25 | 5.3 ± 1.5 | 3 | S/- | S/Ud |
| 305437 | C. chinense | 20 | 0.5 ± 0.4 | 1 | S | S |
| 305455 | C. chinense | 10 | 0.3 ± 0.2 | 1 | S | S |
| 305478 | C. baccatum | 10 | 0.5 ± 0.3 | 1 | R | R |
| Manitta | C. annuum | 100 | 13.7 ± 2.1 | 5 | S | S |
| PBC81 | C. baccatum var. pendulum | 40 | 6.8 ± 1.2 | 4 | R | R |
| AR-Dolgyeoktan | C. sp. | 42 | 6.9 ± 1.6 | 4 | H | H |
| Acc. No. | Origin | Species | Fruit Weight(g) | Fruit Length(cm) | Fruit Width(mm) | Fruit wall Thickness(mm) | Sugar Content (oBrix) |
|---|---|---|---|---|---|---|---|
| 158502 | PER | C. chinense | 21.3 ± 0.8 | 10.2 ± 0.3 | 26.4 ± 0.8 | 3.0 ± 0.5 | 9.1 ± 0.2 |
| 158769 | CHL | C. baccatum var. pendulum | 22 ± 1.6 | 7.9 ± 0.8 | 26.5 ± 0.7 | 1.5 ± 0.1 | 12.9 ± 0.3 |
| 218958 | VEN | C. baccatum var. baccatum | 15.9 ± 1.8 | 9.9 ± 0.4 | 27.1 ± 0.6 | 1.7 ± 0.1 | 15.8 ± 0.6 |
| 229147 | HUN | C. baccatum var. baccatum | 6.4 ± 0.7 | 2.4 ± 0.1 | 21.2 ± 0.8 | 2.4 ± 0.1 | 8.5 ± 0.6 |
| 229200 | HUN | C. chinense | 7.2 ± 0.7 | 6.3 ± 0.3 | 22.2 ± 0.8 | 1.9 ± 0.1 | 7.6 ± 0.1 |
| 240869 | BRA | C. baccatum | 7.7 ± 0.5 | 5.8 ± 0.2 | 12.0 ± 2.2 | 2.0 ± 0.5 | 11.8 ± 0.9 |
| 258953 | UNK | C. baccatum | 1.4 ± 0.1 | 2.6 ± 0.1 | 8.4 ± 0.5 | 1.0 ± 0.1 | 10.3 ± 0.7 |
| 270479 | BRA | C. chinense | 1.3 ± 0.1 | 1.1 ± 0.1 | 14.1 ± 0.1 | 1.4 ± 0.2 | 8.4 ± 0.9 |
| 276470 | CRI | C. frutescens | 6.7 ± 0.1 | 3.2 ± 0.1 | 21.8 ± 0.6 | 1.6 ± 0.3 | 9.3 ± 0.4 |
| 305437 | COL | C. chinense | 8.8 ± 0.6 | 4.8 ± 0.4 | 23.2 ± 1.3 | 2.4 ± 0.2 | 7.8 ± 0.4 |
| 305455 | COL | C. chinense | 4.5 ± 1.0 | 4.5 ± 0.4 | 22.2 ± 2.2 | 1.5 ± 0.1 | 10.2 ± 0.5 |
| 305478 | PER | C. baccatum | 5.4 ± 0.2 | 4.5 ± 0.1 | 18.5 ± 1.3 | 2.7 ± 0.5 | 10.8 ± 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ro, N.-Y.; Sebastin, R.; Hur, O.-S.; Cho, G.-T.; Geum, B.; Lee, Y.-J.; Kang, B.-C. Evaluation of Anthracnose Resistance in Pepper (Capsicum spp.) Genetic Resources. Horticulturae 2021, 7, 460. https://doi.org/10.3390/horticulturae7110460
Ro N-Y, Sebastin R, Hur O-S, Cho G-T, Geum B, Lee Y-J, Kang B-C. Evaluation of Anthracnose Resistance in Pepper (Capsicum spp.) Genetic Resources. Horticulturae. 2021; 7(11):460. https://doi.org/10.3390/horticulturae7110460
Chicago/Turabian StyleRo, Na-Young, Raveendar Sebastin, On-Sook Hur, Gyu-Taek Cho, Bora Geum, Yong-Jik Lee, and Byoung-Cheorl Kang. 2021. "Evaluation of Anthracnose Resistance in Pepper (Capsicum spp.) Genetic Resources" Horticulturae 7, no. 11: 460. https://doi.org/10.3390/horticulturae7110460
APA StyleRo, N.-Y., Sebastin, R., Hur, O.-S., Cho, G.-T., Geum, B., Lee, Y.-J., & Kang, B.-C. (2021). Evaluation of Anthracnose Resistance in Pepper (Capsicum spp.) Genetic Resources. Horticulturae, 7(11), 460. https://doi.org/10.3390/horticulturae7110460





