Dynamic Carotenoid Profiles and Function Analysis of the RrPSY1 Gene in Rosa rugosa Flowers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Carotenoid Extraction and Analysis
2.3. Salt Treatment and qRT-PCR Analysis
2.4. Identification and Bioinformatic Analysis of RrPSY1
2.5. Pigment Complementation Assay
2.6. Subcellular Localization Assay
2.7. Transient Overexpression Transformation in Rose
2.8. Statistical Analysis
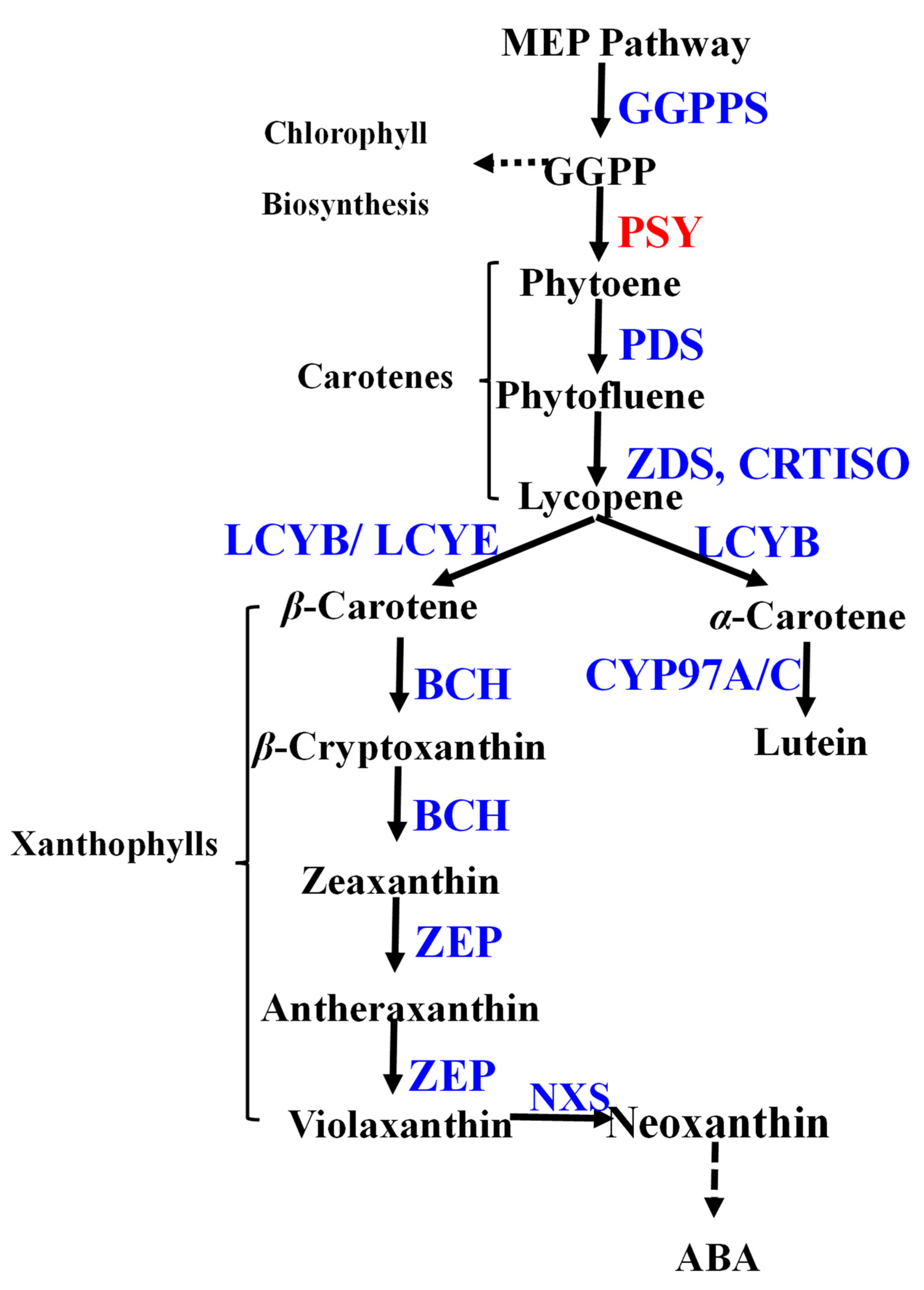
3. Results
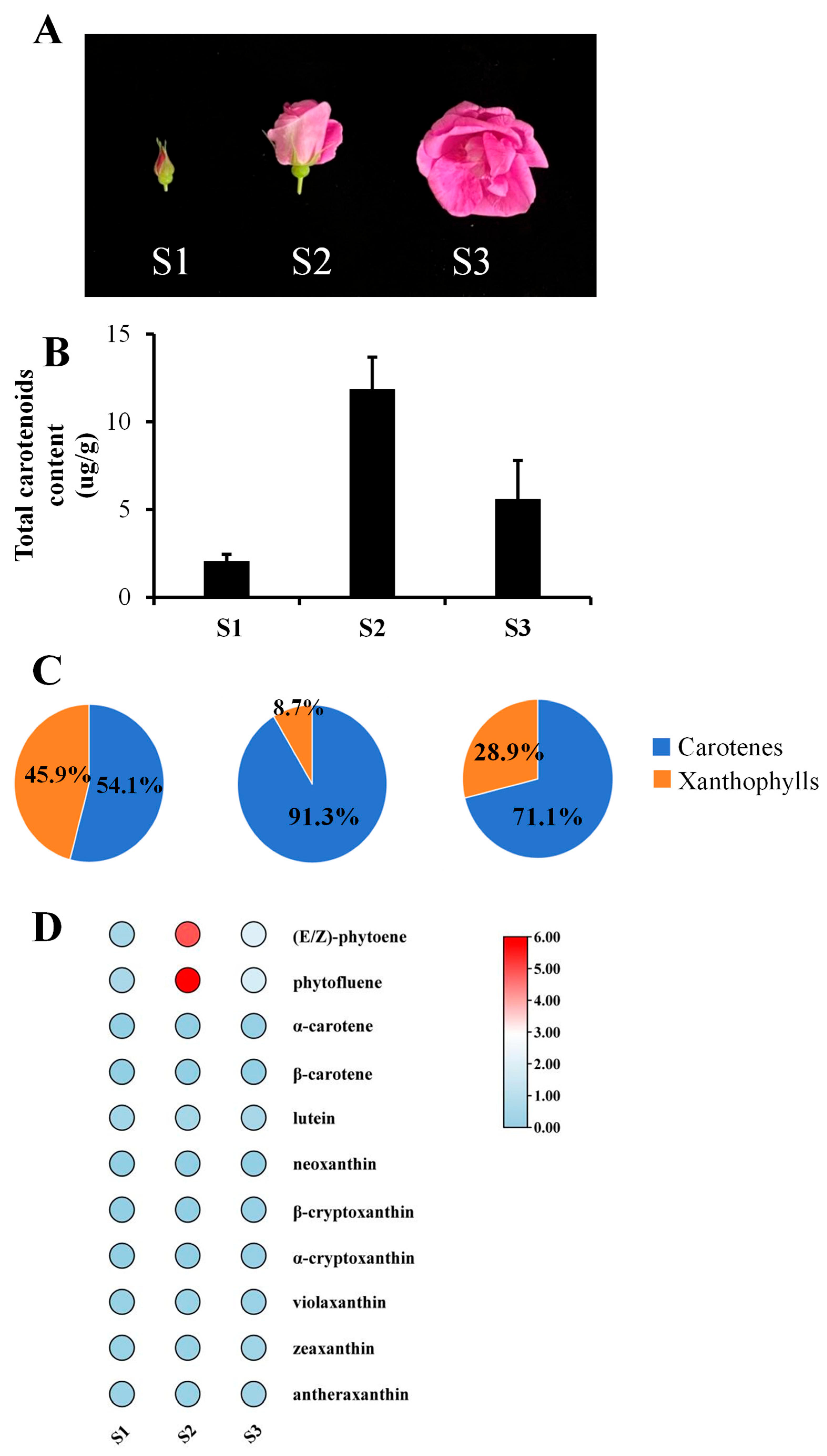
3.1. Dynamic Carotenoid Profiles in R. rugosa Petals During Flower Development
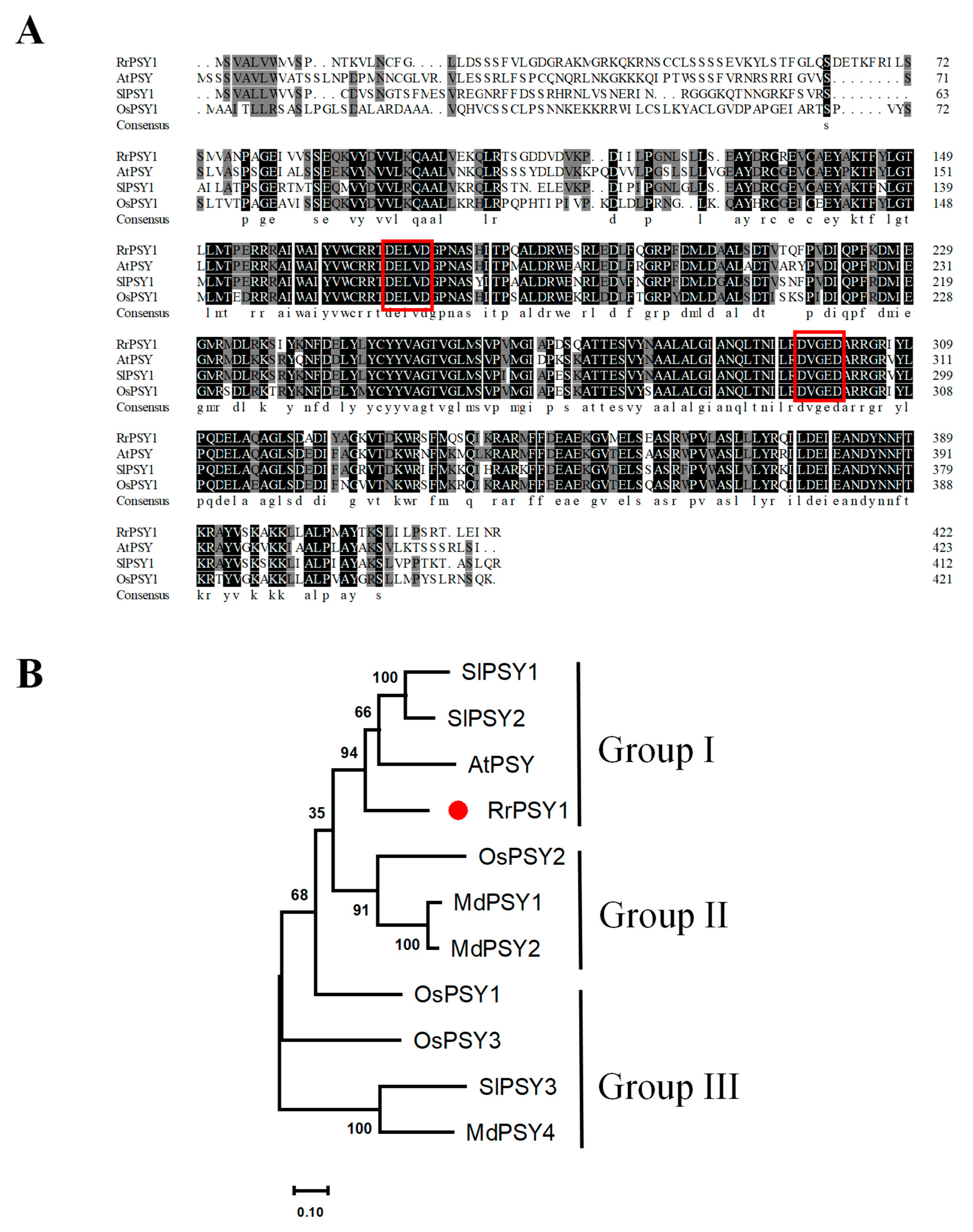
3.2. Identification and Phylogenetic Analysis of the RrPSY1 Gene in R. rugosa
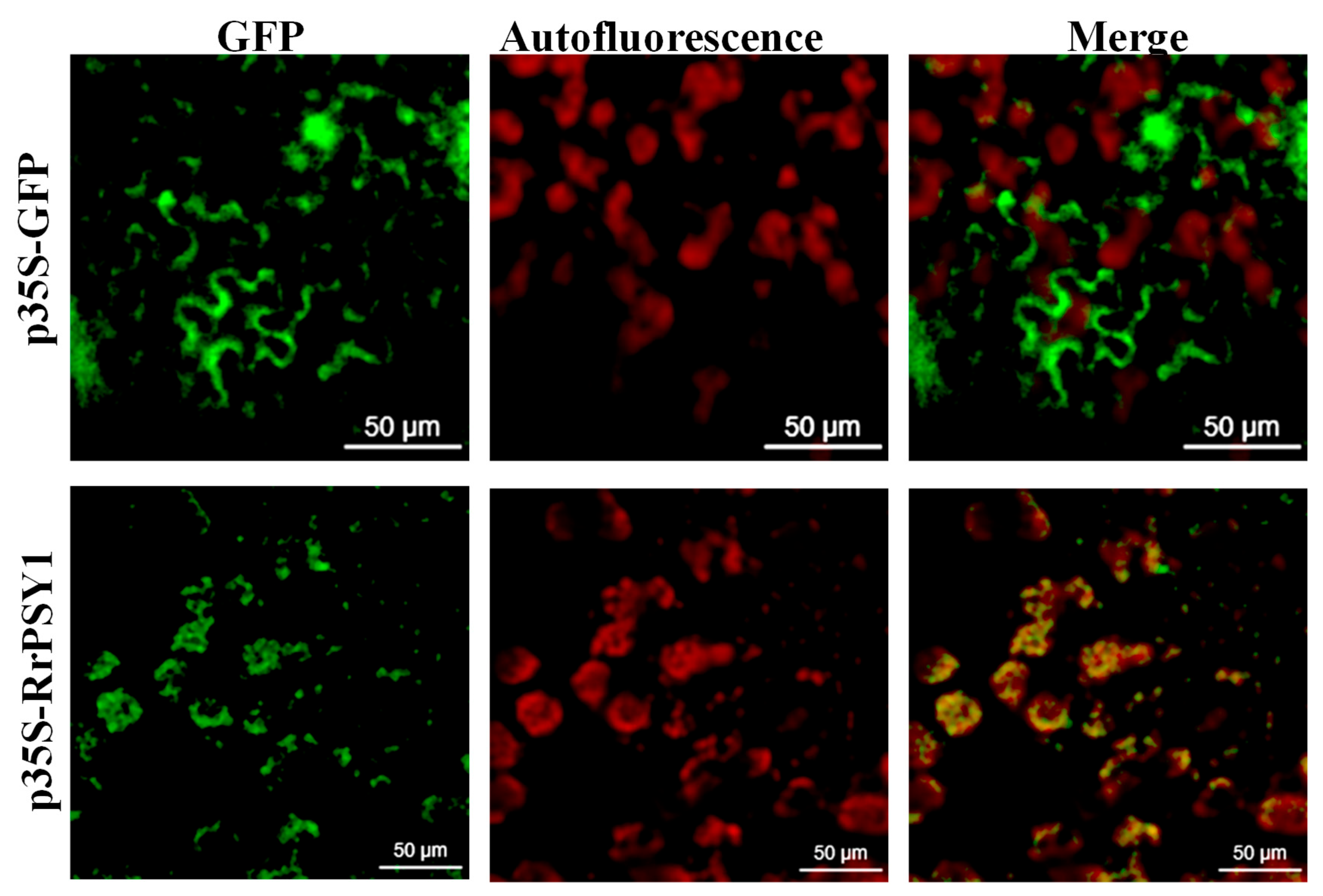
3.3. Subcellular Localization of RrPSY1
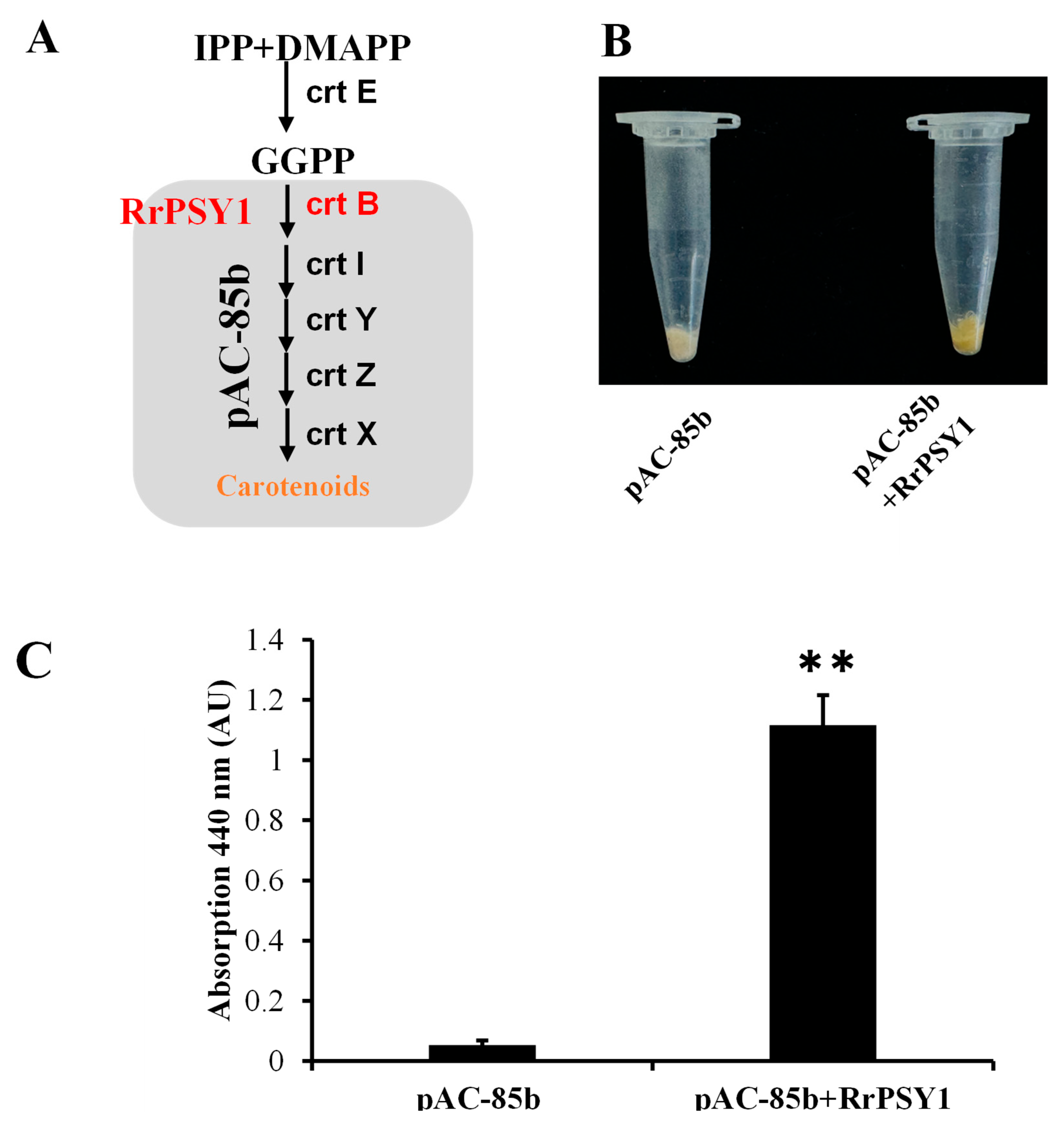
3.4. In Vitro Functional Characterization of RrPSY1
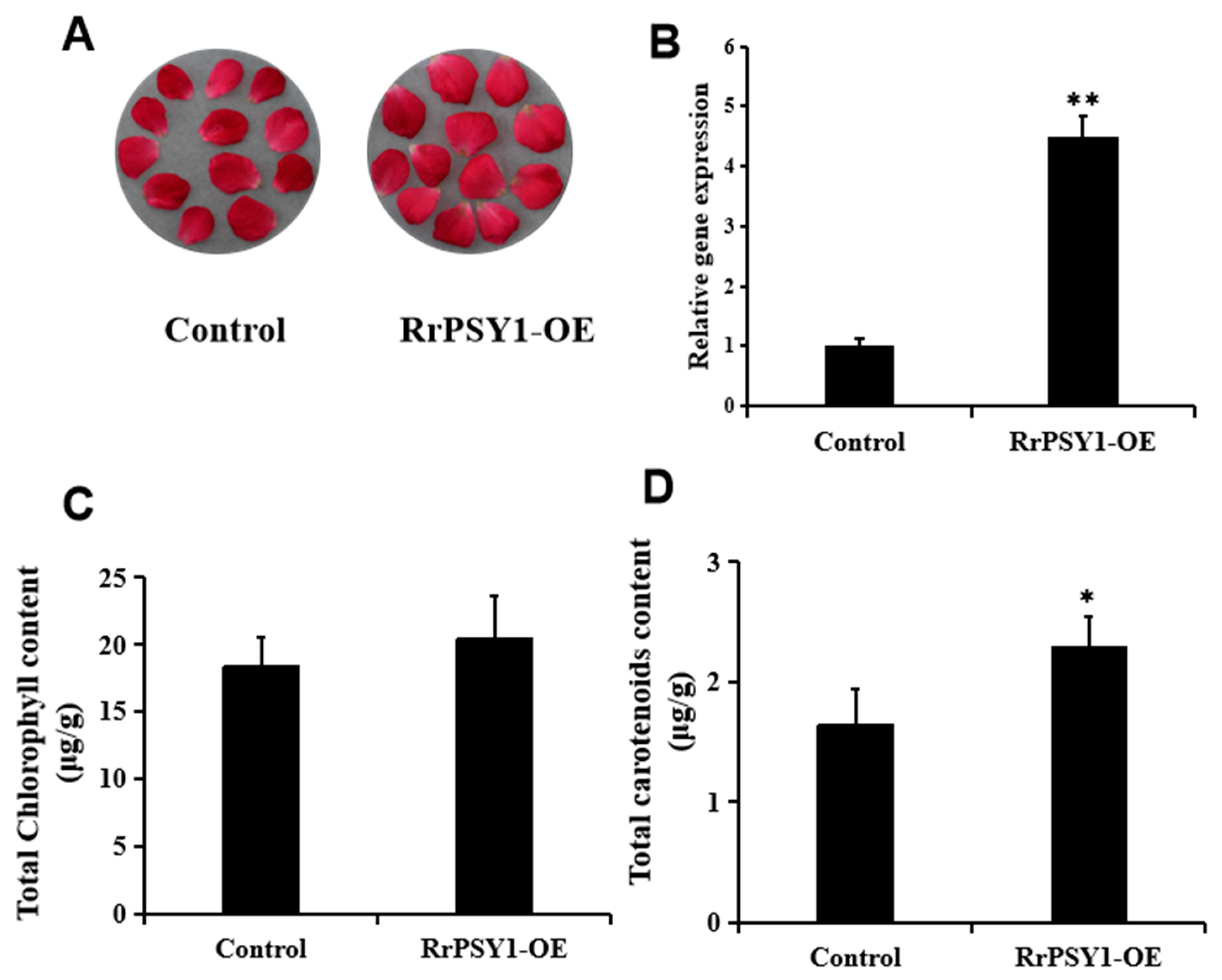
3.5. Transient Overexpression of RrPSY1 in Rose Petals
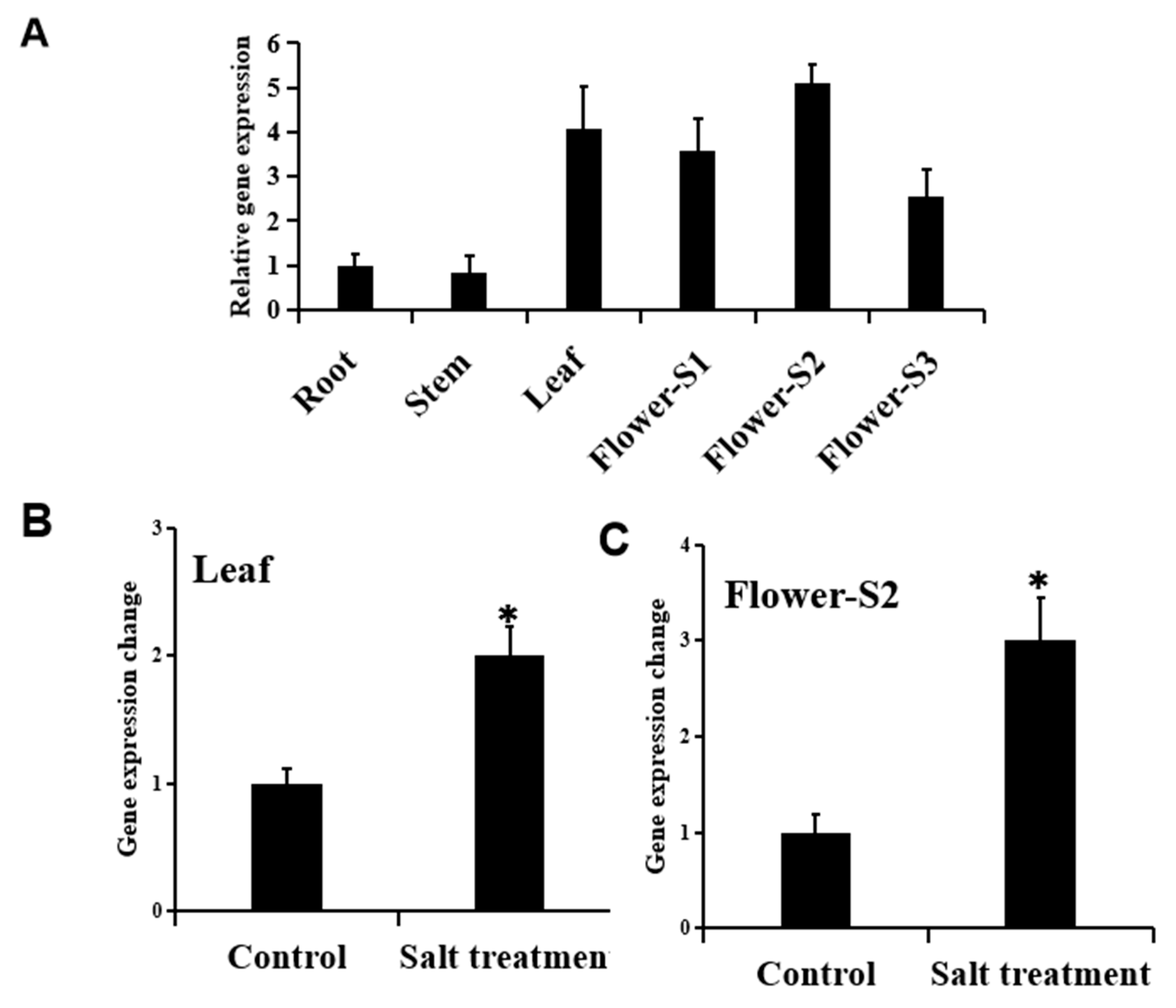
3.6. Expression Analysis of RrPSY1 Under Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2022, 2, 3. [Google Scholar] [CrossRef]
- Rodriguez-Concepcion, M.; Avalos, J.; Bonet, M.L.; Boronat, A.; Gomez-Gomez, L.; Hornero-Mendez, D.; Limon, M.C.; Meléndez-Martínez, A.J.; Olmedilla-Alonso, B.; Palou, A.; et al. A global perspective on carotenoids: Metabolism, biotechnology, and benefits for nutrition and health. Prog. Lipid Res. 2018, 70, 62–93. [Google Scholar] [CrossRef] [PubMed]
- Yabuzaki, J. Carotenoids Database: Structures, chemical fingerprints and distribution among organisms. Database 2017, 2017, bax004. [Google Scholar] [CrossRef]
- Rao, A.V.; Rao, L.G. Carotenoids and human health. Pharmacol. Res. 2007, 55, 207–216. [Google Scholar] [CrossRef]
- Hou, X.; Rivers, J.; León, P.; McQuinn, R.P.; Pogson, B.J. Synthesis and Function of Apocarotenoid Signals in Plants. Trends Plant Sci. 2016, 21, 792–803. [Google Scholar] [CrossRef]
- Moreno, J.C.; Mi, J.; Alagoz, Y.; Al-Babili, S. Plant apocarotenoids: From retrograde signaling to interspecific communication. Plant J. 2021, 105, 351–375. [Google Scholar] [CrossRef]
- Tang, G. Bioconversion of dietary provitamin A carotenoids to vitamin A in humans. Am. J. Clin. Nutr. 2010, 91, 1468S–1473S. [Google Scholar] [CrossRef]
- Van Hoang, D.; Pham, N.M.; Lee, A.H.; Tran, D.N.; Binns, C.W. Dietary Carotenoid Intakes and Prostate Cancer Risk: A Case-Control Study from Vietnam. Nutrients 2018, 10, 70. [Google Scholar] [CrossRef]
- Aziz, E.; Batool, R.; Akhtar, W.; Rehman, S.; Shahzad, T.; Malik, A.; Shariati, M.A.; Laishevtcev, A.; Plygun, S.; Heydari, M.; et al. Xanthophyll: Health benefits and therapeutic insights. Life Sci. 2020, 240, 117104. [Google Scholar] [CrossRef]
- Rodríguez-Concepción, M.; Boronat, A. Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 2002, 130, 1079–1089. [Google Scholar] [CrossRef] [PubMed]
- Barja, M.V.; Rodriguez-Concepcion, M. Plant geranylgeranyl diphosphate synthases: Every (gene) family has a story. aBIOTECH 2021, 2, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Dogbo, O.; Laferriére, A.; D’Harlingue, A.; Camara, B. Carotenoid biosynthesis: Isolation and characterization of a bifunctional enzyme catalyzing the synthesis of phytoene. Proc. Natl. Acad. Sci. USA 1988, 85, 7054–7058. [Google Scholar] [CrossRef]
- Slater, A.; Maunders, M.J.; Edwards, K.; Schuch, W.; Grierson, D. Isolation and characterisation of cDNA clones for tomato polygalacturonase and other ripening-related proteins. Plant Mol. Biol. 1985, 5, 137–147. [Google Scholar] [CrossRef]
- Bartley, G.E.; Viitanen, P.V.; Bacot, K.O.; Scolnik, P.A. A tomato gene expressed during fruit ripening encodes an enzyme of the carotenoid biosynthesis pathway. J. Biol. Chem. 1992, 267, 5036–5039. [Google Scholar] [CrossRef]
- Gupta, P.; Rodriguez-Franco, M.; Bodanapu, R.; Sreelakshmi, Y.; Sharma, R. Phytoene synthase 2 in tomato fruits remains functional and contributes to abscisic acid formation. Plant Sci. 2022, 316, 111177. [Google Scholar] [CrossRef]
- Sun, T.; Yuan, H.; Cao, H.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, L.; Dong, C.; Guo, J.; Jin, L.; Wei, P.; Li, F.; Zhang, X.; Wang, R. Characterization and functional analysis of phytoene synthase gene family in tobacco. BMC Plant Biol. 2021, 21, 32. [Google Scholar] [CrossRef]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rocheford, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Driedonks, N.; Lewis, D.; Shumskaya, M.; Chen, X.; Wurtzel, E.T.; Espley, R.V.; Allan, A.C. The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. BMC Plant Biol. 2015, 15, 185. [Google Scholar] [CrossRef]
- Fraser, P.D.; Schuch, W.; Bramley, P.M. Phytoene synthase from tomato (Lycopersicon esculentum) chloroplasts—Partial purification and biochemical properties. Planta 2000, 211, 361–369. [Google Scholar] [CrossRef]
- Bian, Y.; Pan, J.; Gao, D.; Feng, Y.; Zhang, B.; Song, L.; Wang, L.; Ma, X.; Liang, L. Bioactive metabolite profiles and quality of Rosa rugosa during its growing and flower-drying process. Food Chem. 2024, 450, 139388. [Google Scholar] [CrossRef]
- Chroho, M.; Bouymajane, A.; Oulad El Majdoub, Y.; Cacciola, F.; Mondello, L.; Aazza, M.; Zair, T.; Bouissane, L. Phenolic Composition, Antioxidant and Antibacterial Activities of Extract from Flowers of Rosa damascena from Morocco. Separations 2022, 9, 247. [Google Scholar] [CrossRef]
- Kanani, M.; Chamani, E.; Shokouhian, A.A.; Torabi-Giglou, M. Plant secondary metabolism and flower color changes in damask rose at different flowering development stages. Acta. Physiol. Plant 2021, 43, 55. [Google Scholar] [CrossRef]
- Xu, Y.; Li, M.; Ding, H.; Ma, Y.; Yang, Y.; Feng, L. Comparative effects of humic acid biostimulation on soil properties, growth, and fragrance of Rosa rugosa. Ind. Crops Prod. 2025, 225, 120444. [Google Scholar] [CrossRef]
- Ogata, J.; Kanno, Y.; Itoh, Y.; Tsugawa, H.; Suzuki, M. Anthocyanin biosynthesis in roses. Nature 2005, 435, 757–758. [Google Scholar] [CrossRef]
- Wan, H.; Yu, C.; Han, Y.; Guo, X.; Luo, L.; Pan, H.; Zheng, T.; Wang, J.; Cheng, T.; Zhang, Q. Determination of Flavonoids and Carotenoids and Their Contributions to Various Colors of Rose Cultivars (Rosa spp.). Front. Plant Sci. 2019, 10, 123. [Google Scholar] [CrossRef]
- Wei, G.; Chen, Y.; Wang, M.; Xi, Y.; Xu, Y.; Hussain, H.; Zhu, K.; Xu, Y.; Bai, M.; Wang, J.; et al. Integrative application of metabolomics and transcriptomics provides new insights into carotenoid biosynthesis during Rosa rugosa hips ripening. Food Biosci. 2024, 60, 104422. [Google Scholar] [CrossRef]
- Xu, Y.; Shi, Y.; Zhang, W.; Zhu, K.; Feng, L.; Wang, J. C2H2 Zinc Finger Protein Family Analysis of Rosa rugosa Identified a Salt-Tolerance Regulator, RrC2H2-8. Plants 2024, 13, 3580. [Google Scholar] [CrossRef]
- Wei, G.; Chen, Y.; Wang, J.; Feng, L. Molecular cloning and characterization of farnesyl diphosphate synthase from Rosa rugosa Thunb associated with salinity stress. PeerJ 2024, 12, e16929. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Su, L.; Hu, S.; Xue, J.-Y.; Liu, H.; Liu, G.; Jiang, Y.; Du, J.; Qiao, Y.; Fan, Y.; et al. A chromosome-level genome assembly of rugged rose (Rosa rugosa) provides insights into its evolution, ecology, and floral characteristics. Hortic. Res. 2021, 8, 141. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Jang, S.J.; Jeong, H.B.; Jung, A.; Kang, M.Y.; Kim, S.; Ha, S.H.; Kwon, J.K.; Kang, B.C. Phytoene synthase 2 can compensate for the absence of PSY1 in the control of color in Capsicum fruit. J. Exp. Bot. 2020, 71, 3417–3427. [Google Scholar] [CrossRef] [PubMed]
- Hua, B.; Wu, J.; Han, X.; Bian, X.; Xu, Z.; Sun, C.; Wang, R.; Zhang, W.; Liang, F.; Zhang, H.; et al. Auxin homeostasis is maintained by sly-miR167-SlARF8A/B-SlGH3.4 feedback module in the development of locular and placental tissues of tomato fruits. New Phytol. 2024, 241, 1177–1192. [Google Scholar] [CrossRef] [PubMed]
- Conart, C.; Bomzan, D.P.; Huang, X.Q.; Bassard, J.E.; Paramita, S.N.; Saint-Marcoux, D.; Rius-Bony, A.; Hivert, G.; Anchisi, A.; Schaller, H.; et al. A cytosolic bifunctional geranyl/farnesyl diphosphate synthase provides MVA-derived GPP for geraniol biosynthesis in rose flowers. Proc. Natl. Acad. Sci. USA 2023, 120, e2221440120. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Zhang, R.; Jiang, S.; Wang, H.; Ming, F. The MYB transcription factor RcMYB1 plays a central role in rose anthocyanin biosynthesis. Hortic. Res. 2023, 10, uhad080. [Google Scholar] [CrossRef]
- Zhai, S.; Li, G.; Sun, Y.; Song, J.; Li, J.; Song, G.; Li, Y.; Ling, H.; He, Z.; Xia, X. Genetic analysis of phytoene synthase 1 (Psy1) gene function and regulation in common wheat. BMC Plant Biol. 2016, 16, 228. [Google Scholar] [CrossRef]
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant Salt Stress: Adaptive Responses, Tolerance Mechanism and Bioengineering for Salt Tolerance. Bot. Rev. 2016, 82, 371–406. [Google Scholar] [CrossRef]
- Uarrota, V.G.; Stefen, D.L.V.; Leolato, L.S.; Gindri, D.M.; Nerling, D. Revisiting Carotenoids and Their Role in Plant Stress Responses: From Biosynthesis to Plant Signaling Mechanisms During Stress. In Antioxidants and Antioxidant Enzymes in Higher Plants; Gupta, D.K., Palma, J.M., Corpas, F.J., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 207–232. [Google Scholar]
- Mapelli-Brahm, P.; Meléndez-Martínez, A.J. The colourless carotenoids phytoene and phytofluene: Sources, consumption, bioavailability and health effects. Curr. Opin. Food Sci. 2021, 41, 201–209. [Google Scholar] [CrossRef]
- Engelmann, N.J.; Clinton, S.K.; Erdman, J.W., Jr. Nutritional aspects of phytoene and phytofluene, carotenoid precursors to lycopene. Adv. Nutr. 2011, 2, 51–61. [Google Scholar] [CrossRef]
- Wan, H.; Yu, C.; Han, Y.; Guo, X.; Ahmad, S.; Tang, A.; Wang, J.; Cheng, T.; Pan, H.; Zhang, Q. Flavonols and Carotenoids in Yellow Petals of Rose Cultivar (Rosa ‘Sun City’): A Possible Rich Source of Bioactive Compounds. J. Agric. Food Chem. 2018, 66, 4171–4181. [Google Scholar] [CrossRef]
- Lisboa, M.P.; Canal, D.; Filgueiras, J.P.C.; Turchetto-Zolet, A.C. Molecular evolution and diversification of phytoene synthase (PSY) gene family. Genet. Mol. Biol. 2022, 45, e20210411. [Google Scholar] [CrossRef]
- Pandit, J.; Danley, D.E.; Schulte, G.K.; Mazzalupo, S.; Pauly, T.A.; Hayward, C.M.; Hamanaka, E.S.; Thompson, J.F.; Harwood, H.J., Jr. Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis. J. Biol. Chem. 2000, 275, 30610–30617. [Google Scholar] [CrossRef] [PubMed]
- Welsch, R.; Arango, J.; Bär, C.; Salazar, B.; Al-Babili, S.; Beltrán, J.; Chavarriaga, P.; Ceballos, H.; Tohme, J.; Beyer, P. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. Plant Cell 2010, 22, 3348–3356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fernie, A.R. Metabolons, enzyme–enzyme assemblies that mediate substrate channeling, and their roles in plant metabolism. Plant Commun. 2021, 2, 100081. [Google Scholar] [CrossRef]
- Stauder, R.; Welsch, R.; Camagna, M.; Kohlen, W.; Balcke, G.U.; Tissier, A.; Walter, M.H. Strigolactone Levels in Dicot Roots Are Determined by an Ancestral Symbiosis-Regulated Clade of the PHYTOENE SYNTHASE Gene Family. Front. Plant Sci. 2018, 9, 255. [Google Scholar] [CrossRef] [PubMed]
- Sagawa, J.M.; Stanley, L.E.; LaFountain, A.M.; Frank, H.A.; Liu, C.; Yuan, Y.-W. An R2R3-MYB transcription factor regulates carotenoid pigmentation in Mimulus lewisii flowers. New Phytol. 2016, 209, 1049–1057. [Google Scholar] [CrossRef]
- Meng, Y.; Wang, Z.; Wang, Y.; Wang, C.; Zhu, B.; Liu, H.; Ji, W.; Wen, J.; Chu, C.; Tadege, M.; et al. The MYB Activator WHITE PETAL1 Associates with MtTT8 and MtWD40-1 to Regulate Carotenoid-Derived Flower Pigmentation in Medicago truncatula. Plant Cell 2019, 31, 2751–2767. [Google Scholar] [CrossRef]
- Stanley, L.; Yuan, Y.W. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Y.; Wang, W.; Chen, T.; Tian, S.; Qin, G. Ubiquitination of phytoene synthase 1 precursor modulates carotenoid biosynthesis in tomato. Commun. Biol. 2020, 3, 730. [Google Scholar] [CrossRef]
- Zhou, X.; Welsch, R.; Yang, Y.; Álvarez, D.; Riediger, M.; Yuan, H.; Fish, T.; Liu, J.; Thannhauser, T.W.; Li, L. Arabidopsis OR proteins are the major posttranscriptional regulators of phytoene synthase in controlling carotenoid biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 3558–3563. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, P.; Rao, S.; Zhou, X.; Wrightstone, E.; Lu, S.; Yuan, H.; Yang, Y.; Fish, T.; Thannhauser, T.; et al. Co-chaperoning of chlorophyll and carotenoid biosynthesis by ORANGE family proteins in plants. Mol. Plant 2023, 16, 1048–1065. [Google Scholar] [CrossRef]
- Rao, S.; Cao, H.; O’Hanna, F.J.; Zhou, X.; Lui, A.; Wrightstone, E.; Fish, T.; Yang, Y.; Thannhauser, T.; Cheng, L.; et al. Nudix hydrolase 23 post-translationally regulates carotenoid biosynthesis in plants. Plant Cell 2024, 36, 1868–1891. [Google Scholar] [CrossRef]
- Magnard, J.L.; Roccia, A.; Caissard, J.C.; Vergne, P.; Sun, P.; Hecquet, R.; Dubois, A.; Hibrand-Saint, O.L.; Jullien, F.; Nicole, F.; et al. Biosynthesis of monoterpene scent compounds in roses. Science 2015, 349, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Giuliano, G.; Al-Babili, S. Carotenoid biofortification in crop plants: citius, altius, fortius. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158664. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Y.; Cao, Y.; Chen, Y.; Hussain, H.; Lu, X.; Zhu, K.; Xu, Y.; Feng, L.; Wei, G. Dynamic Carotenoid Profiles and Function Analysis of the RrPSY1 Gene in Rosa rugosa Flowers. Horticulturae 2025, 11, 1137. https://doi.org/10.3390/horticulturae11091137
Yu Y, Cao Y, Chen Y, Hussain H, Lu X, Zhu K, Xu Y, Feng L, Wei G. Dynamic Carotenoid Profiles and Function Analysis of the RrPSY1 Gene in Rosa rugosa Flowers. Horticulturae. 2025; 11(9):1137. https://doi.org/10.3390/horticulturae11091137
Chicago/Turabian StyleYu, Yue, Yazheng Cao, Yudie Chen, Hammad Hussain, Xieyu Lu, Kaikai Zhu, Yong Xu, Liguo Feng, and Guo Wei. 2025. "Dynamic Carotenoid Profiles and Function Analysis of the RrPSY1 Gene in Rosa rugosa Flowers" Horticulturae 11, no. 9: 1137. https://doi.org/10.3390/horticulturae11091137
APA StyleYu, Y., Cao, Y., Chen, Y., Hussain, H., Lu, X., Zhu, K., Xu, Y., Feng, L., & Wei, G. (2025). Dynamic Carotenoid Profiles and Function Analysis of the RrPSY1 Gene in Rosa rugosa Flowers. Horticulturae, 11(9), 1137. https://doi.org/10.3390/horticulturae11091137






