Selection and Validation of Reference Candidate Genes for qRT-PCR Analysis in the Developing Fruit of Phyllanthus emblica L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Selection of Reference Candidate Genes
2.3. Total RNA Extraction and cDNA Synthesis
2.4. qRT-PCR Analysis
2.5. Analysis of Gene Stability
2.6. Verification of Stability of Candidate Reference Genes
2.7. Data Processing
3. Results
3.1. Identification of Candidate Reference Genes
3.2. Primer Specificity and Amplification Efficiency
3.3. Expression Stability Test of the Candidate Reference Genes
PCR Stability Test Using Different Statistical Algorithms
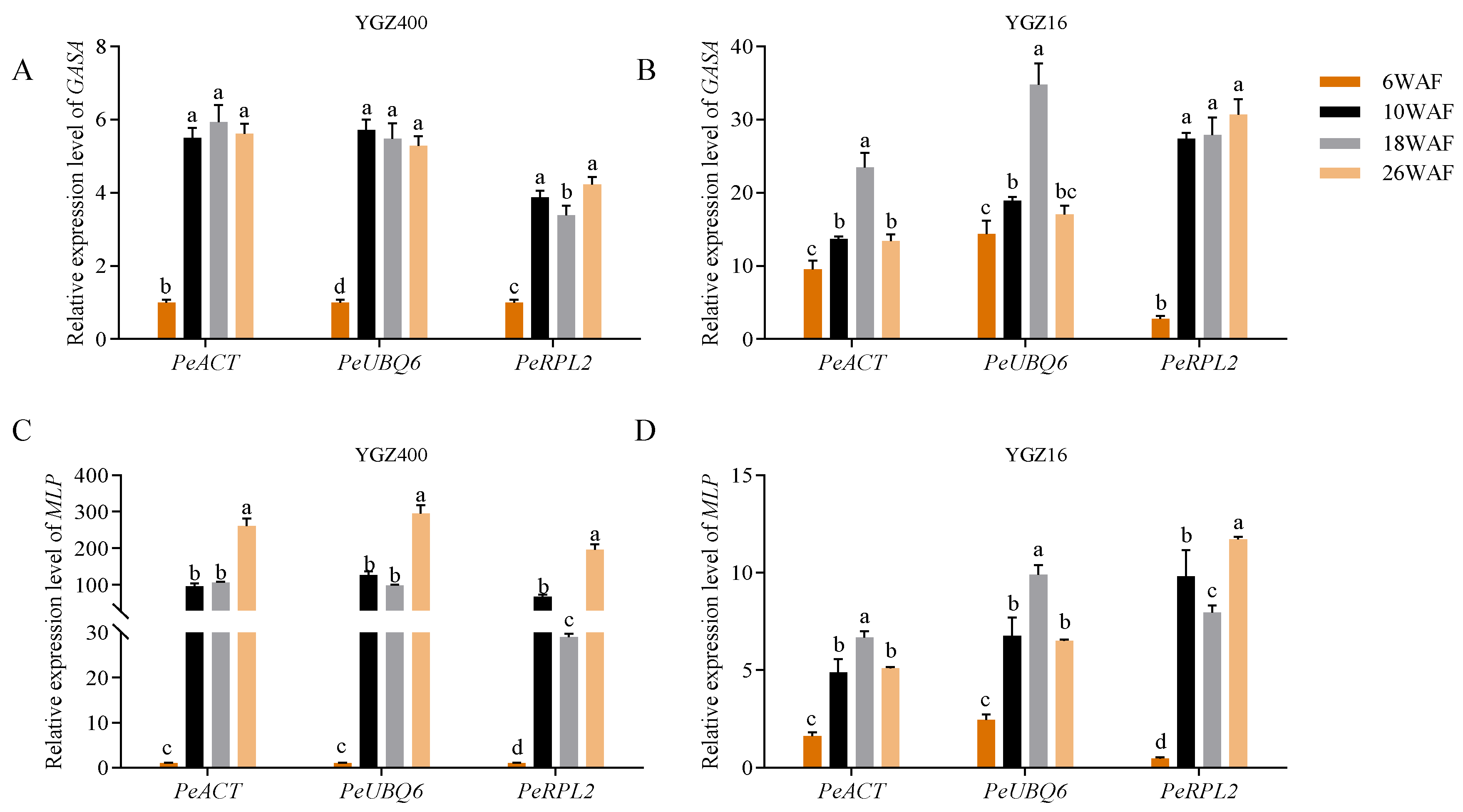
3.4. Validation of Selected Reference Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| qRT-PCR | Quantitative Real-Time Polymerase Chain Reaction |
| GSAS | Gibberellic Acid-Stimulated Arabidopsis |
| MLP | Major Latex-Like Protein |
References
- Hu, M.; Hu, W.; Xia, Z.; Zhou, X.; Wang, W. Validation of reference genes for relative quantitative gene expression studies in Cassava (Manihot esculenta Crantz) by using quantitative real-time PCR. Front. Plant Sci. 2016, 7, 680. [Google Scholar] [CrossRef]
- Huggett, J.; Dheda, K.; Bustin, S.; Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005, 6, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Thellin, O.; Elmoualij, B.; Heinen, E.; Zorzi, W. A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol. Adv. 2009, 27, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.; Hondred, D.; Register, J.C. Keeping qRT-PCR rigorous and biologically relevant. Plant Cell Rep. 2015, 34, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.; Lass, U.; Landt, O.; Nitsche, A.; Laser, J.; Ellerbrok, H.; Pauli, G.; Huhn, D.; Schmidt, C.A. Highly sensitive and specific fluorescence reverse transcription-PCR assay for the pseudogene-free detection of β-Actin transcripts as quantitative reference. Clin. Chem. 1999, 45, 297–300. [Google Scholar] [CrossRef]
- Radonić, A.; Thulke, S.; Mackay, I.M.; Landt, O.; Siegert, W.; Nitsche, A. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004, 313, 856–862. [Google Scholar] [CrossRef]
- Lin, Y.; Lai, Z. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Sci. 2010, 178, 359–365. [Google Scholar] [CrossRef]
- Thellin, O.; Zorzi, W.; Lakaye, B.; De borman, B.; Coumans, B.; Hennen, G.; Grisar, T.; Igout, A.; Heinen, E. Housekeeping genes as internal standards: Use and limits. J. Biotechnol. 1999, 75, 291–295. [Google Scholar] [CrossRef]
- Schmidt, G.W.; Delaney, S.K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Mol. Genet. Genom. 2010, 283, 233–241. [Google Scholar] [CrossRef]
- Park, S.; Kim, Y.; Ji, C.Y.; Park, S.; Jeong, J.C.; Lee, H.; Kwak, S. Stable internal reference genes for the normalization of real-time PCR in different sweetpotato cultivars subjected to abiotic stress conditions. PLoS ONE 2012, 7, e51502. [Google Scholar] [CrossRef]
- Ma, R.; Xu, S.; Zhao, Y.; Xia, B.; Wang, R. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 2016, 7, 536. [Google Scholar] [CrossRef]
- Liu, Y.P.; Zhang, Y.; Liu, F.; Liu, T.; Chen, J.Y.; Fu, G.; Zheng, C.Y.; Su, D.D.; Wang, Y.N.; Zhou, H.K.; et al. Establishment of reference (housekeeping) genes via quantitative real-time PCR for investigation of the genomic basis of abiotic stress resistance in Psammochloa villosa (Poaceae). J. Plant Physiol. 2022, 268, 153575. [Google Scholar] [CrossRef]
- Mao, J.; Li, J.; Wang, Y.; Zhang, Z. Selection and validation of reference genes for qRT-PCR in cultivated octoploid strawberry. Fruit Res. 2024, 4, e010. [Google Scholar] [CrossRef]
- Tang, Q.; Zhou, G.; Liu, S.; Li, W.; Wang, Y.; Xu, G.; Li, T.; Meng, G.; Xue, J. Selection and validation of reference genes for qRT-PCR analysis of gene expression in Tropaeolum majus (Nasturtium). Horticulturae 2023, 9, 1176. [Google Scholar] [CrossRef]
- Suzuki, T.; Higgins, P.J.; Crawford, D.R. Control selection for RNA quantitation. BioTechniques 2000, 29, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Lai, M. Actin, a reliable marker of internal control. Clin. Chim. Acta 2007, 385, 1–5. [Google Scholar] [CrossRef]
- Bustin, S. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. J. Mol. Endocrinol. 2002, 29, 23–39. [Google Scholar] [CrossRef]
- Gutierrez, L.; Mauriat, M.; Pelloux, J.; Bellini, C.; Van Wuytswinkel, O. Towards a systematic validation of references in real-time RT-PCR. Plant Cell 2008, 20, 1734–1735. [Google Scholar] [CrossRef] [PubMed]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1-11. [Google Scholar] [CrossRef] [PubMed]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Wang, Y.; Dai, M.; Cai, D.; Shi, Z. Screening for quantitative real-time PCR reference genes with high stable expression using the mRNA-sequencing data for pear. Tree Genet. Genomes 2019, 15, 54. [Google Scholar] [CrossRef]
- Zhu, L.; Yang, C.; You, Y.; Liang, W.; Wang, N.; Ma, F.; Li, C. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci. Hortic. 2019, 244, 165–171. [Google Scholar] [CrossRef]
- Cheng, Y.; Bian, W.; Pang, X.; Yu, J.; Ahammed, G.J.; Zhou, G.; Wang, R.; Ruan, M.; Li, Z.; Ye, Q.; et al. Genome-wide identification and evaluation of reference genes for quantitative RT-PCR analysis during tomato fruit development. Front. Plant Sci. 2017, 8, 1440. [Google Scholar] [CrossRef]
- Xu, Z.; Dai, J.; Su, W.; Wu, H.; Shah, K.; Xing, L.; Ma, J.; Zhang, D.; Zhao, C. Selection and validation of reliable reference genes for gene expression studies in different genotypes and TRV-infected fruits of peach (Prunus persica L. Batsch) during ripening. Genes 2022, 13, 160. [Google Scholar] [CrossRef] [PubMed]
- Berumen-varela, G.; Palomino-hermosillo, Y.A.; Bautista-rosales, P.U.; Peña-sandoval, G.R.; López-gúzman, G.G.; Balois-morales, R. Identification of reference genes for quantitative real-time PCR in different developmental stages and under refrigeration conditions in soursop fruits (Annona muricata L.). Sci. Hortic. 2020, 260, 108893. [Google Scholar] [CrossRef]
- Gul, M.; Liu, Z.W.; Iahtisham ul haq; Rabail, R.; Faheem, F.; Walayat, N.; Nawaz, A.; Shabbir, M.A.; Munekata, P.E.S.; Lorenzo, J.M.; et al. Functional and nutraceutical significance of amla (Phyllanthus emblica L.): A review. Antioxidants 2022, 11, 816. [Google Scholar] [CrossRef] [PubMed]
- Li, P.H.; Wang, C.W.; Lu, W.C.; Song, T.Y.; Wang, C.C.R. Antioxidant, anti-inflammatory activities, and neuroprotective behaviors of Phyllanthus emblica L. fruit extracts. Agriculture 2022, 12, 588. [Google Scholar] [CrossRef]
- Liu, X.; Ma, Y.; Wan, Y.; Li, Z.; Ma, H. Genetic diversity of Phyllanthus emblica from two different climate type areas. Front. Plant Sci. 2020, 11, 580812. [Google Scholar] [CrossRef]
- Zhang, L.M.; Yuan, J.M.; Pu, T.L.; Qu, W.L.; Lei, X.; Ma, K.H.; Qian, K.J.; Zhao, Q.L.; Liao, C.F.; Jin, J. Chromosome-scale genome assembly of Phyllanthus emblica L. “Yingyu”. DNA Res. 2025, 32, dsaf006. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Czechowski, T.; Stitt, M.; Altmann, T.; Udvardi, M.K.; Scheible, W. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005, 139, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Zhao, Z.; Qian, C.; Sui, Y.; Malik, A.A.; Chen, J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 2010, 399, 257–261. [Google Scholar] [CrossRef]
- Chapman, J.R.; Waldenström, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- Garrido, J.; Aguilar, M.; Prieto, P. Identification and validation of reference genes for RT-qPCR normalization in wheat meiosis. Sci. Rep. 2020, 10, 2726. [Google Scholar] [CrossRef]
- Li, Y.; Yi, H.; Gu, Q.; Zheng, Z.; Zhu, M.; Zha, X.; Zhang, S.; Yang, M. Reference gene selection and gene expression analysis during gall development of Zizania latifolia. Horticulturae 2024, 10, 759. [Google Scholar] [CrossRef]
- Wang, H.; Chen, S.; Jiang, J.; Zhang, F.; Chen, F. Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci. Rep. 2015, 5, 8094. [Google Scholar]
- Guenin, S.; Mauriat, M.; Pelloux, J.; Van Wuytswinkel, O.; Bellini, C.; Gutierrez, L. Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J. Exp. Bot. 2009, 60, 487–493. [Google Scholar] [CrossRef]
- Gentle, A.; Anastasopoulos, F.; Mcbrien, N.A. High-resolution semi-quantitative real-time PCR without the use of a standard curve. BioTechniques 2001, 31, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Maren, N.A.; Kosentka, P.Z.; Liao, Y.; Lu, H.; Duduit, J.R.; Huang, D.; Ashrafi, H.; Zhao, T.; Huerta, A.I.; et al. An optimized protocol for stepwise optimization of real-time RT-PCR analysis. Hortic. Res. 2021, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Wang, H.; Qiu, X.; Yan, H.; Ma, L. Identification and validation of reference genes for qRT-PCR analysis of petal-color-related genes in Rosa praelucens. Genes 2024, 15, 277. [Google Scholar] [CrossRef]
- Lin, Y.; Liu, G.; Rao, Y.; Wang, B.; Tian, R.; Tan, Y.; Peng, T. Identification and validation of reference genes for qRT-PCR analyses under different experimental conditions in Allium wallichii. J. Plant Physiol. 2023, 281, 153925. [Google Scholar] [CrossRef]
- Shivhare, R.; Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 2016, 6, 23036. [Google Scholar] [CrossRef] [PubMed]
- Svitkina, T. The Actin cytoskeleton and Actin-based motility. Cold Spring Harb. Perspect. Biol. 2018, 10, a018267. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Liu, H.; Chong, K.; Xu, Y. Roles of ubiquitination-mediated protein degradation in plant responses to abiotic stresses. Environ. Exp. Bot. 2015, 114, 92–103. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Liu, Q.; Tong, H.; Zhang, T.; Gu, C.; Liu, L.; Huang, S.; Yuan, H. Selection and validation of appropriate reference genes for qRT-PCR analysis of flowering stages and different genotypes of Iris germanica L. Sci. Rep. 2021, 11, 9901. [Google Scholar]
- Zhou, Y.; Zhang, Y.; Mu, D.; Lu, Y.; Chen, W.; Zhang, Y.; Zhang, R.; Qin, Y.; Yuan, J.; Pan, L.; et al. Selection of reference genes in Evodia rutaecarpa var. officinalis and expression patterns of genes involved in its limonin biosynthesis. Plants 2023, 12, 3197. [Google Scholar]
- Zhang, X.; Cui, H.; Ji, X.; Xue, J.; Jia, X.; Li, R. Selection of the optimal reference genes for transcript expression analysis of lipid biosynthesis-related genes in Okra (Abelmoschus esculentus). Sci. Hortic. 2021, 282, 110044. [Google Scholar] [CrossRef]
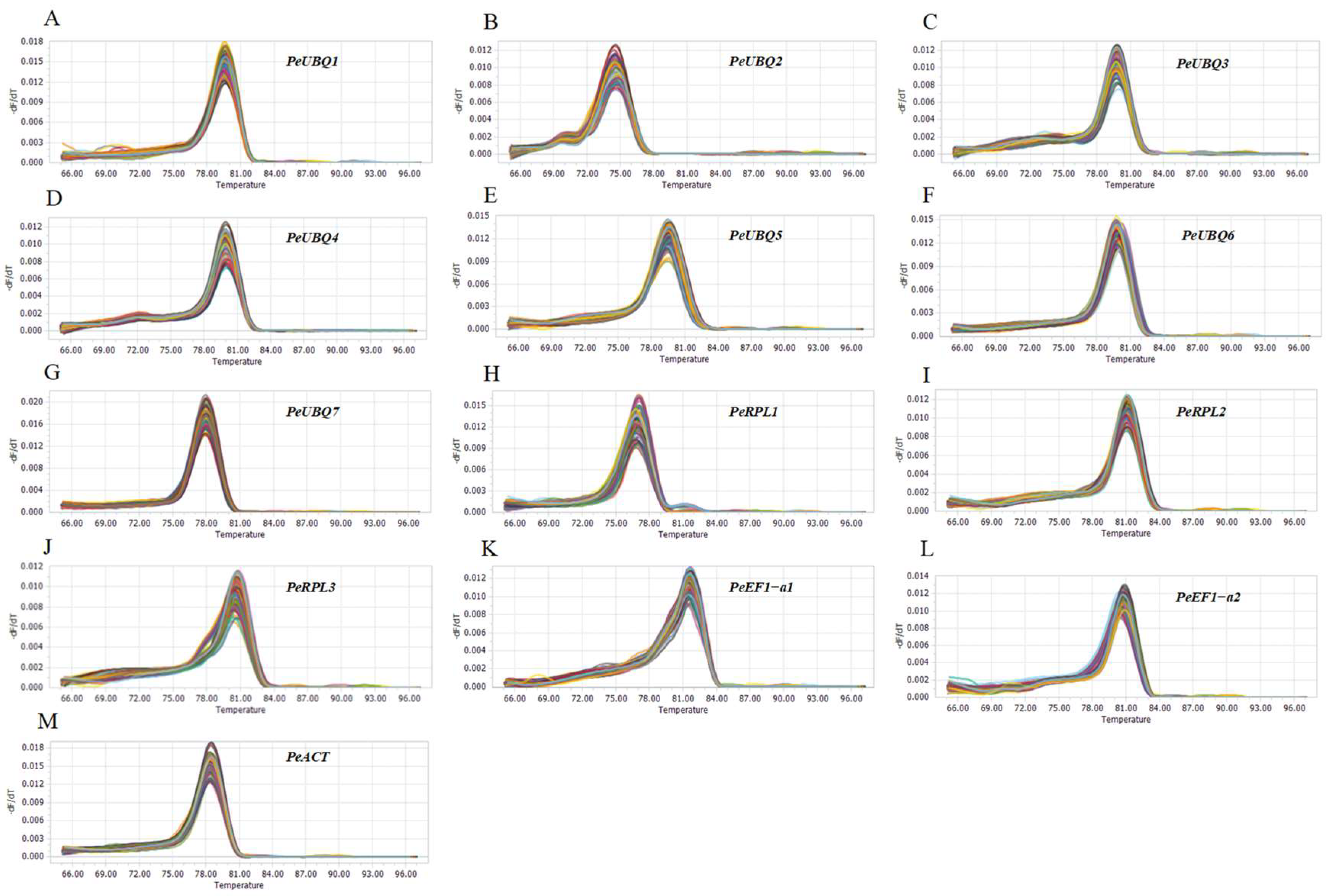
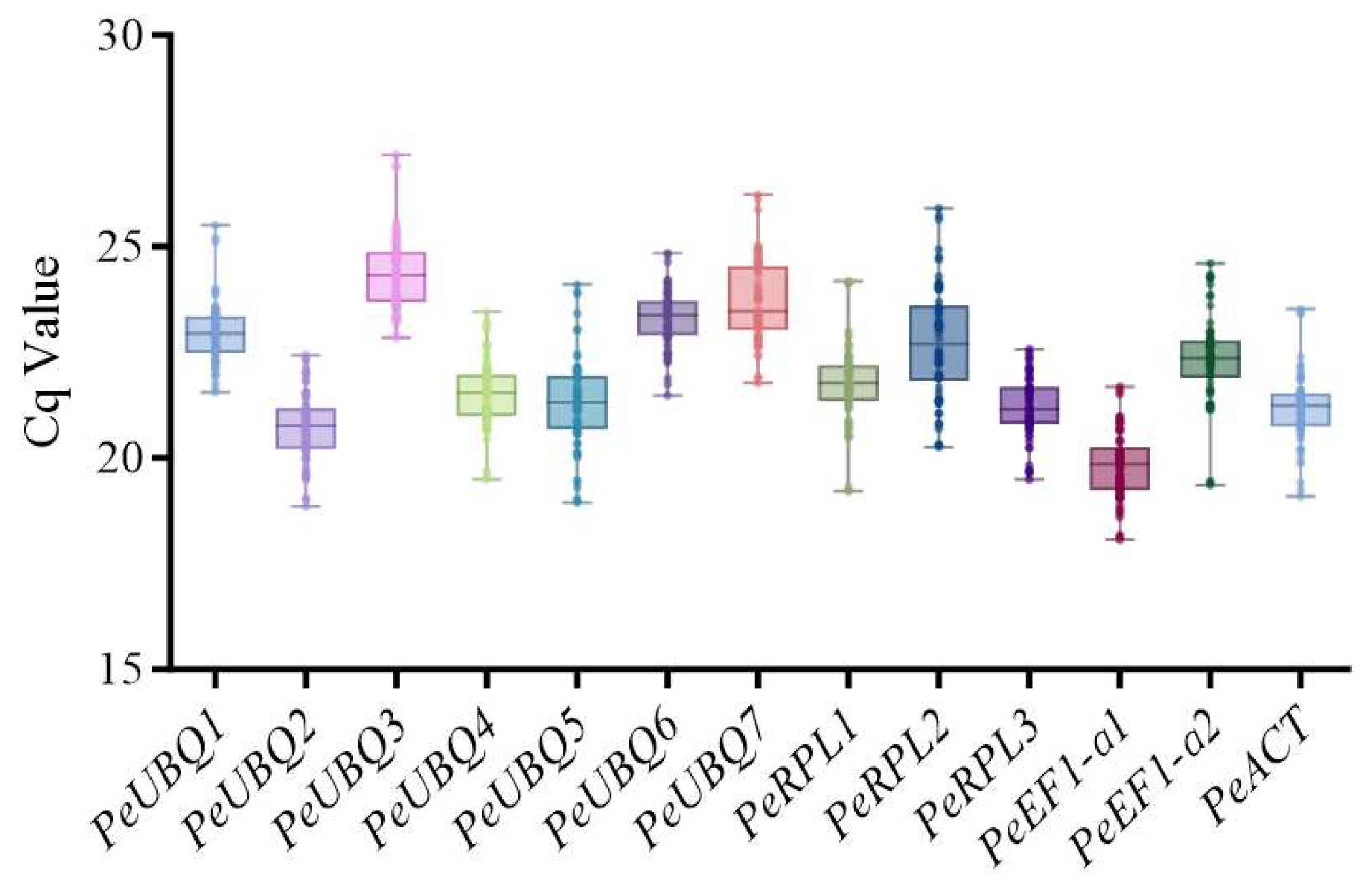
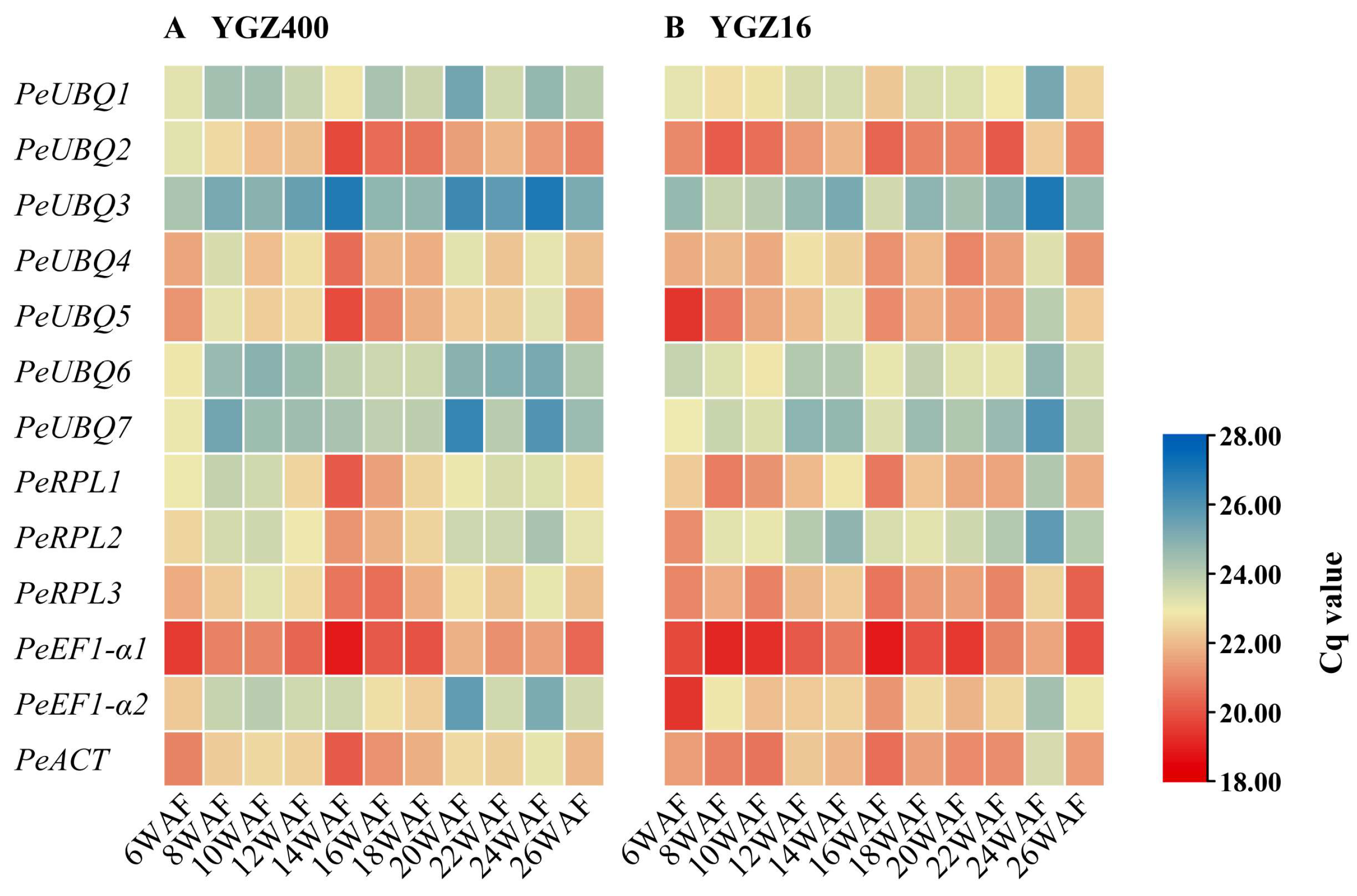
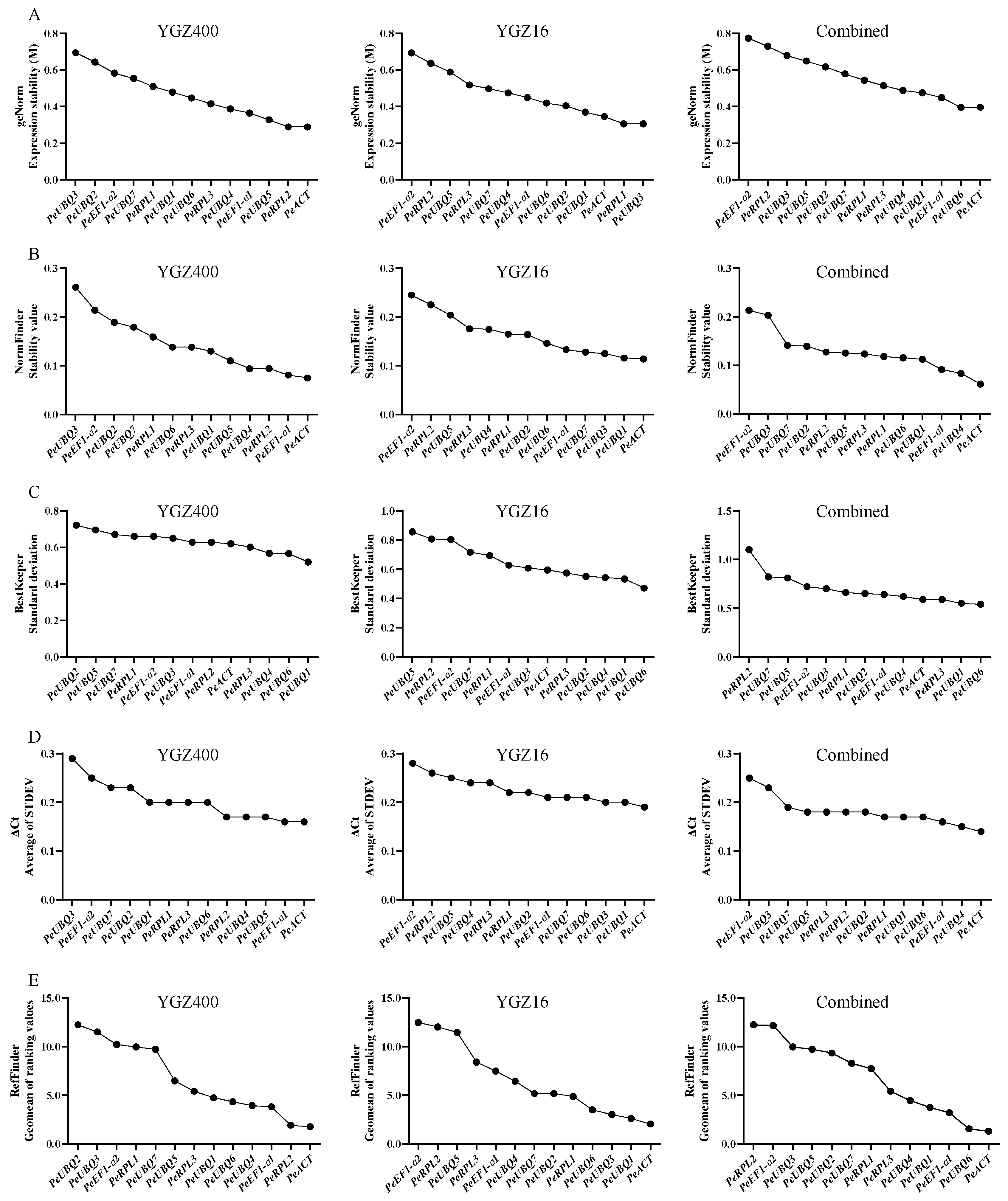

| Gene Name | Gene ID | Arabidopsis Homolog | CDS Length (bp) | Mean TPM | CV |
|---|---|---|---|---|---|
| PeUBQ1 (Non-canonical ubiquitin conjugating enzyme 1) | PeChr11G010870.1 | At3g17000 | 909 | 53.16 | 0.29 |
| PeUBQ2 (Ubiquitin-conjugating enzyme E2) | PeChr09G004080.1 | At3g52560 | 441 | 51.44 | 0.24 |
| PeUBQ3 (E3 ubiquitin ligase) | PeChr22G009860.1 | At1g65040 | 966 | 42.95 | 0.29 |
| PeUBQ4 (Ubiquitin-protein ligases) | PeSTRG.11463.1.p1 | At2g02760 | 459 | 73.41 | 0.29 |
| PeUBQ5 (Ubiquitin-like protein 5) | PeChr02G004390.1 | At5g42300 | 222 | 114.92 | 0.28 |
| PeUBQ6 (Ubiquitin and ubiquitin-like proteins) | PeChr26G003160.2 | At1g31340 | 465 | 36.21 | 0.25 |
| PeUBQ7 (Ubiquitin-like protein) | PeSTRG.21462.1.p1 | At2g17200 | 1701 | 43.27 | 0.20 |
| PeRPL1 (Ribosomal protein RPL1) | PeChr26G007230.1 | At3g09630 | 1227 | 135.42 | 0.29 |
| PeRPL2 (Ribosomal protein L2) | PeSTRG.17299.1.p1 | At4g36130 | 783 | 160.57 | 0.29 |
| PeRPL3 (Ribosomal protein L10) | PeChr02G001910.3 | At1g26910 | 798 | 192.73 | 0.30 |
| PeEF1-α1 (Translation elongation factor 1 alpha) | PeSTRG.12624.1.p1 | At5g60390 | 1350 | 148.27 | 0.24 |
| PeEF1-α2 (Translation elongation factor 1 alpha) | PeSTRG.1197.1.p1 | At5g60390 | 1350 | 217.27 | 0.23 |
| PeACT (Actin depolymerizing factor) | PeSTRG.26181.1.p1 | At3g46010 | 420 | 141.44 | 0.26 |
| Gene Name | Amplification Length/bp | R2 | Efficiency/% |
|---|---|---|---|
| PeUBQ1 | 128 | 0.9991 | 90.27% |
| PeUBQ2 | 80 | 0.9983 | 100.07% |
| PeUBQ3 | 88 | 0.9994 | 102.00% |
| PeUBQ4 | 105 | 0.9966 | 97.74% |
| PeUBQ5 | 89 | 0.9988 | 101.64% |
| PeUBQ6 | 131 | 0.9983 | 109.54% |
| PeUBQ7 | 114 | 0.9992 | 97.67% |
| PeRPL1 | 96 | 0.9997 | 97.17% |
| PeRPL2 | 86 | 0.9997 | 95.24% |
| PeRPL3 | 123 | 0.9974 | 104.69% |
| PeEF1-α1 | 123 | 0.9999 | 101.14% |
| PeEF1-α2 | 140 | 0.9962 | 102.15% |
| PeACT | 106 | 0.9994 | 97.28% |
| Gene Name | YGZ400 | YGZ16 | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | CV/% | Mean | SD | CV/% | Mean | SD | CV/% | |
| PeUBQ1 | 22.72 | 0.64 | 2.81 | 23.20 | 0.78 | 3.37 | 22.96 | 0.75 | 3.27 |
| PeUBQ2 | 20.49 | 0.87 | 4.26 | 20.97 | 0.69 | 3.30 | 20.73 | 0.82 | 3.95 |
| PeUBQ3 | 24.08 | 0.78 | 3.25 | 24.69 | 0.90 | 3.67 | 24.38 | 0.89 | 3.67 |
| PeUBQ4 | 21.15 | 0.73 | 3.46 | 21.90 | 0.69 | 3.14 | 21.53 | 0.80 | 3.72 |
| PeUBQ5 | 20.89 | 0.87 | 4.14 | 21.73 | 1.17 | 5.38 | 21.31 | 1.10 | 5.18 |
| PeUBQ6 | 22.99 | 0.66 | 2.87 | 23.61 | 0.57 | 2.41 | 23.30 | 0.69 | 2.95 |
| PeUBQ7 | 23.25 | 0.88 | 3.78 | 24.19 | 0.87 | 3.60 | 23.72 | 0.99 | 4.17 |
| PeRPL1 | 21.53 | 0.91 | 4.24 | 21.94 | 0.94 | 4.28 | 21.74 | 0.94 | 4.33 |
| PeRPL2 | 21.81 | 0.77 | 3.54 | 23.68 | 1.13 | 4.76 | 22.74 | 1.34 | 5.91 |
| PeRPL3 | 21.00 | 0.77 | 3.65 | 21.39 | 0.68 | 3.17 | 21.19 | 0.75 | 3.52 |
| PeEF1-α1 | 19.60 | 0.77 | 3.92 | 20.00 | 0.80 | 3.99 | 19.80 | 0.80 | 4.05 |
| PeEF1-α2 | 22.40 | 0.91 | 4.06 | 22.27 | 1.20 | 5.36 | 22.34 | 1.06 | 4.72 |
| PeACT | 20.89 | 0.77 | 3.67 | 21.49 | 0.81 | 3.76 | 21.19 | 0.84 | 3.95 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, T.; Yuan, J.; Qu, W.; Liao, C.; Lei, X.; Qian, K.; Zhao, Q.; Shi, L.; Zhang, L.; Liu, A. Selection and Validation of Reference Candidate Genes for qRT-PCR Analysis in the Developing Fruit of Phyllanthus emblica L. Horticulturae 2025, 11, 1054. https://doi.org/10.3390/horticulturae11091054
Pu T, Yuan J, Qu W, Liao C, Lei X, Qian K, Zhao Q, Shi L, Zhang L, Liu A. Selection and Validation of Reference Candidate Genes for qRT-PCR Analysis in the Developing Fruit of Phyllanthus emblica L. Horticulturae. 2025; 11(9):1054. https://doi.org/10.3390/horticulturae11091054
Chicago/Turabian StylePu, Tianlei, Jianmin Yuan, Wenlin Qu, Chengfei Liao, Xiao Lei, Kunjian Qian, Qiongling Zhao, Liangjia Shi, Lumin Zhang, and Aizhong Liu. 2025. "Selection and Validation of Reference Candidate Genes for qRT-PCR Analysis in the Developing Fruit of Phyllanthus emblica L." Horticulturae 11, no. 9: 1054. https://doi.org/10.3390/horticulturae11091054
APA StylePu, T., Yuan, J., Qu, W., Liao, C., Lei, X., Qian, K., Zhao, Q., Shi, L., Zhang, L., & Liu, A. (2025). Selection and Validation of Reference Candidate Genes for qRT-PCR Analysis in the Developing Fruit of Phyllanthus emblica L. Horticulturae, 11(9), 1054. https://doi.org/10.3390/horticulturae11091054






