Abstract
Timely identification of crop conditions is relevant for informed decision-making in precision agriculture. The initial step in determining the conditions that crops require involves isolating the components that constitute them, including the leaves and fruits of the plants. An alternative method for conducting this separation is to utilize intelligent digital image processing, wherein plant elements are labeled for subsequent analysis. The application of Deep Learning algorithms offers an alternative approach for conducting segmentation tasks on images obtained from complex environments with intricate patterns that pose challenges for separation. One such application is semantic segmentation, which involves assigning a label to each pixel in the processed image. This task is accomplished through training various models of Convolutional Neural Networks. This paper presents a comparative analysis of semantic segmentation performance using a convolutional neural network model with different backbone architectures. The task focuses on pixel-wise classification into three categories: leaves, fruits, and background, based on images of semi-hydroponic tomato crops captured in greenhouse settings. The main contribution lies in identifying the most efficient backbone-UNet combination for segmenting tomato plant leaves and fruits under uncontrolled conditions of lighting and background during image acquisition. The Convolutional Neural Network model UNet is is implemented with different backbones to use transfer learning to take advantage of the knowledge acquired by other models such as MobileNet, VanillaNet, MVanillaNet, ResNet, VGGNet trained with the ImageNet dataset, in order to segment the leaves and fruits of tomato plants. Highest percentage performance across five metrics for tomato plant fruit and leaves segmentation is the MVanillaNet-UNet and VGGNet-UNet combination with and respectively. A comparison of the best results of semantic segmentation versus those obtained with a color-dominant segmentation method optimized with a greedy algorithm is presented.
1. Introduction
Agriculture is a fundamental task for human subsistence, since a large part of the consumed food is produced by this economic activity [1,2]. In field work, the farmer’s experience has been fundamental in the irrigation and fertilization processes, in the search for diseases and pests, which can lead to inefficient processes and therefore, low productivity. During the XXI century, the agricultural industry has experienced great technological development in search of improving performance in terms of food production and cost reduction, since it is estimated that by 2050 the world population will be about 10 billion according to the forecast of the United Nations, which poses a great challenge for agriculture [3].
In the last three decades, the use of Artificial Intelligence (AI) and other areas of technology have created an area of knowledge called Precision Agriculture (PA). One of its main objectives is to provide crops with the necessary resources for their adequate development, improving the cost-benefit ratio [4,5,6].
In order to achieve the objectives of PA, farmers utilize various technological advancements, among which is Deep Learning (DL), this involves employing different algorithms to train models of Convolutional Neural Networks (CNN) [7]. These have been applied in different fields, for example in medicine for the classification of various cell types [8,9,10], face recognition [11,12], identification of objects in different environments [13], in agricultural tasks, in pest and disease detection [14,15,16], leaces identification [17,18] and estimation of nutrients present in plant leaves [19], to name a few.
This article presents the results of the semantic segmentation process done with the CNN model UNet [20] using the knowledge of the models MobileNet [21], MVanillaNet, VanillaNet [22], ResNet [23] and VGGNet [24] in what is known as Transfer Learning (TL) [25], with aims of classifying the pixels of the images in the classes of leaves, fruits and background. This work contributes to measure the effectiveness of taking advantage of the knowledge previously acquired by the models used as backbone extracted from large amounts of data and using specialized hardware that are not available. The UNet model is used with different backbones to segment leaves and fruits of tomato crops, which provides the opportunity to test this type of models with more than one class and with images of crops in real production environments.
The organization of the paper is as follows: Section 2 presents a series of works related to segmentation applied to agriculture. Section 3 describes the previous work done with the dataset images, presents the models used and details how they were trained. The results obtained by the trained models are described and discussed in Section 4. A comparison of results between the most successful CNN and a greedy algorithm is reported in Section 5 and Section 6 contains the conclusions of the research.
2. Related Works
In agricultural research, numerous studies employ Computer Vision (CV) techniques for segmentation tasks. For instance, Xu [26] introduced a method to extract color and texture characteristics from tomato plant leaves, employing histograms and Fourier transforms. Castillo-Martínez [27] presented a color index-based thresholding approach for background and foreground segmentation in plant images, leveraging two color indexes related to the green color of plants. Lin [28] proposed a detection algorithm based on color, depth, and shape information for detecting spherical or cylindrical fruits on plants. Fan [29] employed color information, image features in a segmentation method based on gray-centered red-green-blue color space to label apple pixels. Image segmentation methods using Computer Vision (CV) techniques achieve competitive results comparable to those of DL approaches.
CNN are advanced in DL algorithms designed for image processing. They extract spatial features through convolutional layers, enabling sophisticated pattern recognition and in-depth image analysis [30]. In recent years, various models have emerged to address diverse needs. They share a common trait: a growing number of trainable parameters and the adoption of different activation functions. This versatility enables their application across various fields and contexts, marking significant progress in machine learning. In 1998, LeNet-5 [31] introduced convolution kernels as activation functions and layer grouping, boasting 60 thousand parameters. In 2012, AlexNet [32] emerged with 60 million parameters, pioneering the use of ReLU and Dropout. The introduction of VGG-16 [24] in 2014 featured 138 million parameters, while Inception-v1 [33] marked the first implementation of layer blocks in a CNN.
In PA, DL is harnessed to address various challenges across various crop types. Zheng et al. [34] curated a dataset comprising more than 31,000 images from diverse capture devices, encompassing 31 crop classes for classification and detection tasks. Mukti [35] applied TL to ResNet50-based CNN models for pest detection, offering comparative insights into various CNN architectures. Additionally, Mohanty [16] employed AlexNet and GoogleNet for disease detection, exploring different training configurations. CNN UNet has been used in the healthcare field for the segmentation of different elements of interest. Weng [8] used UNet in combination with a NAS algorithm for the segmentation of elements in magnetic resonance imaging, computed tomography and others. Guan [36] proposed a modified CNN architecture called Fully Dense UNet for removing artifacts from 2D PAT images reconstructed from sparse data using images of type photoacoustic tomography.
CNNs have been used for semantic segmentation of different types of crops. Ni [37] worked with blueberry crops to determine the degree of maturity and the amount of fruits, reporting an effectiveness greater than in accuracy using a Mask RCNN model. Majeed [38] implemented the models FCN, VGG-16 and SegNet-VGG-16 to segment the stems of grape plants in order to have a control parameter on their growth. Kang [39] performed real-time segmentation of the fruit and branches of apple trees, using a pyramid-type model for segmentation. Jin [40] utilized a model VNCC to separate the leaves and stalks of corn plants. Addressing more complex segmentation challenges, Liu [41] tackled cucumber segmentation within greenhouses, where color similarity between leaves and fruits complicates the process, utilizing a Mask RCNN model. The UNet model has been tested mainly in the medical field, however, it has been tested in different contexts, for example in landslides [42], Roy [43] proposed UNet and an Enhanced UNet adaptation for detecting rotten fruit parts via semantic segmentation, in agricultural fields in disease [44] detection tasks and exact identification of crop rows [45].
The segmentation of elements that compose tomato plants has been approached with different algorithms, from CV segmentation algorithms to the adaptation of CNN models. Arakeri [46] developed a sorting system that involves hardware and software to sort tomatoes according to their ripeness, the hardware aims at capturing images and the software employs segmentation algorithms based on thresholds determined by CV algorithms. Dhakshina [47] presented a work to classify tomatoes in 3 stages using CV algorithms, in the first stage he classifies the elements that are tomatoes from those that are not, in the second stage he classifies them as ripe and unripe and in the last stage he detects defects in the tomato fruits. Guerra [48] used the similarity of the RGB model and the dominance of the green and red color present in tomato crops to develop a method for leaves and fruit segmentation. Wan [49] proposed a procedure to assess the maturity of fresh supermarket tomatoes at three levels, developing a threshold segmentation algorithm based on the RGB color model.
The use of DL algorithms to work with tomato plants addresses different problems such as disease detection, pests identification and segmentation. Tran [50] utilized Inception-ResNet v2 and Autoencoder models to predict nutrient deficiencies in tomato crops. Ponce [19] proposed a CNN model called CNN+AHN which can be configured with different parameters to estimate nutritional deficiencies in tomato plants. Muhammad [51] used the CNN EfficientNet model and the UNet to perform segmentation of tomato leaf images in controlled environments, with varying illumination. The models were also applied to classify the leaves as healthy or unhealthy. Deng [52] proposes a modified U-Net model for tomato leaves disease detection, called the Cross Layer Attention Fusion Mechanism combined with a Multiscale Convolution Module (MC-UNet). This approach uses images of leaves that were cut from the plants and placed on a high-contrast background, achieving segmentation efficiencies above . Similarly Badiger [53] addresses tomato leaves disease classification using a combination of different convolutional neural network (CNN) models at various stages. His approach includes the use of U-Net trained with an optimization technique known as Gradient-Golden Search.
In DL, the standard procedure involves training and test data sharing the same input feature map and distribution. However, generating well-matched training, Validation, and test datasets is resource-intensive in both time and cost. To address this, TL leverages knowledge acquired in one domain for application in another, reducing the need for extensive data collection and training [25]. Wulandhari [30] implemented TL with the ResNet model to detect the deficit of macronutrients such as nitrogen, phosphorus and potassium in okra plants.
The papers related to the segmentation of tomato crop elements have in common that they are implemented and tested in laboratory environments with controlled lighting conditions, controlled shooting angles, and a background that provides high contrast [50,51,52,53]. Most of the models used in the literature are implemented with the objective of segmenting a single class, either leaves or fruits. The results presented in this research are determined by combining several backbones with the UNet model to segment three classes in images of real tomato crop environments.
3. Materials and Methods
Training CNN models requires a labeled image dataset for semantic segmentation, where each class is manually annotated. Additionally, specialized hardware, such as GPUs, is recommended to ensure efficient processing and optimal performance.
3.1. Computer Characteristics
The CNN model experiments were conducted on a computer configured with the following hardware and software components:
- Manufacturer: ASUSTeK COMPUTER INC., Taipei, China
- Model: X510UNR
- Processor: Intel® Core™ i7-8550U CPU @ 1.80 GHz × 8.
- RAM: 16 GB.
- Video card: NVIDIA® GeForce® 150MX.
3.2. Software Characteristics
The following software resources were used to develop and run the UNet CNN model:
- Operating system: Ubuntu 22.04.2 LTS 64 bits.
- Cuda tool kit: 10.1.243.
- Cudnn: 7.6.5.
- Tensorflow: 2.4.1.
- Keras: 2.4.3.
- Opencv: 3.4.2.
It is worth noting that knowledge acquired through training available online was used to implement TL.
3.3. Dataset
The CNN UNet model with different backbones were trained and tested using a dataset of digital images of tomato plants cultivated under semi-hydroponic greenhouse conditions. This dataset is publicly available for download at the following link: https://www.kaggle.com/datasets/andrewmvd/tomato-detection (accessed on 10 July 2024). The dataset has a total of 895 images, an example of which is shown in Figure 1a. The site hosting the dataset does not provide information on factors such as lighting conditions, capture device, or type of lighting. However, it is evident from the images that they were taken in various environments, at different times of day, and under natural sunlight.
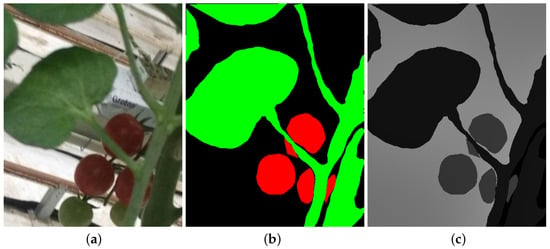
Figure 1.
Dataset images. (a) Example of an image of the dataset. (b) Mask with the tomato leaves and fruit. (c) Contrasted image for training.
From the original set of images, 340 were randomly selected, which corresponds to of the total images that make up the dataset. The selected images were randomly separated into three different subsets: 180 for training, 60 for Validation, and 100 for testing.
3.3.1. Labeling
Labeling objects of interest in an image is a prerequisite for training and effectively using CNN models. In this work, the selected images were labeled using the Computer Vision Annotation Tool (CVAT), which is available at the following link https://www.cvat.ai/ (accessed on 6 January 2025). For the segmentation of tomato plant leaves and fruits, it is necessary to define two distinct labels in the CVAT annotation task—one for each object of interest—since any region not manually labeled is automatically considered background. In this case, the Draw new polygon tool was used to annotate each object, assigning it to the corresponding label. The number of points used in each polygon was determined based on the complexity of the object being labeled.
It is important to mention that the labeling process must be performed on all images used in the experimental section; this requires a large amount of time to perform. Figure 2 shows the CVAT graphical interface in the labeling process of one of the images to be processed.
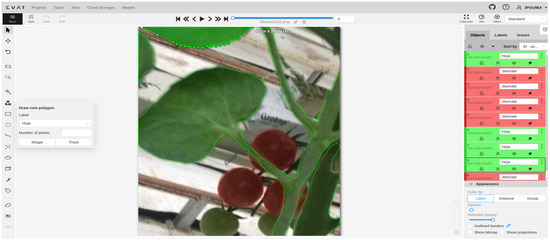
Figure 2.
CVAT labeling interface for the draw new polygon tool.
Figure 1b is the labeling done on Figure 1a, in which the pixels that form the leaves are marked in green, those that form the fruits are in red and the background of the image in black.
Another relevant aspect emphasizing the significance of labeling tomato plant leaves and fruits is the utilization of resultant masks to evaluate segmentation model efficiency using diverse performance metrics. From the 340 selected images, a total of 68 million pixels were labeled across the three classes: as background, as leaves, and as fruit (see Figure 3).
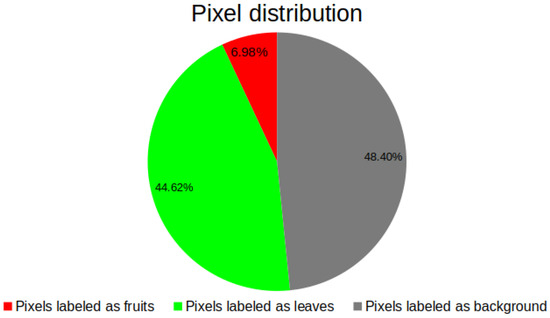
Figure 3.
Pixel distribution in leaves, fruits and background classes.
The dataset exhibits significant class imbalance, a crucial factor in metric selection for performance evaluation. Although model performance could theoretically improve with expanded training data, this makes it necessary to have several performance metrics to determine which combination of backbone with the UNet has the best performance.
3.3.2. Data Preprocessing
The images is in the Training, Validation, and Test sets, along with their corresponding masks, have a resolution of pixels. To facilitate the training process, image dimensions are needed to be adjusted according to the input requirements of the CNN models. For the UNet model, dimensions of pixels are necessary.
In addition to image labeling, it is necessary to generate a grayscale mask by assigning a specific value (ranging from 0 to 255) to each class. This enables the model to learn and later classify the pixels of the test images during the training and evaluation phases. For the case study presented here there are three classes: leaves, fruits and background. Figure 1c shows the result of labeling the image in Figure 1b.
In the case of preprocessing of the selected images, it consists of two stages: the first of them consists of making a size adjustment using the resize function of the Opencv library, specifying only the size of , this process is performed on the original image Figure 1a and its mask Figure 1b. The second stage consists of generating the image in Figure 1c, for this a pixel by pixel scan is performed of the reduction of the image in Figure 1b and assigning a value of 0 to each black pixel, a value of 1 to the green ones and a value of 2 to the red ones.
3.4. Metrics Performance
It is essential to measure the performance of the semantic segmentation performed by the CNN models. In this work, the following metrics were used: Accuracy, Precision, Recall, F1-Score [54] and IoU [55]. To calculate these metrics, four fundamental concepts must be identified: True Positives (TP), True Negatives (TN), False Positives (FP) and False Negatives (FN).
The Accuracy metric means the total number of correctly classified elements (pixels) relative to the total number of classifications made by the model, as defined by the nest equation:
Precision, another metric, indicates the total number of true positives ones in relation to the number of predicted positives ones, defined by the equation:
The Recall metric reflects the number of true positives classified by the model relative to the total number of positive classifications, defined as:
F1-Score combines Precision and Recall, defined by:
Lastly, IoU measures the ratio of true positives to the total number of correctly classified items, defined by
3.5. Network Architecture Model
The training process when TL is used aims to preserve the knowledge acquired by the model used as a backbone to identify different types of elements, it is important to mention that most of these models are trained with data and hardware that are complicated to obtain and manage. The last layer of the models is trained to classify the data according to the interest of the project, in this case leaves and fruits of tomato crops using the CNN UNet model.
3.5.1. UNet
In 2015, the UNet model was developed to perform segmentation tasks using convolutional layers, primarily for medical images [20]. The UNet comprises two stages: the first one is responsible for encoding the information within the image to comprehend the contextual understanding, achieved through consecutive layers of convolution followed by max-pooling operations. The second stage aims to symmetrically expand the encoded information from the first stage to accurately localize pixels through a series of transposed convolutions. Refer to Figure 4 for the UNet architecture.
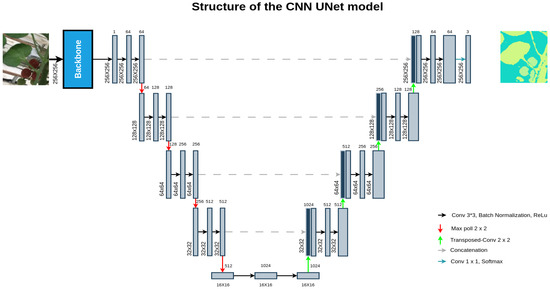
Figure 4.
Structure of the CNN UNet with transfer learning.
3.5.2. Transfer Learning
This work aims to evaluate the efficiency of the UNet model in segmenting tomato plant leaves and fruits. To achieve this, knowledge transfer was tested using five CNN models: VGG, ResNet, MobileNet, Vanilla, and MiniVanilla. These backbones are downloaded from the official Keras website accessible from https://keras.io/api/applications/ (accessed on 24 January 2025).
3.6. Training Module
Most of the studies referenced in Section 2, which work with tomato plants, only perform leaves segmentation in images with a controlled background, which facilitates training and improves the results of the tested CNN models.
The objective of the training process is to allow the CNN model UNet to learn and subsequently label the pixels of the images in the dataset in uncontrolled conditions, with regard to lighting and image capture angle, similar to a farmer’s workday, in our case, this means assigning each of them to one of three classes: leaves, fruit or background.
During the training process two sets of images were used for training, the training set with 180 items and the Validation sets with 60 items, each with their respective images with their corresponding labeling. The parameters with which the UNet model was trained are as follows:
- encoder_freeze is set to True.
- Optimizer: Adam.
- Learning rate: 0.01
- Loss: Categorical_crossemtropy
- Batch size: 16
- Epoch: 75.
4. Results and Discussion
The aim of this research is to assess the effectiveness of the UNet with different backbones for implemented transfer learning models in segmenting leaves and fruits of tomato plants. With the configuration data outlined in Section 3.6, the UNet model is trained and tested.
The training duration for the UNet model was approximately 5 h approximately in each combination of the CNN UNet and backbone model, during which it achieved metric Accuracies higher than with four of the five backbones implemented in training, this as showed in the Figure 5.
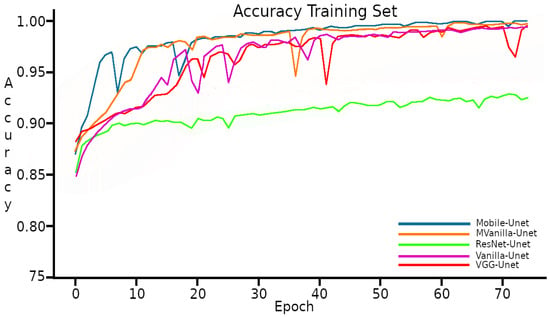
Figure 5.
Accuracy with the Training set.
The Figure 6 shows the performance of accuracy during training with the Validation Set, the five implemented models reached of accuracy.
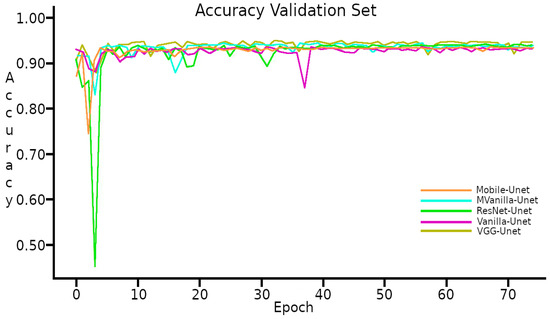
Figure 6.
Accuracy with the Validation set.
The learning curves in Figure 5 and Figure 6 exhibit a typical behavior observed in training processes, with the model demonstrating a better performance on the Training set compared to the Validation set. It is worth noting that the results are closely aligned with each other.
The Loss function resulting from the learning process used in the CNNS UNet model with the different backbones tested is less than in four out of the five combinations implemented; its behavior is shown graphically in Figure 7.
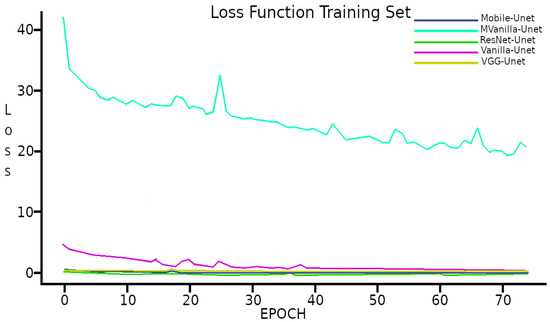
Figure 7.
Loss function with Training set.
In the case of the Validation Set, the Loss Function shows a behavior with several peaks and varied final value. See Figure 8.
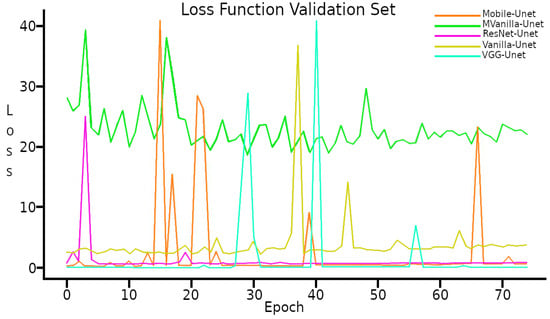
Figure 8.
Loss function with Validation set.
The behavior of the Loss function with the Validation set shows peaks during the 75 training epochs, which may be due to the low number of elements in the Validation set or errors in the labeling of the images.
Figure 9 displays three randomly selected images from the Test set to show examples the segmentation results of leaves and fruits across the UNet and backbone combinations.

Figure 9.
Randomly selected images from Test set to illustrate segmentation result.
The trained UNet model processed and segmented 100 images, forming the Test set. Figure 10 shows the results of semantic segmentation performed by the UNet with different backbones by separating the pixels that make up the leaves of tomato plants from five randomly selected images.

Figure 10.
Semantic segmentation of leaves and fruits pixels in Figure 9. (a) Mask of leaves. (b) MobileNet-UNet. (c) MVanillaNet-UNet. (d) ResNet-UNet. (e) VanillaNet-UNet. (f) VGGNet-UNet.
The leaves and fruit classes in the training and validation sets of the tomato plants are unbalanced, with a disproportionate number of samples between classes of leaves and fruits, in addition to the background class, what is described at the end of this Section 3.3.1. The F1-Score metric is widely used to determine the effectiveness of an AI model in which the classes are unbalanced; it has the particularity of combining the Accuracy and Recall metrics. Finally, the IoU metric is the one that gives us an idea of which generates results most similar to true labeling of each image.
In the examples of semantic segmentation of leaves and fruits in the images in Figure 10, the results delivered by the combination of UNet with the different backbone tested are shown, being qualitatively difficult to detect significant differences with the naked eye. Therefore, the selection of the best combination will be carried out using performance metrics.
A quantitative method for evaluating the performance of semantic segmentation is essential. Metrics such as Accuracy, Precision, Recall, F1-Score, and IoU are used to assess the effectiveness of the selected CNN models in segmenting tomato plant leaves and fruits. Table 1 shows the quantitative results of the segmentation of the image leaves in Figure 10 with UNet model with different backbone.

Table 1.
Efficiency of the implemented models in segmenting pixels of leaves in images Test set.
Figure 11 presents the performance metric graphs for leaves segmentation, revealing a similar performance across the different UNet combinations. The highest average performance is observed with the VGGNet-UNet combination.
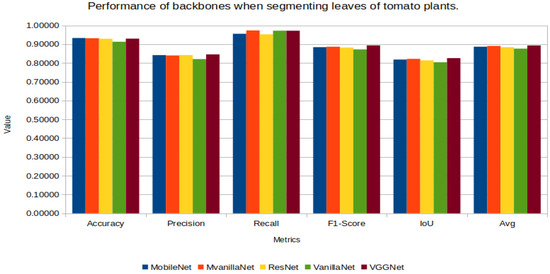
Figure 11.
UNet’s performance with the different backbones in leaves segmentation.
Figure 12 shows consistent performance across all five metrics. The Accuracy metric shows minimal variability, although the other metrics require interpretation due to class imbalance. The Recall metric shows a median close to 0.95, revealing a high recall rate. The F1-Score demonstrated a balance between precision and completeness. However, the Precision metric showed greater dispersion, leading to false positives in some images. The IoU metric suggests a satistfactory level of overlap.
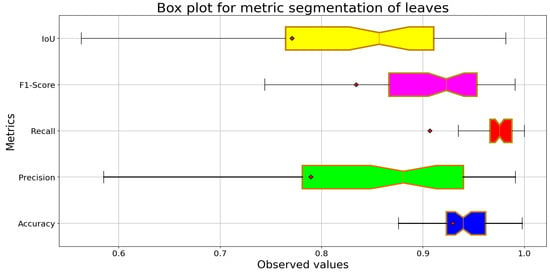
Figure 12.
Box plot for leaves segmentation with VGGNet-UNet.
The quantitative results of the segmentation of the pixels that form the tomato leaves of the images of the Test set are listed in Table 2.

Table 2.
Efficiency of the implemented models in segmenting pixels of fruits in images Test set.
Figure 13 shows the graphs of the results of the five performance metrics when segmenting the pixels of fruits, showing a very similar yield with different combinations, with the highest average being the combination MVanillaNet-UNet.
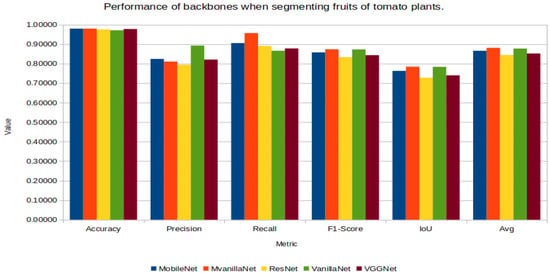
Figure 13.
UNet’s performance with the different backbones in fruit segmentation.
Figure 14 shows that the model achieves positive overall performance, with high values for Accuracy and Recall, reflecting an adequate ability to detect the fruits. The Precision metric shows greater variability, indicating that the model incurs false positives in some cases. The IoU metric presents an acceptable median but with significant dispersion, reflecting challenges in correctly delimiting the fruits’ spatial distribution.
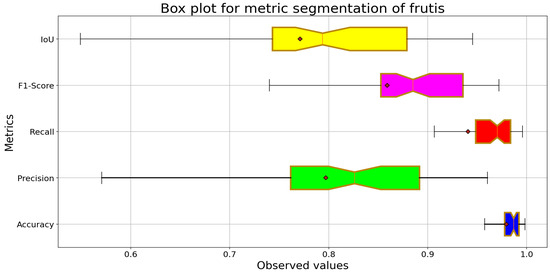
Figure 14.
Box plot for leaves segmentation with MvanillaNet-UNet.
As the data in Table 1 and Table 2 show, all the combinations tested have similar performances, but there are some combinations that perform better in leaves segmentation than in fruit segmentation. The efficiency of the models depends on factors such as depth, architecture, spatial context management, class balance and Loss function.
The results presented here demonstrate both statistical validity and practical promise.
5. Comparative of Results with a Greedy-Type Optimization Algorithm
In [56] Guerra presents the results of a Greedy Algorithm (GA) for the optimization of leaves and fruit segmentation of tomato plants based on the dominance of green color for leaves and red for fruits. The method consists of sequential search of an value that maximizes the dominance of the natural colors present in the leaves and fruits for the correct classification of the pixels in one of the two classes. The implementation of the GA is shown in the Algorithm 1.
| Algorithm 1 Greedy Algorithm for the optimization of color dominance segmentation. |
| function GA() |
| while do |
| if then |
| end if |
| end while |
| return |
| end function |
The GA receives three parameters: the first is IM, representing the set of images to be segmented; the second is i, which denotes the initial value of the parameter used to maximize the results of color dominance-based segmentation [48]; and the third is d, the increment applied to in each iteration of the algorithm. The reported data indicate that an initial value of , with an increment of per iteration, yields optimal results in the GA. It is important to note that the algorithm must be executed separately for leaves segmentation and for fruit segmentation.
Table 3 and Table 4 show the comparison between the results obtained by the best combination of UNet and those reported by GA.

Table 3.
Comparison of VGGNet-UNet vs. GA results for leaves segmentation.

Table 4.
Comparison of MVanillaNet-UNet vs. GA results for fruits segmentation.
The results of the segmentation of the leaves, the GA obtains better results in the performance metrics used, surpassing the VGGNet-UNet by in the final average.
In the case of fruit segmentation the MVanillaNet-UNet combination significantly outperforms the reported GA by in the final average.
Figure 15 contains the graphs of the five metrics and their average leaves and fruit segmentation of the tomato plants of the compared methods.
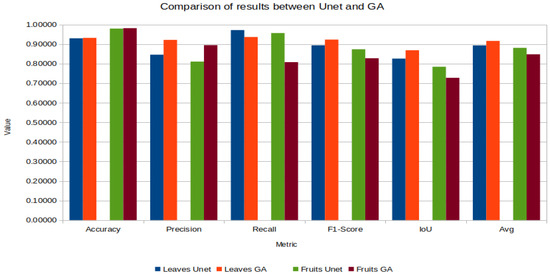
Figure 15.
Graphical comparison of results between UNet and GA.
6. Conclusions
The application of models in agriculture is advancing efficiency and productivity, directly supporting the needs of an increasingly demanding society. In this study, the UNet model, integrated with various backbone architectures, was trained and tested for semantic segmentation of tomato plant leaves and fruits cultivated under semi-hydroponic greenhouse conditions. As mentioned in the abstract, the most significant contribution is the fact of testing the segmentation of tomato plant leaves in uncontrolled environments, which increases the complexity of the segmentation. Another aspect that differentiates this work from others is the fact that the same model is used to segment the fruits of tomato plants in comparison with the papers mentioned in Section 2.
In terms of training time, the UNet model with different backbone required approximately 5 hours to complete the training process. Performance was evaluated using Accuracy, Precision, Recall, F1-Score and IoU metrics on the 100 images of the Test set. The VGGNet-UNet model outperformed than the other four combinations in the average of the five metrics employed in the task of segmenting the leaves of the tomato plants, the MVanillaNet-UNet combination delivers results similar to the best. The MVanillaNet-UNet combination gives the best results in the classification of the pixels that form the fruits of the tomato plant, the combination that is closest in terms of the average of the five metrics is VanillaNet-UNet. In the case of having to select only one model for the segmentation of leaves and fruits of tomato plants, the chosen combination will be the MVanillaNet-UNet, since the average of both segmentation is superior to the other four.
An opportunity to improve the performance of the models in labeling the pixels corresponding to leaves and fruits of tomato crops—based on the evaluation metrics used—is to increase the number of images in the Training and Validation sets. This can be achieved either by acquiring more images or by applying data augmentation techniques to create variations of the original images through rotations, noise addition, and cropping. Furthermore, dataset quality could be enhanced by refining the manual annotation process used to create masks for the training, validation, and test sets. The current class imbalance negatively impacts UNet and backbone architecture performance, as predictions are biased toward dominant classes (leaves and background). This issue can be mitigated through oversampling of fruit-dense images or by acquiring additional images where fruits are more prevalent. This can be achieved through cross-checking by multiple annotators, supported by guidance from agricultural experts. Another way to enhance the model’s performance is by tuning its hyperparameters. For example, reducing the learning rate, increasing the number of training epochs, or experimenting with alternative loss functions such as Sparse Categorical Crossentropy, Dice Loss, or Focal Loss could lead to better results. The use of early stopping mechanisms helps prevent overfitting by continuously monitoring the model’s performance on the Validation set. When a decline in performance is detected, the training process is halted to preserve the model’s generalization capability.
In the comparison with the GA, an improvement in performance was observed when segmenting the leaves, bringing the results closer to those reported in the paper. In the case of fruit segmentation, the MVanilla-UNet produced better results than the GA, demonstrating the potential of CNNs for such tasks in PA. Testing additional models or adjusting training parameters can further enhance segmentation performance. The key advantage of knowledge transfer in deep learning over algorithms like GA is its ability to generalize learned information during training, enabling adaptation to similar tasks.
A vast amount of knowledge is available on the Internet through various CNN models that have been trained on extremely large and diverse datasets, using computational resources that exceed the capabilities of most researchers. This highlights the relevance of testing such acquired knowledge in diverse contexts, including agricultural applications. Compared to other methods reported in the literature, where leaves are isolated from the plant and their natural environment, this approach applies transfer learning (TL) under uncontrolled conditions, making it more representative of the actual conditions faced by farmers. This increases the potential for the technology to be effectively integrated into real-world agricultural practices and food production systems.
The application of various DL methods offers an opportunity to elevate agriculture to the next level, promoting sustainability by reducing environmental impact without compromising the food productivity needed to support a growing global population. The results of this work can be applied to the detection of pests, diseases or unwanted conditions depending on the type of each crop, allowing correct decisions to be made at the right time.
Future Work
The next phase of this research will focus on increasing the size of the image dataset employed for training, a task that requires a significant time investment for image annotation. To support this goal, our strategy includes enhancing the accuracy of existing labels and generating additional annotations. Using these new labels, we will evaluate the most effective UNet-backbone combinations for classifying fruit maturity levels, aiming to enable harvest estimation and the early detection of unfavorable crop conditions. Furthermore, we intend to investigate modifications to UNet and other semantic segmentation architectures and assess their performance across various crop types. The successful execution of these objectives will necessitate an expansion of our present computing resources. In another line of research we will work with lighter models that can be implemented in mobile devices like Fast-SCNN and BiSeNet among others.
Author Contributions
Conceptualization, J.P.G.I. and F.J.C.d.l.R.; methodology, J.P.G.I. and F.J.C.d.l.R.; software, J.P.G.I. and J.R.H.V.; validation, J.P.G.I. and F.J.C.d.l.R.; formal analysis, J.P.G.I. and F.J.C.d.l.R.; investigation, J.P.G.I. writing—original draft preparation, J.P.G.I. and F.J.C.d.l.R.; writing—review and editing, J.P.G.I. and F.J.C.d.l.R.; supervision, F.J.C.d.l.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Acknowledgments
It is important to thank the Institutions and companies that made the development of this work possible by allocating resources of different kinds to carry it out. Secretaria de Ciencias, Humanidades, Tecnología e Innovación (SECIHTI), Centro de Investigaciones en Óptica A.C. (CIO) and Instituto Tecnológico de Estudios Superiores de Zamora (ITESZ).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Awasthi, Y. Press “A” for Artificial Intelligence in Agriculture: A Review. Int. J. Inform. Vis. 2020, 4, 112–116. [Google Scholar] [CrossRef]
- Ray, P.P. Internet of things for smart agriculture: Technologies, practices and future direction. J. Ambient. Intell. Smart Environ. 2017, 9, 395–420. [Google Scholar] [CrossRef]
- FAO. Our Approach|Food Systems|Food and Agriculture Organization of the United Nations. 2023. Available online: http://www.fao.org/food-systems/our-approach/en/ (accessed on 5 January 2025).
- Gebbers, R.; Adamchuk, V.I. Precision agriculture and food security. Science 2010, 327, 828–831. [Google Scholar] [CrossRef] [PubMed]
- Pierce, F.J.; Nowak, P. Aspects of precision agriculture. Adv. Agron. 1999, 67, 1–68. [Google Scholar]
- Patrício, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef]
- Weng, Y.; Zhou, T.; Li, Y.; Qiu, X.; Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; et al. NAS-Unet: Neural architecture search for medical image segmentation. IEEE Access 2019, 7, 21420–21428. [Google Scholar] [CrossRef]
- Huang, H.; Lin, L.; Tong, R.; Hu, H.; Zhang, Q.; Iwamoto, Y.; Han, X.; Chen, Y.W.; Wu, J. UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. In Proceedings of the ICASSP, IEEE International Conference on Acoustics, Speech and Signal Processing—Proceedings, Virtual, 4–9 May 2020; IEEE: New York, NY, USA, 2020; Volume 2020, pp. 1055–1059. [Google Scholar] [CrossRef]
- Esteva, A.; Chou, K.; Yeung, S.; Naik, N.; Madani, A.; Mottaghi, A.; Liu, Y.; Topol, E.; Dean, J.; Socher, R. Deep learning-enabled medical computer vision. NPJ Digit. Med. 2021, 4, 1–9. [Google Scholar] [CrossRef]
- Lu, Z.; Jiang, X.; Kot, A. Deep Coupled ResNet for Low-Resolution Face Recognition. IEEE Signal Process. Lett. 2018, 25, 526–530. [Google Scholar] [CrossRef]
- Mandal, B.; Okeukwu, A.; Theis, Y. Masked Face Recognition using ResNet-50. arXiv 2021, arXiv:2104.08997. [Google Scholar]
- Guo, Y.; Liu, Y.; Georgiou, T.; Lew, M.S. A review of semantic segmentation using deep neural networks. Int. J. Multimed. Inf. Retr. 2018, 7, 87–93. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.; Park, D. A Robust Deep-Learning-Based Detector for Real-Time Tomato Plant Diseases and Pests Recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef]
- Sladojevic, S.; Arsenovic, M.; Anderla, A.; Culibrk, D.; Stefanovic, D. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Comput. Intell. Neurosci. 2016, 2016, 3289801. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, S.P.; Hughes, D.; Salathe, M. Using Deep Learning for Image-Based Plant Disease Detection. Front. Plant Sci. 2016, 7, 215232. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Misra, A.K. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Hall, D.; McCool, C.; Dayoub, F.; Sünderhauf, N.; Upcroft, B. Evaluation of features for leaf classification in challenging conditions. In Proceedings of the 2015 IEEE Winter Conference on Applications of Computer Vision, WACV 2015, Waikoloa, HI, USA, 5–9 January 2015; pp. 797–804. [Google Scholar] [CrossRef]
- Ponce, H.; Cevallos, C.; Espinosa, R.; Gutierrez, S. Estimation of Low Nutrients in Tomato Crops Through the Analysis of Leaf Images Using Machine Learning. J. Artif. Intell. Technol. 2021, 1, 131–137. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. UNet: Convolutional Networks for Biomedical Image Segmentation; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Chen, H.; Wang, Y.; Guo, J.; Tao, D. VanillaNet: The Power of Minimalism in Deep Learning. Adv. Neural Inf. Process. Syst. 2023, 36, 7050–7064. [Google Scholar]
- McNeely-White, D.; Beveridge, J.R.; Draper, B.A. Inception and ResNet features are (almost) equivalent. Cogn. Syst. Res. 2020, 59, 312–318. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Xu, G.; Zhang, F.; Ghafoor Shah, S.; Ye, Y.; Mao, H.; Shah, S.G.; Ye, Y.; Mao, H. Use of leaf color images to identify nitrogen and potassium deficient tomatoes. Pattern Recognit. Lett. 2011, 32, 1584–1590. [Google Scholar] [CrossRef]
- Castillo-Martínez, M.; Gallegos-Funes, F.J.; Carvajal-Gámez, B.E.; Urriolagoitia-Sosa, G.; Rosales-Silva, A.J. Color index based thresholding method for background and foreground segmentation of plant images. Comput. Electron. Agric. 2020, 178, 105783. [Google Scholar] [CrossRef]
- Lin, G.; Tang, Y.; Zou, X.; Xiong, J.; Fang, Y. Color-, depth-, and shape-based 3D fruit detection. Precis. Agric. 2020, 21, 1–17. [Google Scholar] [CrossRef]
- Fan, P.; Lang, G.; Yan, B.; Lei, X.; Guo, P.; Liu, Z.; Yang, F. A Method of Segmenting Apples Based on Gray-Centered RGB Color Space. Remote Sens. 2021, 13, 1211. [Google Scholar] [CrossRef]
- Wulandhari, L.A.; Gunawan, A.A.S.; Qurania, A.; Harsani, P.; Triastinurmiatiningsih; Tarawan, F.; Hermawan, R.F. Plant nutrient deficiency detection using deep convolutional neural network. ICIC Express Lett. 2019, 13, 971–977. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2323. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; Volume 2016, pp. 2818–2826. [Google Scholar] [CrossRef]
- Zheng, Y.Y.; Kong, J.L.; Jin, X.B.; Wang, X.Y.; Su, T.L.; Zuo, M. Cropdeep: The crop vision dataset for deep-learning-based classification and detection in precision agriculture. Sensors 2019, 19, 1058. [Google Scholar] [CrossRef]
- Mukti, I.Z.; Biswas, D. Transfer Learning Based Plant Diseases Detection Using ResNet50. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology, EICT 2019, Khulna, Bangladesh, 20–22 December 2019; Institute of Electrical and Electronics Engineers Inc.: New York, NY, USA, 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Guan, S.; Khan, A.A.; Sikdar, S.; Chitnis, P.V. Fully Dense UNet for 2-D Sparse Photoacoustic Tomography Artifact Removal. IEEE J. Biomed. Health Inform. 2020, 24, 568–576. [Google Scholar] [CrossRef]
- Ni, X.; Li, C.; Jiang, H.; Takeda, F. Deep learning image segmentation and extraction of blueberry fruit traits associated with harvestability and yield. Hortic. Res. 2020, 7, 110. [Google Scholar] [CrossRef]
- Majeed, Y.; Karkee, M.; Zhang, Q.; Fu, L.; Whiting, M.D. Determining grapevine cordon shape for automated green shoot thinning using semantic segmentation-based deep learning networks. Comput. Electron. Agric. 2020, 171, 105308. [Google Scholar] [CrossRef]
- Kang, H.; Chen, C. Fruit detection and segmentation for apple harvesting using visual sensor in orchards. Sensors 2019, 19, 4599. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Ma, Q.; Xu, K.; Hu, T.; Liu, J.; Pang, S.; Guan, H.; et al. Separating the Structural Components of Maize for Field Phenotyping Using Terrestrial LiDAR Data and Deep Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2020, 58, 2644–2658. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, D.; Jia, W.; Ji, W.; Ruan, C.; Sun, Y. Cucumber fruits detection in greenhouses based on instance segmentation. IEEE Access 2019, 7, 139635–139642. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, T.; Hu, K.; Zhang, Y.; Yu, X.; Li, Y. A Deep Learning Semantic Segmentation Method for Landslide Scene Based on Transformer Architecture. Sustainability 2022, 14, 16311. [Google Scholar] [CrossRef]
- Roy, K.; Chaudhuri, S.S.; Pramanik, S. Deep learning based real-time Industrial framework for rotten and fresh fruit detection using semantic segmentation. Microsyst. Technol. 2021, 27, 3365–3375. [Google Scholar] [CrossRef]
- Wang, H.; Ding, J.; He, S.; Feng, C.; Zhang, C.; Fan, G.; Wu, Y.; Zhang, Y. MFBP-UNet: A Network for Pear Leaf Disease Segmentation in Natural Agricultural Environments. Plants 2023, 12, 3209. [Google Scholar] [CrossRef]
- Guo, Z.; Geng, Y.; Wang, C.; Xue, Y.; Sun, D.; Lou, Z.; Chen, T.; Geng, T.; Quan, L. InstaCropNet: An efficient Unet-Based architecture for precise crop row detection in agricultural applications. Artif. Intell. Agric. 2024, 12, 85–96. [Google Scholar] [CrossRef]
- Arakeri, M.; Lakshmana, B. Computer Vision Based Fruit Grading System for Quality Evaluation of Tomato in Agriculture industry. Procedia Comput. Sci. 2016, 79, 426–433. [Google Scholar] [CrossRef]
- Dhakshina Kumar, S.; Esakkirajan, S.; Bama, S.; Keerthiveena, B. A microcontroller based machine vision approach for tomato grading and sorting using SVM classifier. Microprocess. Microsyst. 2020, 76, 103090. [Google Scholar] [CrossRef]
- Guerra Ibarra, J.P.; Cuevas, F.J. Segmentation of Leaves and Fruits of Tomato Plants by Color Dominance. AgriEngineering 2023, 5, 1846–1864. [Google Scholar] [CrossRef]
- Wan, P.; Toudeshki, A.; Tan, H.; Ehsani, R. A methodology for fresh tomato maturity detection using computer vision. Comput. Electron. Agric. 2018, 146, 43–50. [Google Scholar] [CrossRef]
- Tran, T.T.; Choi, J.W.; Le, T.T.H.; Kim, J.W. A comparative study of deep CNN in forecasting and classifying the macronutrient deficiencies on development of tomato plant. Appl. Sci. 2019, 9, 1601. [Google Scholar] [CrossRef]
- Chowdhury, M.E.H.; Rahman, T.; Khandakar, A.; Ayari, M.A.; Khan, A.U.; Khan, M.S.; Al-Emadi, N.; Reaz, M.B.I.; Islam, M.T.; Ali, S.H.M. Automatic and Reliable Leaf Disease Detection. AgriEngineering 2021, 3, 294–312. [Google Scholar] [CrossRef]
- Deng, Y.; Xi, H.; Zhou, G.; Chen, A.; Wang, Y.; Li, L.; Hu, Y. An Effective Image-Based Tomato Leaf Disease Segmentation Method Using MC-UNet. Plant Phenomics 2023, 5, 49. [Google Scholar] [CrossRef]
- Badiger, M.; Mathew, J.A. Tomato plant leaf disease segmentation and multiclass disease detection using hybrid optimization enabled deep learning. J. Biotechnol. 2023, 374, 101–113. [Google Scholar] [CrossRef]
- Davis, J.; Goadrich, M. The relationship between Precision-Recall and ROC curves. In Proceedings of the 23rd International Conference on Machine Learning, Pittsburgh, PA, USA, 25–29 June 2006; pp. 233–240. [Google Scholar]
- Yu, J.; Xu, J.; Chen, Y.; Li, W.; Wang, Q.; Yoo, B.; Han, J.J. Learning Generalized Intersection Over Union for Dense Pixelwise Prediction. In Proceedings of Machine Learning Research, Proceedings of the 38th International Conference on Machine Learning, Virtual, 18–24 July 2021; Meila, M., Zhang, T., Eds.; PMLR: New York, NY, USA, 2021; Volume 139, pp. 12198–12207. [Google Scholar]
- Guerra Ibarra, J.P.; Cuevas de la Rosa, F. Optimization of Color Dominance Factor by Greedy Algorithm for Leaves and Fruit Segmentation of Tomato Plants. In Pattern Recognition; Mezura-Montes, E., Acosta-Mesa, H.G., Carrasco-Ochoa, J.A., Martínez-Trinidad, J.F., Olvera-López, J.A., Eds.; Springer: Cham, Switzerland, 2024; pp. 200–209. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).