Evolutionary Forces Shaping Trans-Species Polymorphisms in Genus Cucumis
Abstract
1. Introduction
2. Materials and Methods
2.1. Species Sampling
2.2. Variant Calling
2.3. Population Genetic Analysis
2.4. Identification and Statistic of Trans-Species Polymorphisms
2.5. Demographic Analysis
2.6. Introgression Analysis
2.7. Identification of Trans-Species Polymorphisms Under Long-Term Balancing Selection
3. Results
3.1. Trans-Species Polymorphisms Between Cucumber and Melon
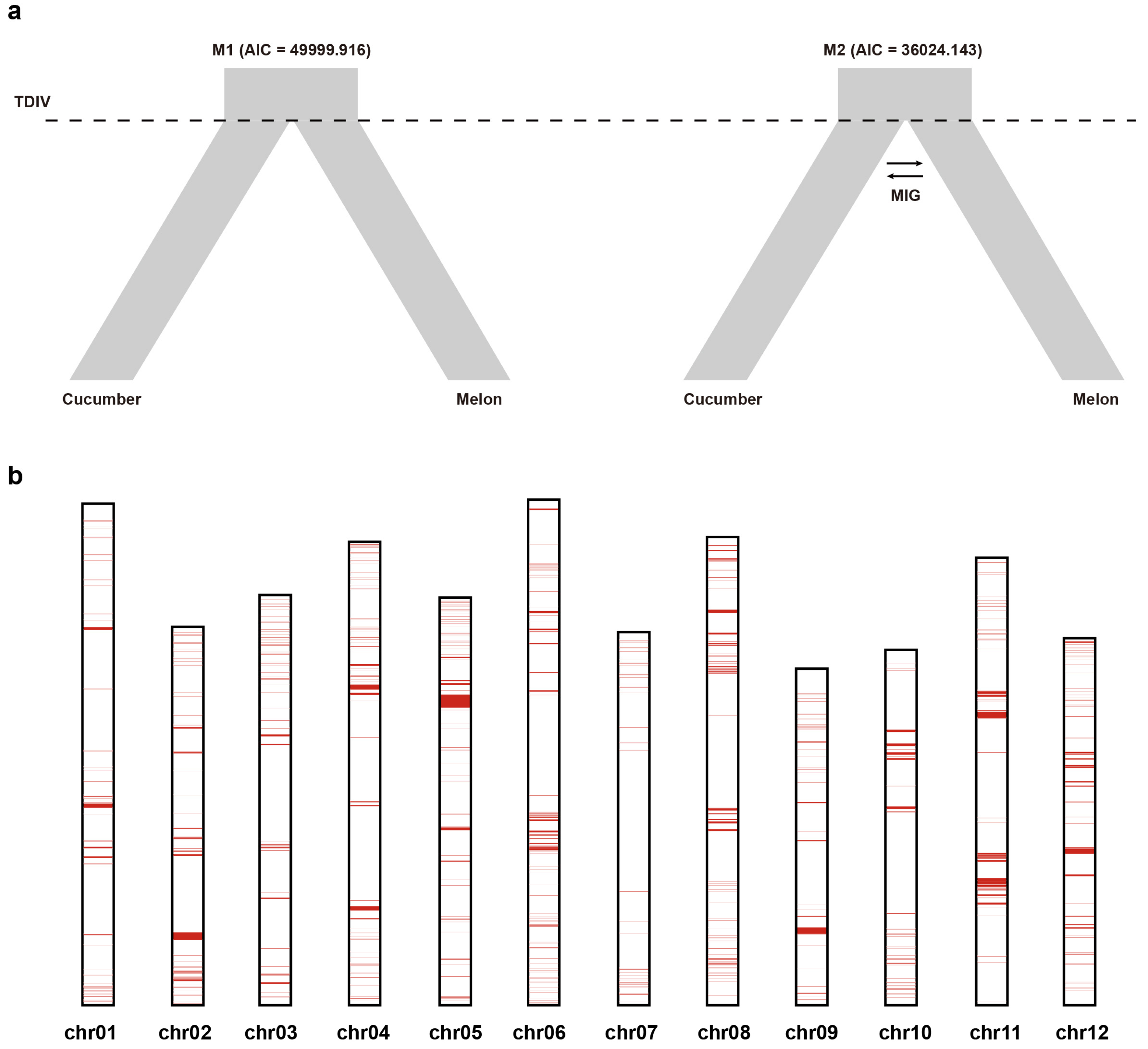
3.2. Demographic History of the Two Species
3.3. Trans-Species Polymorphisms Under Balancing Selection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cornetti, L.; Fields, P.D.; Du Pasquier, L.; Ebert, D. Long-term balancing selection for pathogen resistance maintains trans-species polymorphisms in a planktonic crustacean. Nat. Commun. 2024, 15, 5333. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.S.; Karram, M.; Bass, D.J.; Doceti, M.; Becker, D.; Nunez, J.C.; Ratan, A.; Bergland, A.O. Trans-Specific Polymorphisms Between Cryptic Daphnia Species Affect Fitness and Behavior. Mol. Ecol. 2025, 34, e17632. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, L.; Zhang, Z.; Li, M.; Wang, D.; Zhang, X.; Xi, Z.; Keefover-Ring, K.; Smart, L.B.; DiFazio, S.P. Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 2020, 225, 1370–1382. [Google Scholar] [CrossRef] [PubMed]
- Roux, C.; Pauwels, M.; Ruggiero, M.-V.; Charlesworth, D.; Castric, V.; Vekemans, X. Recent and ancient signature of balancing selection around the S-locus in Arabidopsis halleri and A. lyrata. Mol. Biol. Evol. 2012, 30, 435–447. [Google Scholar] [CrossRef]
- Ségurel, L.; Thompson, E.E.; Flutre, T.; Lovstad, J.; Venkat, A.; Margulis, S.W.; Moyse, J.; Ross, S.; Gamble, K.; Sella, G. The ABO blood group is a trans-species polymorphism in primates. Proc. Natl. Acad. Sci. USA 2012, 109, 18493–18498. [Google Scholar] [CrossRef]
- Tellini, N.; De Chiara, M.; Mozzachiodi, S.; Tattini, L.; Vischioni, C.; Naumova, E.S.; Warringer, J.; Bergström, A.; Liti, G. Ancient and recent origins of shared polymorphisms in yeast. Nat. Ecol. Evol. 2024, 8, 761–776. [Google Scholar] [CrossRef]
- Bitarello, B.D.; Brandt, D.Y.; Meyer, D.; Andrés, A.M. Inferring balancing selection from genome-scale data. Genome Biol. Evol. 2023, 15, evad032. [Google Scholar] [CrossRef]
- Aqil, A.; Speidel, L.; Pavlidis, P.; Gokcumen, O. Balancing selection on genomic deletion polymorphisms in humans. Elife 2023, 12, e79111. [Google Scholar] [CrossRef]
- Fijarczyk, A.; Babik, W. Detecting balancing selection in genomes: Limits and prospects. Mol. Ecol. 2015, 24, 3529–3545. [Google Scholar] [CrossRef]
- Těšický, M.; Vinkler, M. Trans-species polymorphism in immune genes: General pattern or MHC-restricted phenomenon? J. Immunol. Res. 2015, 2015, 838035. [Google Scholar] [CrossRef] [PubMed]
- Bechsgaard, J.S.; Castric, V.; Charlesworth, D.; Vekemans, X.; Schierup, M.H. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol. Biol. Evol. 2006, 23, 1741–1750. [Google Scholar] [CrossRef]
- Guo, Y.-L.; Zhao, X.; Lanz, C.; Weigel, D. Evolution of the S-locus region in Arabidopsis relatives. Plant Physiol. 2011, 157, 937–946. [Google Scholar] [CrossRef]
- Delph, L.F.; Kelly, J.K. On the importance of balancing selection in plants. New Phytol. 2014, 201, 45–56. [Google Scholar] [CrossRef]
- Le Veve, A.; Burghgraeve, N.; Genete, M.; Lepers-Blassiau, C.; Takou, M.; De Meaux, J.; Mable, B.K.; Durand, E.; Vekemans, X.; Castric, V. Long-term balancing selection and the genetic load linked to the self-incompatibility locus in Arabidopsis halleri and A. lyrata. Mol. Biol. Evol. 2023, 40, msad120. [Google Scholar] [CrossRef] [PubMed]
- Lija, M.; Beevy, S.S. A Review on the diversity of Melon. Plant Sci. Today 2021, 8, 995–1003. [Google Scholar] [CrossRef]
- Schaefer, H.; Heibl, C.; Renner, S.S. Gourds afloat: A dated phylogeny reveals an Asian origin of the gourd family (Cucurbitaceae) and numerous oversea dispersal events. Proc. R. Soc. B Biol. Sci. 2009, 276, 843–851. [Google Scholar] [CrossRef] [PubMed]
- Vanneste, K.; Baele, G.; Maere, S.; Van de Peer, Y. Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous–Paleogene boundary. Genome Res. 2014, 24, 1334–1347. [Google Scholar] [CrossRef]
- Barrera-Redondo, J.; Ibarra-Laclette, E.; Vázquez-Lobo, A.; Gutierrez-Guerrero, Y.T.; de la Vega, G.S.; Piñero, D.; Montes-Hernández, S.; Lira-Saade, R.; Eguiarte, L.E. The genome of Cucurbita argyrosperma (silver-seed gourd) reveals faster rates of protein-coding gene and long noncoding RNA turnover and neofunctionalization within Cucurbita. Mol. Plant 2019, 12, 506–520. [Google Scholar] [CrossRef]
- Li, H.-T.; Yi, T.-S.; Gao, L.-M.; Ma, P.-F.; Zhang, T.; Yang, J.-B.; Gitzendanner, M.A.; Fritsch, P.W.; Cai, J.; Luo, Y. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 2019, 5, 461–470. [Google Scholar] [CrossRef]
- Xie, D.; Xu, Y.; Wang, J.; Liu, W.; Zhou, Q.; Luo, S.; Huang, W.; He, X.; Li, Q.; Peng, Q. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019, 10, 5158. [Google Scholar] [CrossRef]
- Guo, X.; Fang, D.; Sahu, S.K.; Yang, S.; Guang, X.; Folk, R.; Smith, S.A.; Chanderbali, A.S.; Chen, S.; Liu, M. Chloranthus genome provides insights into the early diversification of angiosperms. Nat. Commun. 2021, 12, 6930. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef]
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Gao, P.; Zhu, Q.; Zhu, Z.; Liu, H.; Wang, X.; Weng, Y.; Gao, M.; Luan, F. Resequencing of 297 melon accessions reveals the genomic history of improvement and loci related to fruit traits in melon. Plant Biotechnol. J. 2020, 18, 2545–2558. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Chai, S.; Yang, Z.; Zhang, Q.; Xin, H.; Xu, Y.; Lin, S.; Chen, X.; Yao, Z. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nat. Commun. 2022, 13, 682. [Google Scholar] [CrossRef]
- Oren, E.; Dafna, A.; Tzuri, G.; Halperin, I.; Isaacson, T.; Elkabetz, M.; Meir, A.; Saar, U.; Ohali, S.; La, T. Pan-genome and multi-parental framework for high-resolution trait dissection in melon (Cucumis melo). Plant J. 2022, 112, 1525–1542. [Google Scholar] [CrossRef]
- Castanera, R.; Ruggieri, V.; Pujol, M.; Garcia-Mas, J.; Casacuberta, J.M. An improved melon reference genome with single-molecule sequencing uncovers a recent burst of transposable elements with potential impact on genes. Front. Plant Sci. 2020, 10, 1815. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Chen, N. Using Repeat Masker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinform. 2004, 5, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Cingolani, P. Variant annotation and functional prediction: SnpEff. In Variant Calling: Methods and Protocols; Springer: New York, NY, USA, 2012; pp. 289–314. [Google Scholar]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Excoffier, L.; Marchi, N.; Marques, D.A.; Matthey-Doret, R.; Gouy, A.; Sousa, V.C. fastsimcoal2: Demographic inference under complex evolutionary scenarios. Bioinformatics 2021, 37, 4882–4885. [Google Scholar] [CrossRef]
- Ossowski, S.; Schneeberger, K.; Lucas-Lledó, J.I.; Warthmann, N.; Clark, R.M.; Shaw, R.G.; Weigel, D.; Lynch, M. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 2010, 327, 92–94. [Google Scholar] [CrossRef]
- Malinsky, M.; Matschiner, M.; Svardal, H. Dsuite-Fast D-statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 2021, 21, 584–595. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Xiao, F.; Zheng, Y.; Liu, G.; Zhuang, Y.; Wang, Z.; Zhang, Y.; He, J.; Fu, C.; Lin, H. Pamp-induced secreted peptide 3 modulates salt tolerance through receptor-like kinase 7 in plants. Plant Cell 2022, 34, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Liu, J.; Li, X.; Amanullah, S.; Lu, X.; Zhang, M.; Zhang, Y.; Luan, F.; Liu, H.; Wang, X. Transcriptomic analysis of Fusarium oxysporum stress-induced pathosystem and screening of Fom-2 interaction factors in contrasted melon plants. Front. Plant Sci. 2022, 13, 961586. [Google Scholar] [CrossRef]
- Steidele, C.; Stam, R. Multi-omics approach highlights differences between RLP classes in Arabidopsis thaliana. BMC Genomics 2021, 22, 557. [Google Scholar] [CrossRef]
- Kratochwil, C.F.; Kautt, A.F.; Nater, A.; Härer, A.; Liang, Y.; Henning, F.; Meyer, A. An intronic transposon insertion associates with a trans-species color polymorphism in Midas cichlid fishes. Nat. Commun. 2022, 13, 296. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Han, T.-S.; Chen, X.; Chen, J.-F.; Zou, Y.-P.; Li, Z.-W.; Xu, Y.-C.; Guo, Y.-L. Long-term balancing selection contributes to adaptation in Arabidopsis and its relatives. Genome Biol. 2017, 18, 217. [Google Scholar] [CrossRef]
- Wang, B.; Mitchell-Olds, T. Balancing selection and trans-specific polymorphisms. Genome Biol. 2017, 18, 231. [Google Scholar] [CrossRef]
- Zhou, C.; Zhang, Q.; Chen, Y.; Huang, J.; Guo, Q.; Li, Y.; Wang, W.; Qiu, Y.; Guan, W.; Zhang, J. Balancing selection and wild gene pool contribute to resistance in global rice germplasm against planthopper. J. Integr. Plant Biol. 2021, 63, 1695–1711. [Google Scholar] [CrossRef]
- Fournier-Level, A.; Korte, A.; Cooper, M.D.; Nordborg, M.; Schmitt, J.; Wilczek, A.M. A map of local adaptation in Arabidopsis thaliana. Science 2011, 334, 86–89. [Google Scholar] [CrossRef]
- Hancock, A.M.; Brachi, B.; Faure, N.; Horton, M.W.; Jarymowycz, L.B.; Sperone, F.G.; Toomajian, C.; Roux, F.; Bergelson, J. Adaptation to climate across the Arabidopsis thaliana genome. Science 2011, 334, 83–86. [Google Scholar] [CrossRef]
- Li, Y.; Wang, N.; Feng, J.; Liu, Y.; Wang, H.; Deng, S.; Dong, W.; Liu, X.; Lv, B.; Sun, J. Enhancing genetic transformation efficiency in cucurbit crops through AtGRF5 overexpression: Mechanistic insights and applications. J. Integr. Plant Biol. 2025. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, X.; Liu, Y.; Li, Y.; Hu, M.; Yu, T.; Yu, Q.; Wang, H.; Chen, X.; Chai, S.; Xu, K. Evolutionary Forces Shaping Trans-Species Polymorphisms in Genus Cucumis. Horticulturae 2025, 11, 452. https://doi.org/10.3390/horticulturae11050452
Su X, Liu Y, Li Y, Hu M, Yu T, Yu Q, Wang H, Chen X, Chai S, Xu K. Evolutionary Forces Shaping Trans-Species Polymorphisms in Genus Cucumis. Horticulturae. 2025; 11(5):452. https://doi.org/10.3390/horticulturae11050452
Chicago/Turabian StyleSu, Xiaofeng, Yi Liu, Yueting Li, Minghe Hu, Tao Yu, Qing Yu, Huilin Wang, Xinxiu Chen, Sen Chai, and Kuipeng Xu. 2025. "Evolutionary Forces Shaping Trans-Species Polymorphisms in Genus Cucumis" Horticulturae 11, no. 5: 452. https://doi.org/10.3390/horticulturae11050452
APA StyleSu, X., Liu, Y., Li, Y., Hu, M., Yu, T., Yu, Q., Wang, H., Chen, X., Chai, S., & Xu, K. (2025). Evolutionary Forces Shaping Trans-Species Polymorphisms in Genus Cucumis. Horticulturae, 11(5), 452. https://doi.org/10.3390/horticulturae11050452




