Genome-Wide Identification of GATA Transcription Factors in Acorus and Expression Patterns Under Abiotic Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification, Classification and Phylogenetic Analysis of A. calamus and A. gramineus GATA Transcription Factors
2.2. Conserved Motifs, Domains, and Multiple Sequence Alignment
2.3. Chromosomal Localization and Collinearity Analysis
2.4. Cis-Elements and Protein–Protein Interaction Analysis
2.5. Plant Drought and Salt Stress Treatment
2.6. Gene Expression Patterns and Quantitative Fluorescence Validation Under Abiotic Stress
3. Results
3.1. Identification and Characteristics Analysis of GATAs
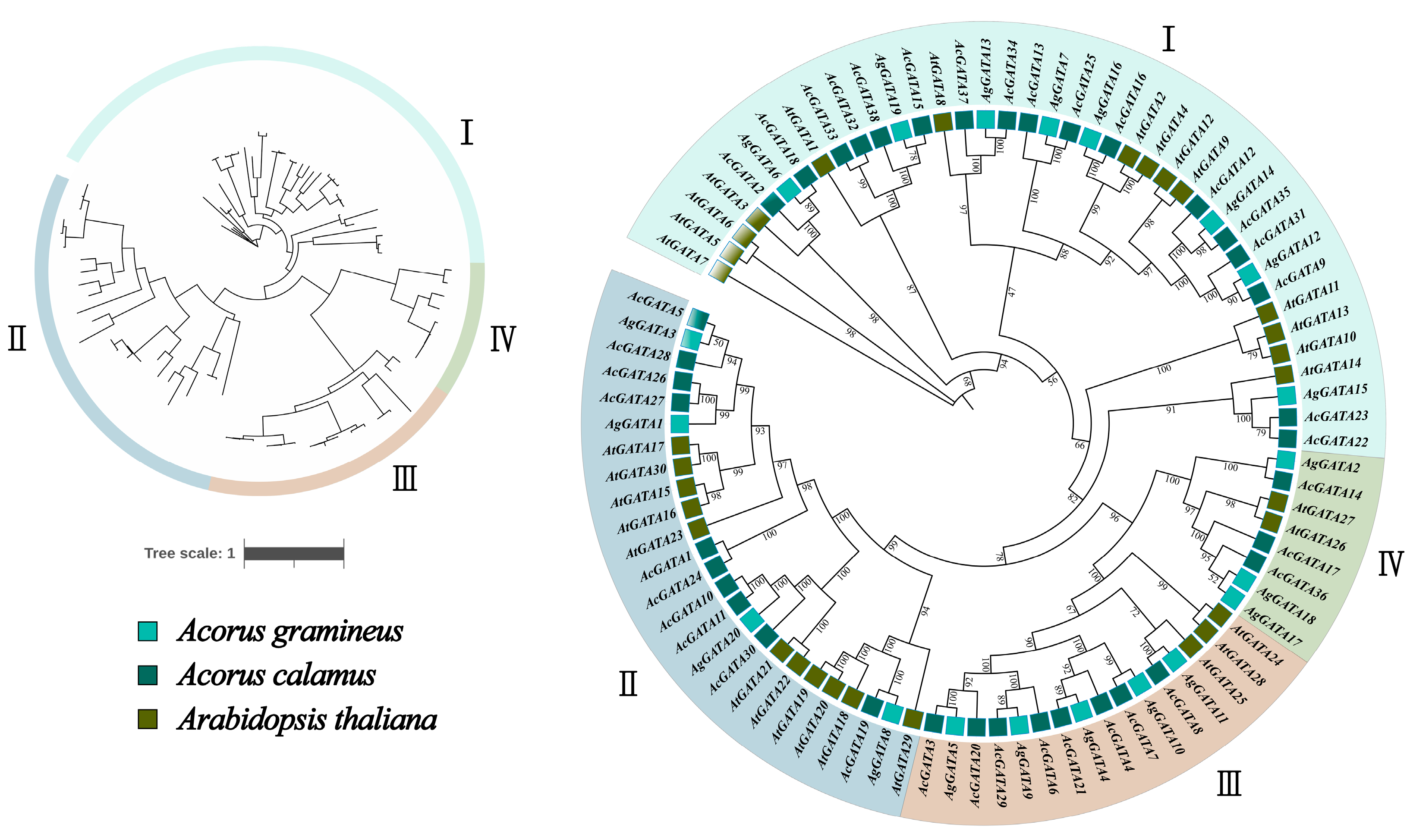
3.2. Phylogenetic Analysis of AcGATAs and AgGATAs
3.3. Analysis of Conserved Motifs and Domains in GATA Proteins
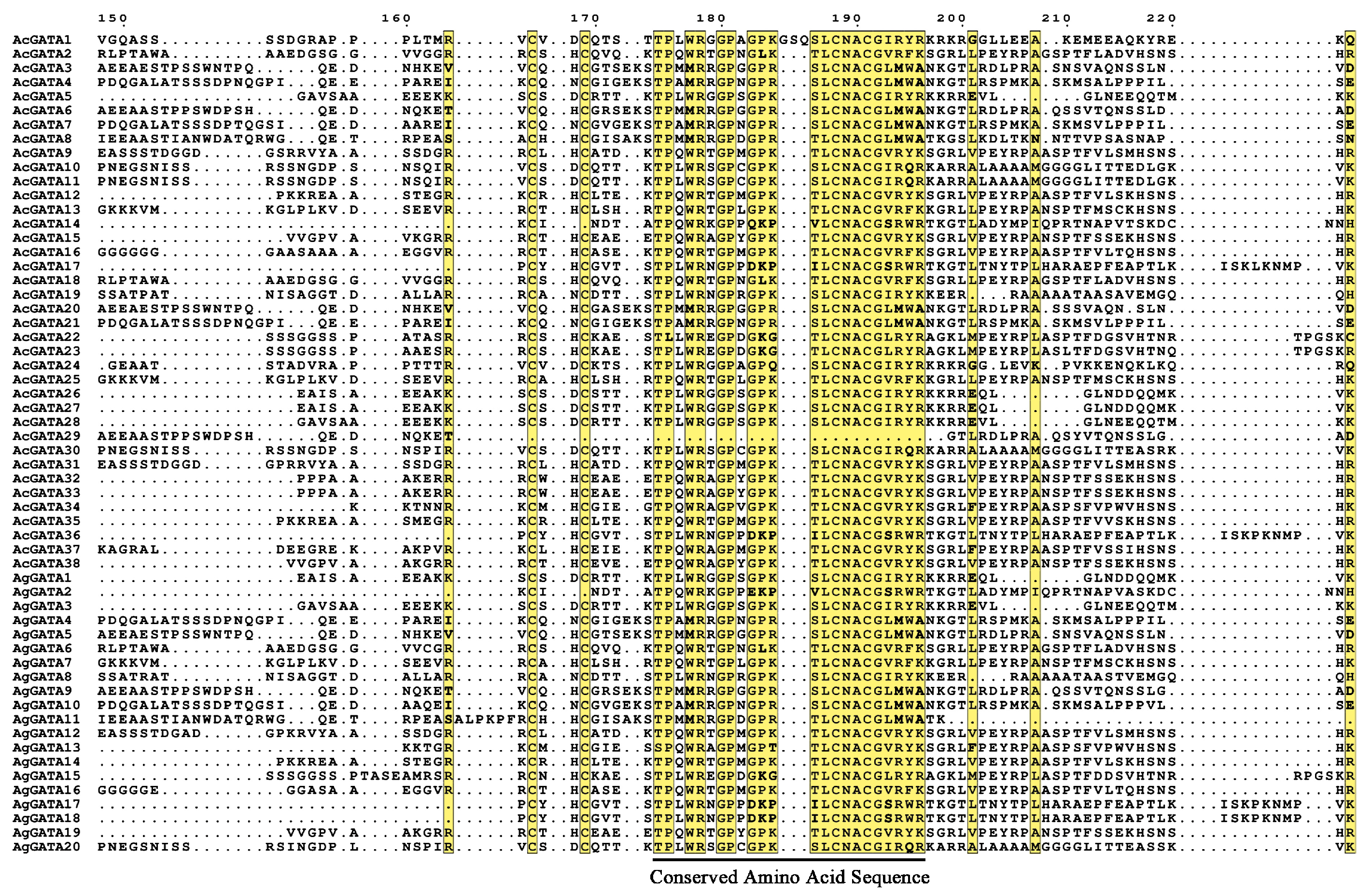
3.4. Multiple Sequence Analysis of AcGATAs and AgGATAs
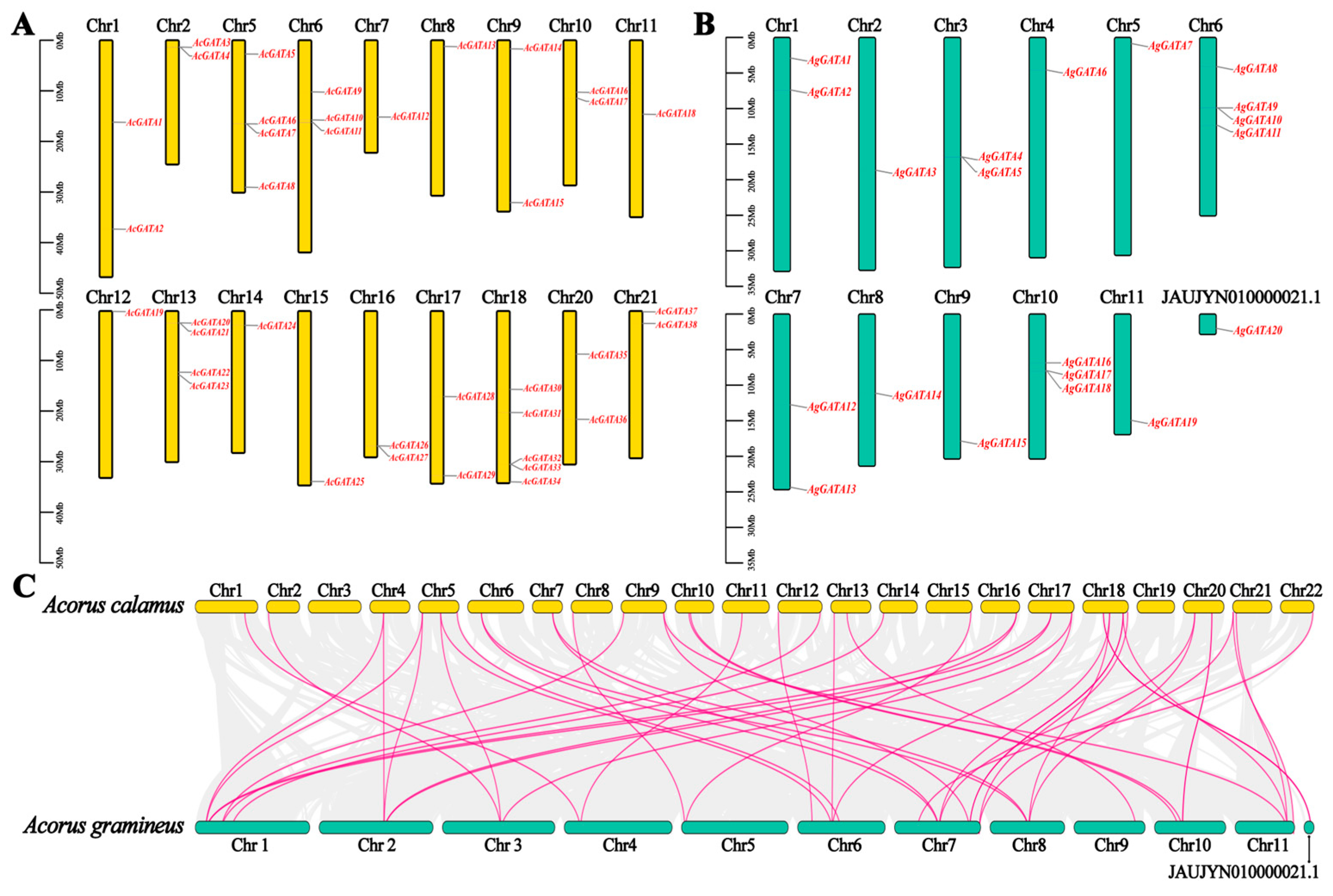
3.5. Localization and Collinearity Analysis of AcGATAs and AgGATAs
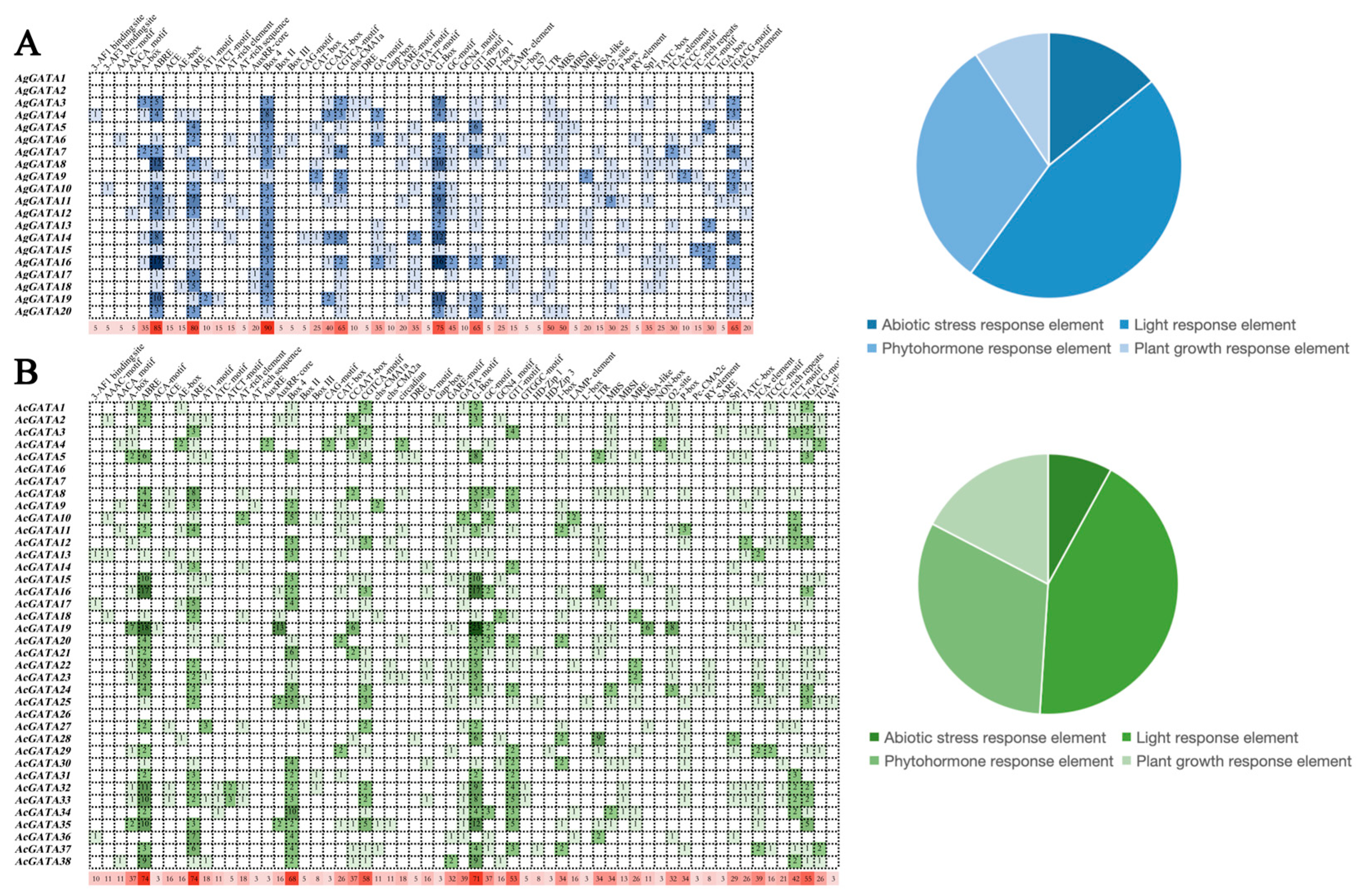
3.6. Analysis of Cis-Acting Elements on Promoters
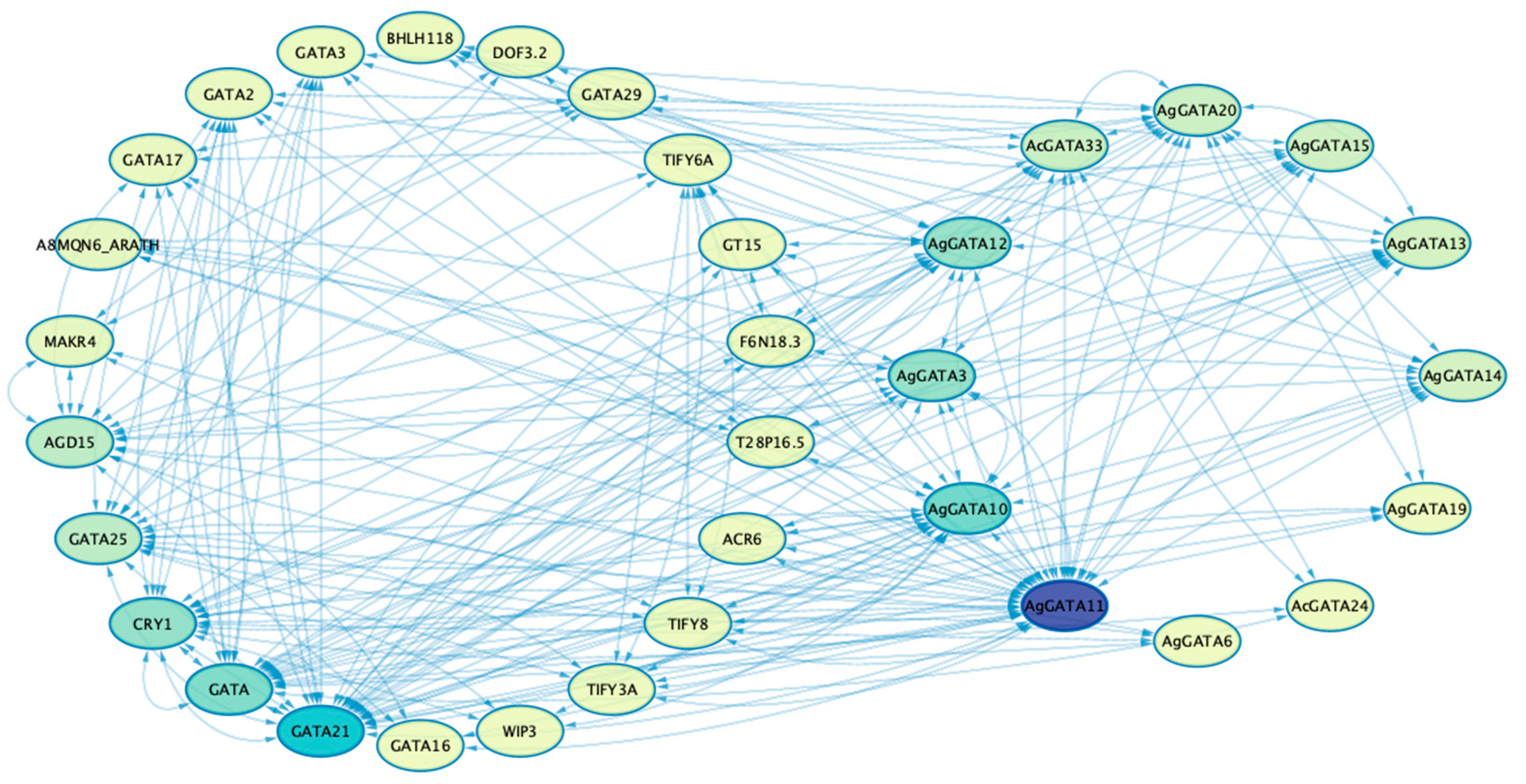
3.7. Protein–Protein Interaction and Structure Analysis
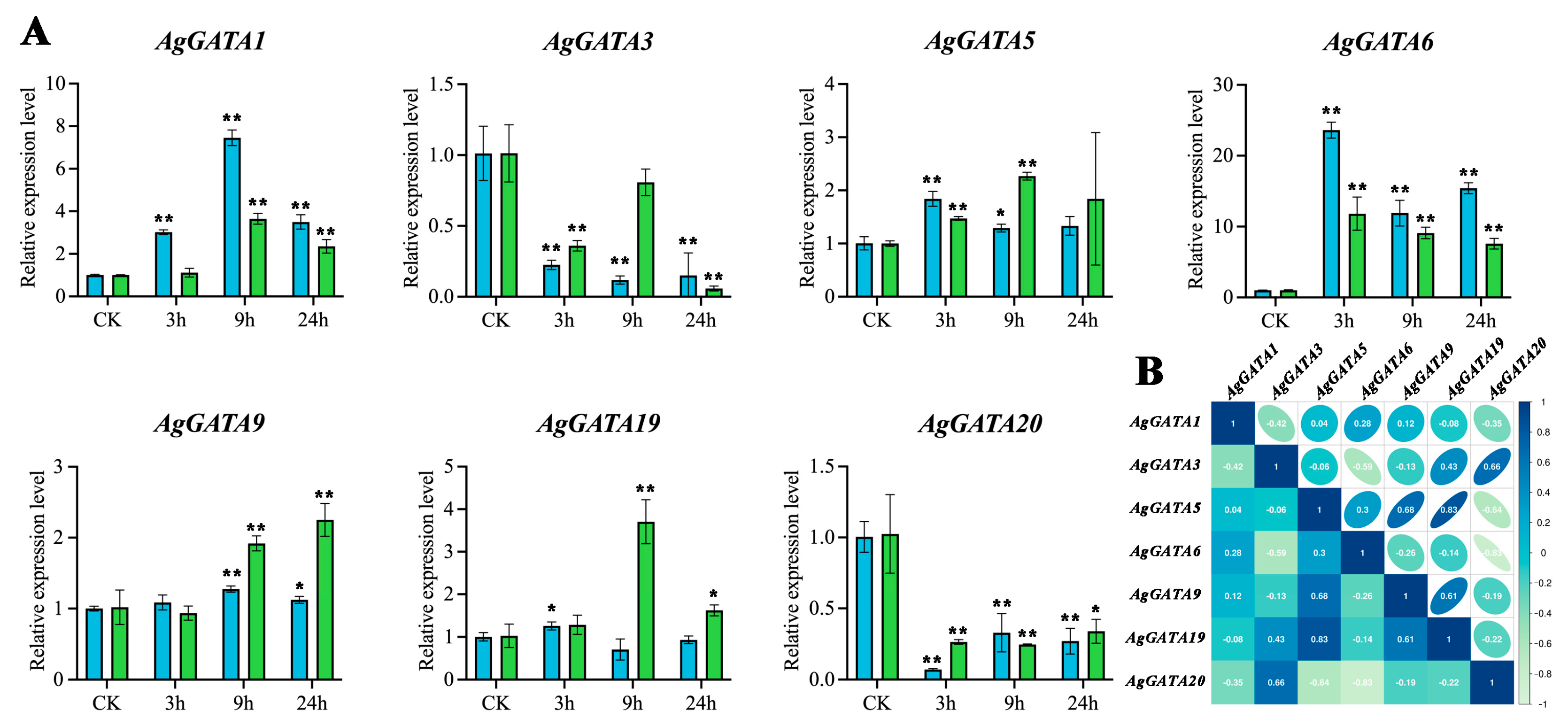
3.8. AgGATA Gene Responds to Abiotic Stress
3.9. Quantitative Real-Time PCR Under Abiotic Stress in A. gramineus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Du, B.; Haensch, R.; Alfarraj, S.; Rennenberg, H. Strategies of plants to overcome abiotic and biotic stresses. Biol. Rev. Camb. Philos. Soc. 2024, 99, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Weidemüller, P.; Kholmatov, M.; Petsalaki, E.; Zaugg, J.B. Transcription factors: Bridge between cell signaling and gene regulation. Proteomics 2021, 21, e2000034. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Guo, Y.; Chen, Y.; Wu, D.; Jiang, L. Genome-wide identification, phylogenetic and expression pattern analysis of GATA family genes in Brassica napus. BMC Plant Biol. 2020, 20, 543. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bates, D.L.; Dey, R.; Chen, P.-H.; Machado, A.C.D.; Laird-Offringa, I.A.; Rohs, R.; Chen, L. DNA Binding by GATA Transcription Factor Suggests Mechanisms of DNA Looping and Long-Range Gene Regulation. Cell Rep. 2012, 2, 1197–1206. [Google Scholar] [CrossRef]
- Patient, R.K.; McGhee, J.D. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 2002, 12, 416–422. [Google Scholar] [CrossRef]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol. 2004, 134, 1718–1732. [Google Scholar] [CrossRef]
- Kim, M.; Xi, H.; Park, J. Genome-wide comparative analyses of GATA transcription factors among 19 Arabidopsis ecotype genomes: Intraspecific characteristics of GATA transcription factors. PLoS ONE 2021, 16, e0252181. [Google Scholar] [CrossRef]
- Daniel-Vedele, F.; Caboche, M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 1993, 240, 365–373. [Google Scholar] [CrossRef]
- Chen, H.; Shao, H.; Li, K.; Zhang, D.; Fan, S.; Li, Y.; Han, M. Genome-wide identification, evolution, and expression analysis of GATA transcription factors in apple (Malus × domestica Borkh.). Gene 2017, 627, 460–472. [Google Scholar] [CrossRef]
- Zhang, C.; Hou, Y.; Hao, Q.; Chen, H.; Chen, L.; Yuan, S.; Shan, Z.; Zhang, X.; Yang, Z.; Qiu, D.; et al. Genome-wide survey of the soybean GATA transcription factor gene family and expression analysis under low nitrogen stress. PLoS ONE 2015, 10, e0125174. [Google Scholar] [CrossRef]
- An, Y.; Zhou, Y.; Han, X.; Shen, C.; Wang, S.; Liu, C.; Yin, W.; Xia, X. The GATA transcription factor GNC plays an important role in photosynthesis and growth in poplar. J. Exp. Bot. 2020, 71, 1969–1984. [Google Scholar] [CrossRef] [PubMed]
- Manfield, I.W.; Devlin, P.F.; Jen, C.H.; Westhead, D.R.; Gilmartin, P.M. Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 2007, 143, 941–958. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.M.; Zhang, Y.; Signorelli, T.; Zhao, R.; Zhu, T.; Rothstein, S. Genetic analysis of Arabidopsis GATA transcription factor gene family reveals a nitrate-inducible member important for chlorophyll synthesis and glucose sensitivity. Plant J. 2005, 44, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.H.; Zubo, Y.O.; Tapken, W.; Kim, H.J.; Lavanway, A.M.; Howard, L.; Pilon, M.; Kieber, J.J.; Schaller, G.E. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012, 160, 332–348. [Google Scholar] [CrossRef]
- Zhao, Y.; Medrano, L.; Ohashi, K.; Fletcher, J.C.; Yu, H.; Sakai, H.; Meyerowitz, E.M. HANABA TARANU is a GATA transcription factor that regulates shoot apical meristem and flower development in Arabidopsis. Plant Cell 2004, 16, 2586–2600. [Google Scholar] [CrossRef]
- Zhang, K.; Jia, L.; Yang, D.; Hu, Y.; Njogu, M.K.; Wang, P.; Lu, X.; Yan, C. Genome-Wide Identification, Phylogenetic and Expression Pattern Analysis of GATA Family Genes in Cucumber (Cucumis sativus L.). Plants 2021, 10, 1626. [Google Scholar] [CrossRef]
- Zhang, X.; Ma, J.; Yang, S.; Yao, W.; Zhang, N.; Hao, X.; Xu, W. Analysis of GATA transcription factors and their expression patterns under abiotic stress in grapevine (Vitis vinifera L.). BMC Plant Biol. 2023, 23, 611. [Google Scholar] [CrossRef]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic Stresses Cause Differential Regulation of Alternative Splice Forms of GATA Transcription Factor in Rice. Front. Plant Sci. 2017, 8, 1944. [Google Scholar] [CrossRef]
- Ravindran, P.; Verma, V.; Stamm, P.; Kumar, P.P. A Novel RGL2–DOF6 Complex Contributes to Primary Seed Dormancy in Arabidopsis thaliana by Regulating a GATA Transcription Factor. Mol. Plant 2017, 10, 1307–1320. [Google Scholar] [CrossRef]
- Hudson, D.; Guevara, D.R.; Hand, A.J.; Xu, Z.; Hao, L.; Chen, X.; Zhu, T.; Bi, Y.-M.; Rothstein, S.J. Rice Cytokinin GATA Transcription Factor1 Regulates Chloroplast Development and Plant Architecture. Plant Physiol. 2013, 162, 132–144. [Google Scholar] [CrossRef]
- Lu, G.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Bi, Y.M.; Rothstein, S.J. Overexpression of OsGATA12 regulates chlorophyll content, delays plant senescence and improves rice yield under high density planting. Plant Mol. Biol. 2017, 94, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Liu, P.; Yang, Y.S.; Chen, W.R. Phytoremediation of urban wastewater by model wetlands with ornamental hydrophytes. J. Environ. Sci. 2007, 19, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Shu, H.; Zhang, S.; Lei, Q.; Zhou, J.; Ji, Y.; Luo, B.-S.; Hong, L.; Li, F.; Liu, B.; Long, C. Ethnobotany of Acorus in China. Acta Soc. Bot. Pol. 2018, 87, 1–14. [Google Scholar] [CrossRef]
- Cheng, Z.; Shu, H.; Zhang, S.; Luo, B.; Gu, R.; Zhang, R.; Ji, Y.; Li, F.; Long, C. From Folk Taxonomy to Species Confirmation of Acorus (Acoraceae): Evidences Based on Phylogenetic and Metabolomic Analyses. Front. Plant Sci. 2020, 11, 965. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, D.; Hu, Z.; Zhang, A.; Li, F.; Zhang, J.; Wu, Y.; Tang, Y. Tolerance and physiological responses of sweet flag (Acorus calamus L.) under nitrite stress during wastewater treatment. Ecol. Eng. 2018, 122, 107–111. [Google Scholar] [CrossRef]
- Zhang, S.H.; Zhang, S.Y.; Li, G. Acorus calamus root extracts to control harmful cyanobacteria blooms. Ecol. Eng. 2016, 94, 95–101. [Google Scholar] [CrossRef]
- Ma, Y.-Y.; Chen, Y.-T.; Hu, X.-X.; Ma, Q.-R.; Feng, T.-C.; Feng, G.-L.; Ma, D. The 2022 record-breaking high temperature in China: Sub-seasonal stepwise enhanced characteristics, possible causes and its predictability. Adv. Clim. Change Res. 2023, 14, 651–659. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil salinity and its associated effects on soil microorganisms, greenhouse gas emissions, crop yield, biodiversity and desertification: A review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef]
- Singh, A. Soil salinization management for sustainable development: A review. J. Environ. Manag. 2021, 277, 111383. [Google Scholar] [CrossRef]
- Khojasteh, D.; Haghani, M.; Nicholls, R.J.; Moftakhari, H.; Sadat-Noori, M.; Mach, K.J.; Fagherazzi, S.; Vafeidis, A.T.; Barbier, E.; Shamsipour, A.; et al. The evolving landscape of sea-level rise science from 1990 to 2021. Commun. Earth Environ. 2023, 4, 257. [Google Scholar] [CrossRef]
- Ma, L.; Liu, K.-W.; Li, Z.; Hsiao, Y.-Y.; Qi, Y.; Fu, T.; Tang, G.-D.; Zhang, D.; Sun, W.-H.; Liu, D.-K.; et al. Diploid and tetraploid genomes of Acorus and the evolution of monocots. Nat. Commun. 2023, 14, 3661. [Google Scholar] [CrossRef] [PubMed]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. Trimal: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef]
- Benmrid, B.; Ghoulam, C.; Ammar, I.; Nkir, D.; Saidi, R.; Staropoli, A.; Iacomino, G.; ELhajjami, E.; Cheto, S.; Geistlinger, J.; et al. Drought-tolerant rhizobacteria with predicted functional traits enhanced wheat growth and P uptake under moderate drought and low P-availability. Microbiol. Res. 2024, 285, 127795. [Google Scholar] [CrossRef]
- Cheng, S.; Fan, S.; Yang, C.; Hu, W.; Liu, F. Proteomics revealed novel functions and drought tolerance of Arabidopsis thaliana protein kinase ATG1. BMC Biol. 2025, 23, 48. [Google Scholar] [CrossRef]
- Lu, X.; Zhou, X.; Cao, Y.; Zhou, M.; McNeil, D.; Liang, S.; Yang, C. RNA-seq Analysis of Cold Drought Responsive Transcriptomes of Zea mays ssp. mexicana, L. Front. Plant Sci. 2017, 8, 136. [Google Scholar] [CrossRef]
- Sha, M.; Fu, X.; Bai, R.; Zhong, Z.; Jiang, H.; Li, F.; Yang, S. YOLOv8-licorice: A lightweight salt-resistance detection method for licorice based on seed germination state. Front. Plant Sci. 2024, 15, 1474321. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotechnol. Softw. Int. Rep. 2000, 1, 270–272. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rai, N.; Rai, S.P.; Sarma, B.K. Chickpea defense against dual stresses of salt and Fusarium wilt is enhanced through selected bHLH transcription factors carrying the bHLH-MYC_N domain. Plant Physiol. Biochem. 2025, 218, 109314. [Google Scholar] [CrossRef]
- Li, M.; Fan, D.; Wen, Z.; Meng, J.; Li, P.; Cheng, T.; Zhang, Q.; Sun, L. Genome-wide identification of the Dof gene family: How it plays a part in mediating cold stress response in Prunus mume. Plant Physiol. Biochem. 2024, 217, 109215. [Google Scholar] [CrossRef]
- Chen, Q.; Dai, R.; Shuang, S.; Zhang, Y.; Huo, X.; Shi, F.; Zhang, Z. Genome-wide investigation of the TIFY transcription factors in alfalfa (Medicago sativa L.): Identification, analysis, and expression. BMC Plant Biol. 2024, 24, 840. [Google Scholar] [CrossRef]
- Duan, Z.; Zhang, Y.; Tu, J.; Shen, J.; Yi, B.; Fu, T.; Dai, C.; Ma, C. The Brassica napus GATA transcription factor BnA5.ZML1 is a stigma compatibility factor. J. Integr. Plant Biol. 2020, 62, 1112–1131. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Schröder, P.M.; Blaby-Haas, C.E. Plant GATA Factors: Their Biology, Phylogeny, and Phylogenomics. Annu. Rev. Plant Biol. 2022, 73, 123–148. [Google Scholar] [CrossRef]
- Zheng, L.; Tang, L.; Li, J. Genome-wide identification of the GATA gene family in melon (Cucumis melo) and analysis of their expression characteristics under biotic and abiotic stresses. Front. Plant Sci. 2024, 15, 1462924. [Google Scholar] [CrossRef]
- Wang, Y.; Li, X.; Mo, Y.; Jiang, C.; Zhou, Y.; Hu, J.; Zhang, Y.; Lv, J.; Zhao, K.; Lu, Z. Identification and expression profiling of SmGATA genes family involved in response to light and phytohormones in eggplant. Front. Plant Sci. 2024, 15, 1415921. [Google Scholar] [CrossRef]
- Yu, R.; Chang, Y.; Chen, H.; Feng, J.; Wang, H.; Tian, T.; Song, Y.; Gao, G. Genome-wide identification of the GATA gene family in potato (Solanum tuberosum L.) and expression analysis. J. Plant Biochem. Biotechnol. 2022, 31, 37–48. [Google Scholar] [CrossRef]
- Feng, X.; Yu, Q.; Zeng, J.; He, X.; Liu, W. Genome-wide identification and characterization of GATA family genes in wheat. BMC Plant Biol. 2022, 22, 372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zou, X.; Huang, Z.; Fan, S.; Qun, G.; Liu, A.; Gong, J.; Li, J.; Gong, W.; Shi, Y.; et al. Genome-wide identification and analysis of the evolution and expression patterns of the GATA transcription factors in three species of Gossypium genus. Gene 2019, 680, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Ranftl, Q.L.; Bastakis, E.; Klermund, C.; Schwechheimer, C. LLM-Domain Containing B-GATA Factors Control Different Aspects of Cytokinin-Regulated Development in Arabidopsis thaliana. Plant Physiol. 2016, 170, 2295–2311. [Google Scholar] [CrossRef]
- Saidi, A.; Hajibarat, Z.; Hajibarat, Z. Phylogeny, gene structure and GATA genes expression in different tissues of solanaceae species. Biocatal. Agric. Biotechnol. 2021, 35, 102015. [Google Scholar] [CrossRef]
- Dar, N.A.; Amin, I.; Wani, W.; Wani, S.A.; Shikari, A.B.; Wani, S.H.; Masoodi, K.Z. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 2017, 11, 106–111. [Google Scholar] [CrossRef]
- Chaffai, R.; Ganesan, M.; Cherif, A. Plant Hormones and Their Regulatory Roles in Abiotic Stress Response. In Plant Adaptation to Abiotic Stress: From Signaling Pathways and Microbiomes to Molecular Mechanisms; Springer Nature: Singapore, 2024; pp. 249–274. [Google Scholar]
- Muhammad Aslam, M.; Waseem, M.; Jakada, B.H.; Okal, E.J.; Lei, Z.; Saqib, H.S.A.; Yuan, W.; Xu, W.; Zhang, Q. Mechanisms of Abscisic Acid-Mediated Drought Stress Responses in Plants. Int. J. Mol. Sci. 2022, 23, 1084. [Google Scholar] [CrossRef]
- Zheng, K.; Lv, M.; Qian, J.; Lian, Y.; Liu, R.; Huo, S.; Rehman, O.U.; Lin, Q.; Zhou, Z.; Liu, X.; et al. Identification and Characterization of the DOF Gene Family in Phoebe bournei and Its Role in Abiotic Stress-Drought, Heat and Light Stress. Int. J. Mol. Sci. 2024, 25, 11147. [Google Scholar] [CrossRef]
- Lu, S.; Sun, Y.; Liu, X.; Wang, F.; Luan, S.; Wang, H. The SlbHLH92 transcription factor enhances salt stress resilience by fine-tuning hydrogen sulfide biosynthesis in tomato. Int. J. Biol. Macromol. 2024, 282 Pt 5, 137294. [Google Scholar] [CrossRef]


| Gene ID | Gene Name | Primer Name | Sequence |
|---|---|---|---|
| KAK1280067.1 | AgGATA1 | 1-F | GGAGGTGGAGCGAACTTGAACTG |
| AgGATA1 | 1-R | AGCATGGATGAATCCGCAAGAGAG | |
| KAK1277125.1 | AgGATA3 | 3-F | GGAGAAGAAGAGCTGCTCGGATTG |
| AgGATA3 | 3-R | CACAGAGACTTTGGCCCAGATGG | |
| KAK1276896.1 | AgGATA5 | 5-F | GAACTCCAACCACCACGATGACTG |
| AgGATA5 | 5-R | CTCTGATCCATCCATGCCGTTGAC | |
| KAK1272898.1 | AgGATA6 | 6-F | GTCATCCTCATCCACCACCACAAC |
| AgGATA6 | 6-R | GCCTTGGCTTCCTCTGCTTCTTC | |
| KAK1268328.1 | AgGATA9 | 9-F | AACTTGAACCGCCACGATGACTG |
| AgGATA9 | 9-R | ATCCGTCCACGCCGTCCATC | |
| KAK1260277.1 | AgGATA19 | 19-F | ATTGCGAGGCGGAGGAGACG |
| AgGATA19 | 19-R | CCGGCCCGACTTATACCTGACC | |
| KAK1258291.1 | AgGATA20 | 20-F | CCGATCAGAGTCTGCTCAGATTGC |
| AgGATA20 | 20-R | GCCCTTCTCGCCTTCCTTTGTC | |
| KAK1276548.1 | AgACT1 | ACT-F | GCTTCTTCCATCGTGTCTCGTCTC |
| AgACT1 | ACT-R | TGTCCTTCCCAGTCCCTTGAGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, L.; Zhou, C.-Y.; Gong, C.-Y.; He, Y.; Chang, M.-L.; Fu, H.-H.; Liu, Z.-J.; Chen, S.-P. Genome-Wide Identification of GATA Transcription Factors in Acorus and Expression Patterns Under Abiotic Stress. Horticulturae 2025, 11, 410. https://doi.org/10.3390/horticulturae11040410
Ma L, Zhou C-Y, Gong C-Y, He Y, Chang M-L, Fu H-H, Liu Z-J, Chen S-P. Genome-Wide Identification of GATA Transcription Factors in Acorus and Expression Patterns Under Abiotic Stress. Horticulturae. 2025; 11(4):410. https://doi.org/10.3390/horticulturae11040410
Chicago/Turabian StyleMa, Liang, Cheng-Yuan Zhou, Chen-Yu Gong, Yao He, Meng-Lin Chang, Hou-Hua Fu, Zhong-Jian Liu, and Shi-Pin Chen. 2025. "Genome-Wide Identification of GATA Transcription Factors in Acorus and Expression Patterns Under Abiotic Stress" Horticulturae 11, no. 4: 410. https://doi.org/10.3390/horticulturae11040410
APA StyleMa, L., Zhou, C.-Y., Gong, C.-Y., He, Y., Chang, M.-L., Fu, H.-H., Liu, Z.-J., & Chen, S.-P. (2025). Genome-Wide Identification of GATA Transcription Factors in Acorus and Expression Patterns Under Abiotic Stress. Horticulturae, 11(4), 410. https://doi.org/10.3390/horticulturae11040410








