Cytological, Physiological and Genotyping-by-Sequencing Analysis Revealing Dynamic Variation of Leaf Color in Ginkgo biloba L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Leaf Anatomical Characters
2.2.1. Light Microscope Observation
2.2.2. Scanning Electron Microscope Observation
2.2.3. Transmission Electron Microscopic Observation
2.3. Determination of Physiological and Secondary Metabolite
2.4. Genotyping-by-Sequencing
2.5. Data Processing
3. Results
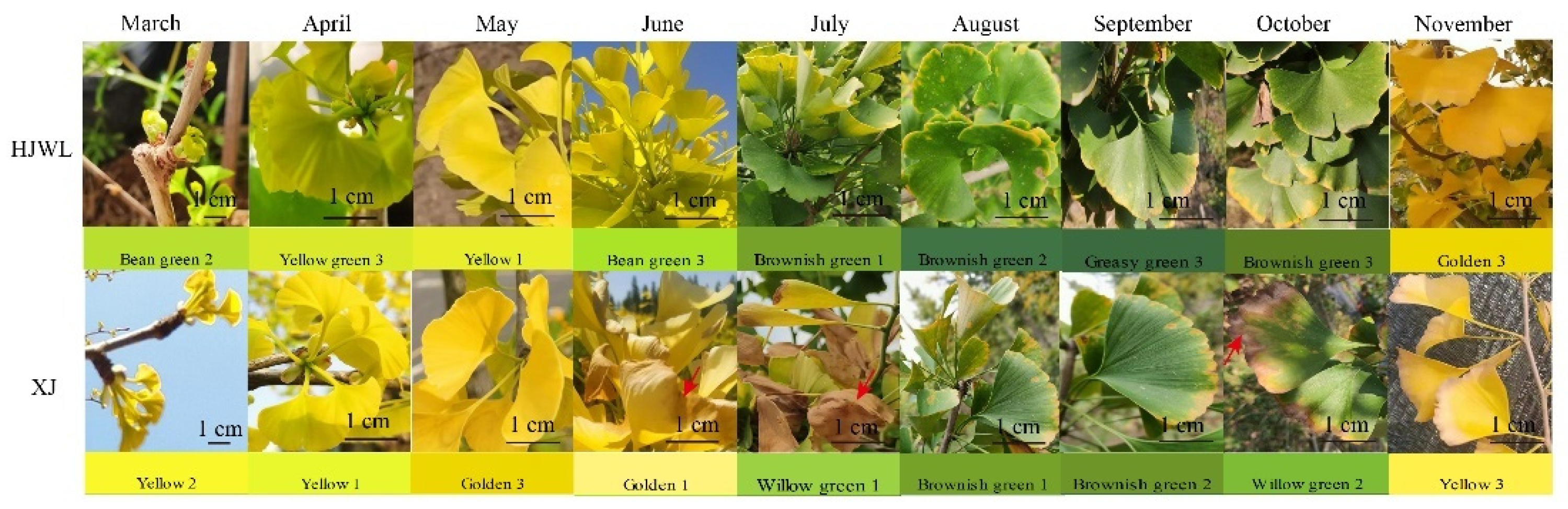
3.1. Seasonal Dynamic Changes in Ginkgo Leaf Color
3.2. Seasonal Dynamic Changes in Pigment Content in Ginkgo Leaves
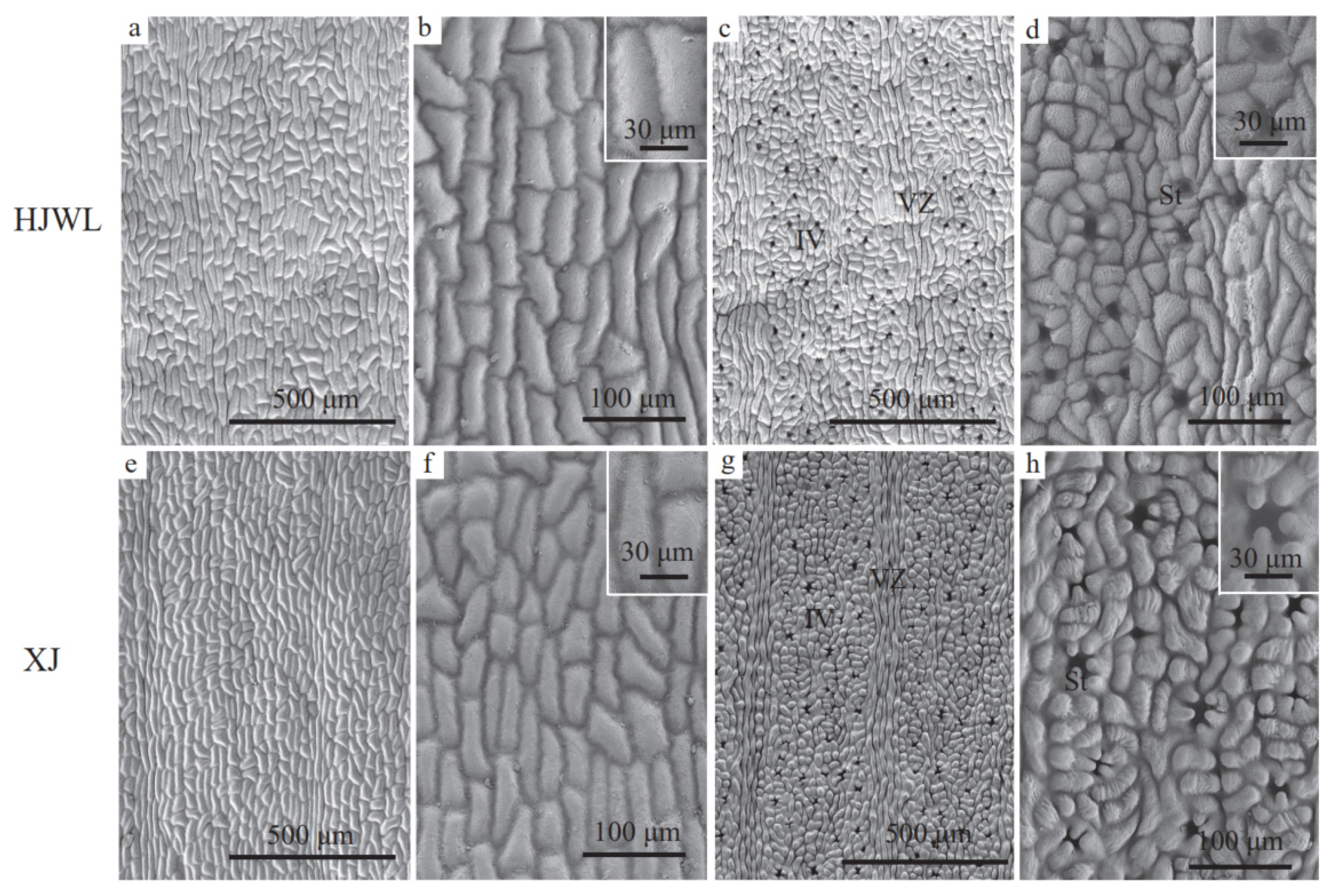
3.3. Phenotypic Characteristics in HJWL Leaves
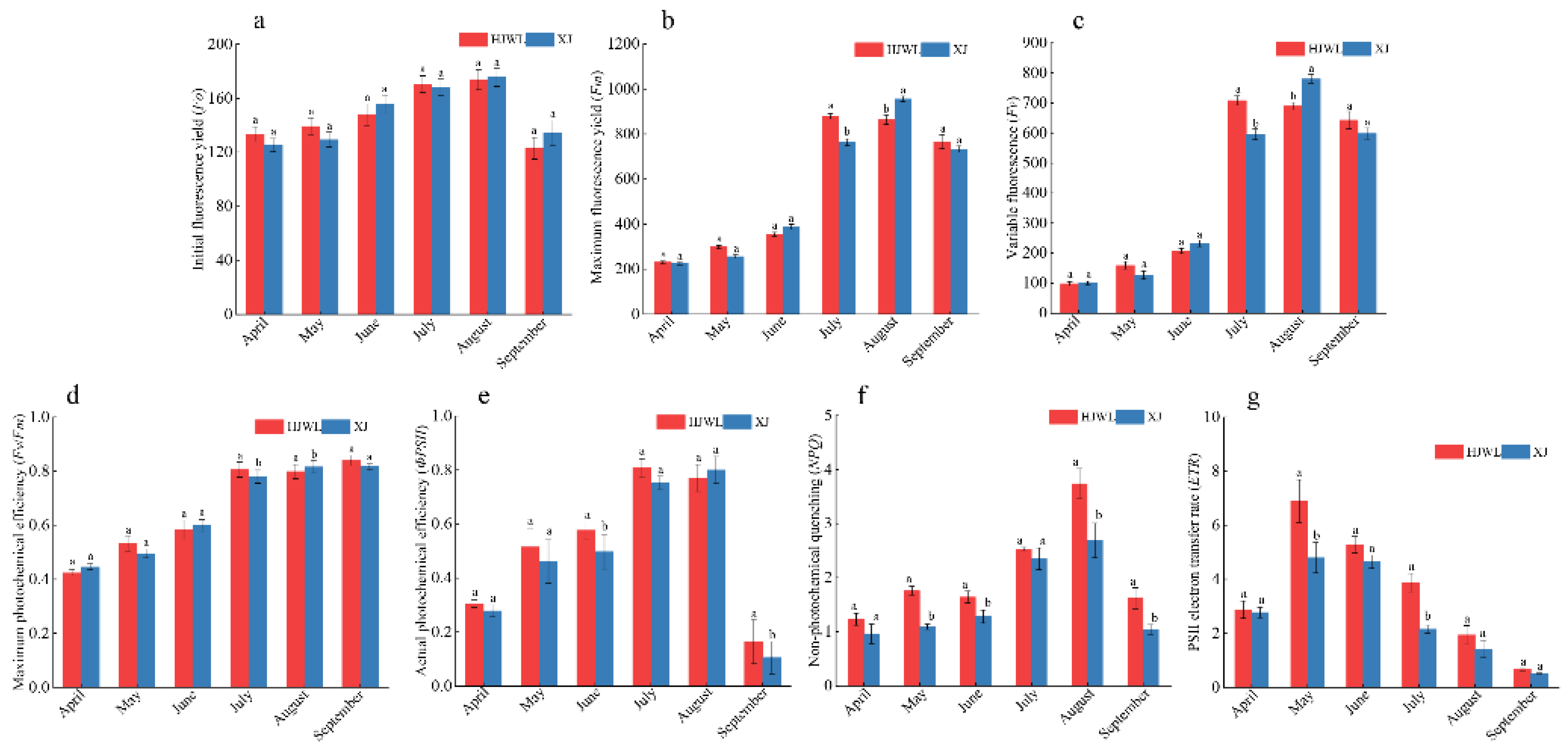
3.4. Chlorophyll Fluorescence Parameters of Ginkgo
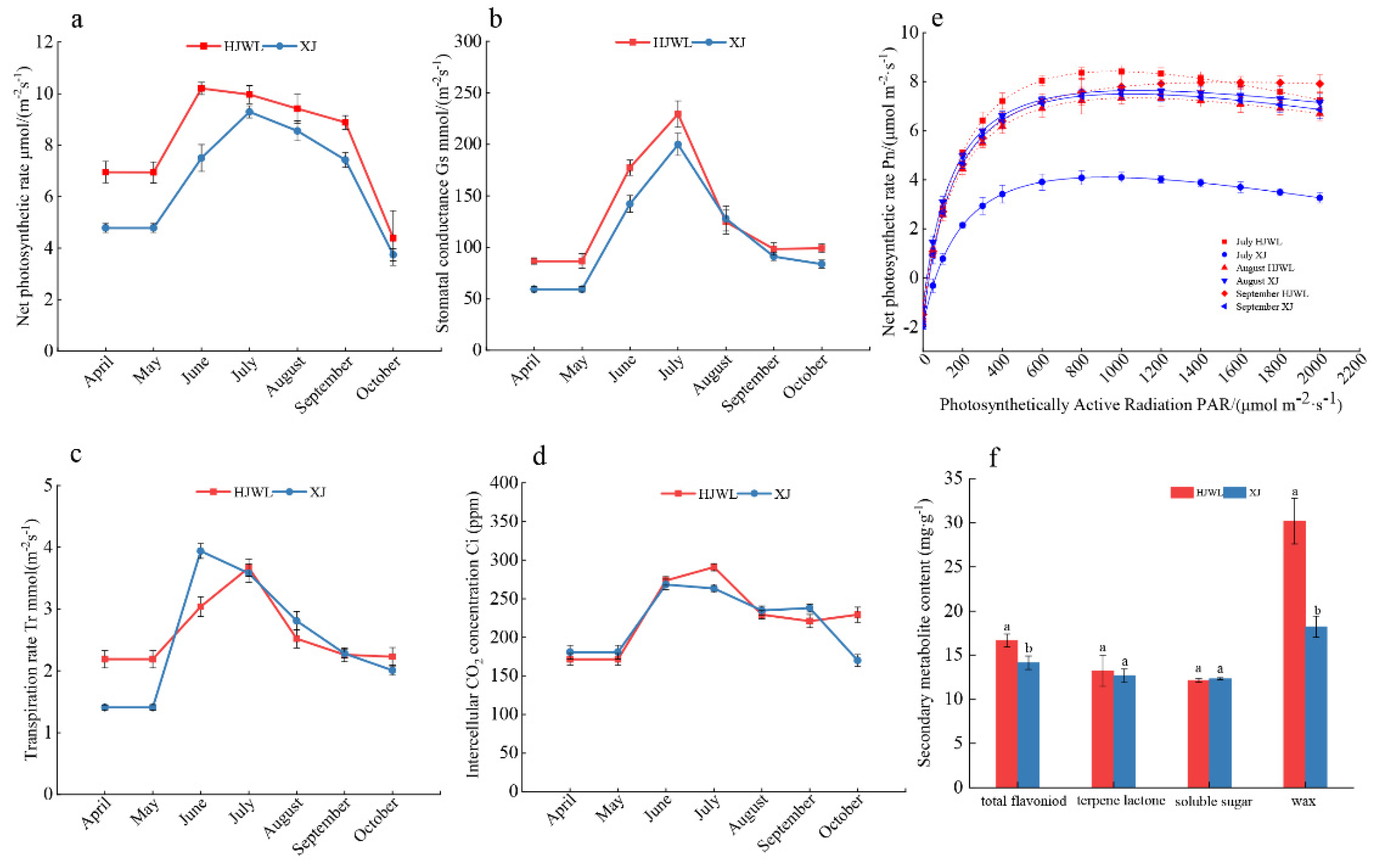
3.5. Comparison of Photosynthetic Physiology and Metabolites
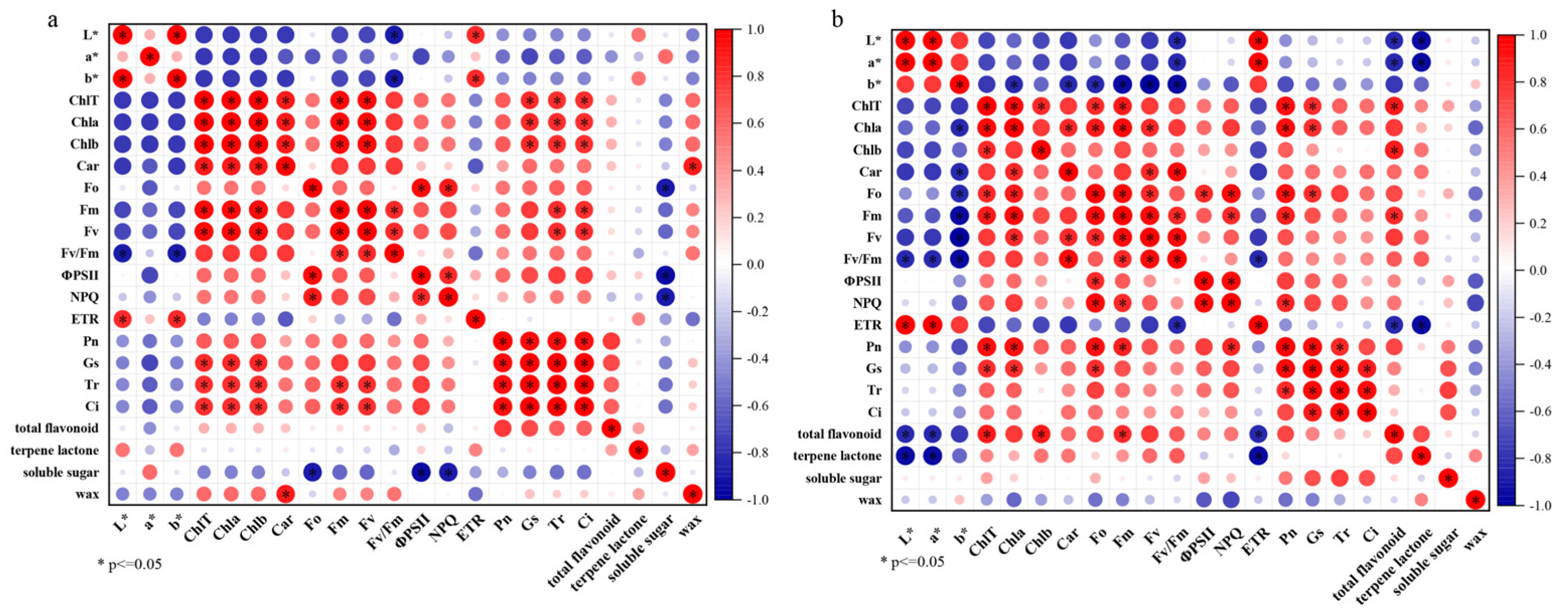
3.6. Correlation Analysis Between Physiological Indexes
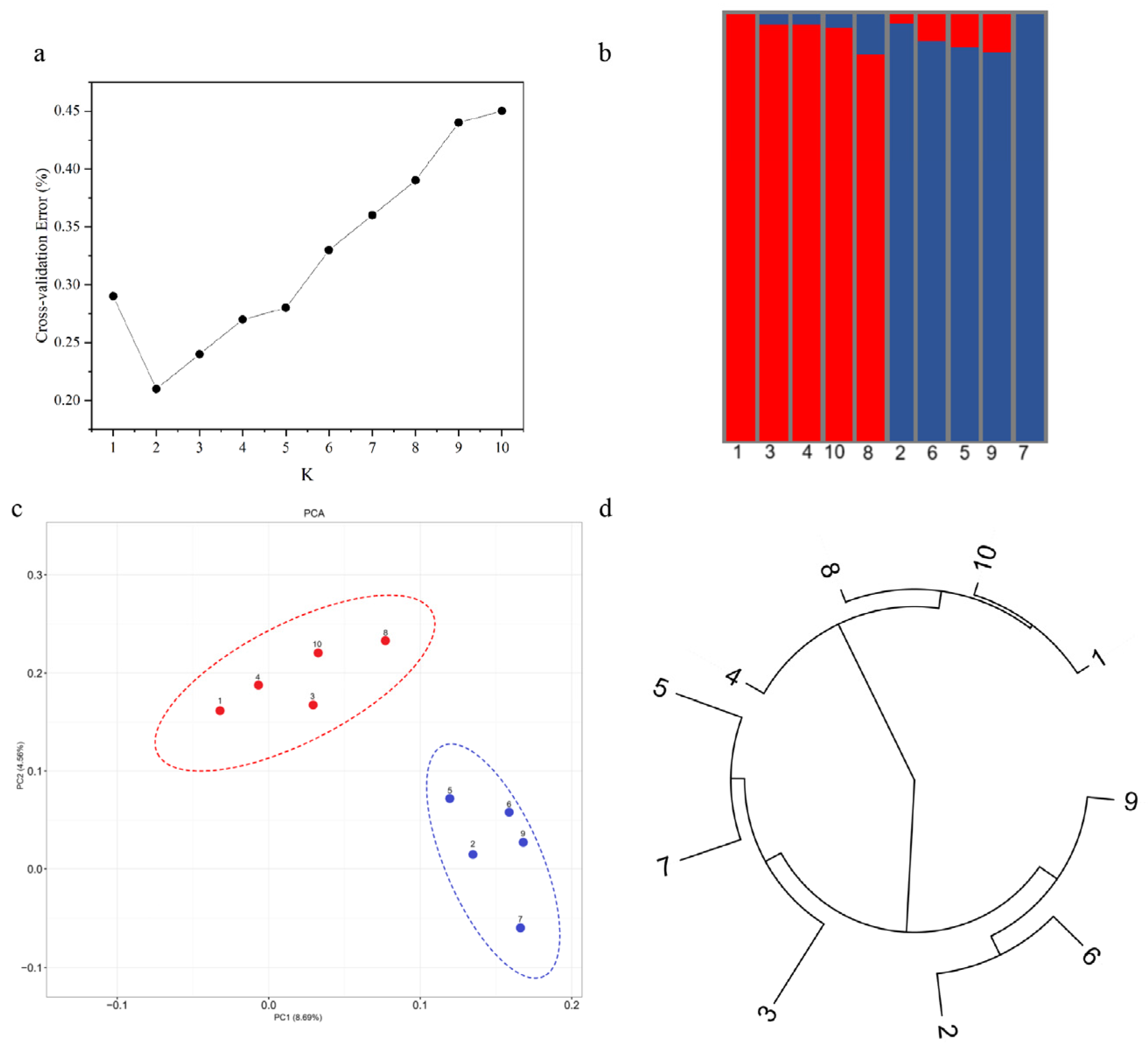
3.7. GBS Simplified Genome Analysis
4. Discussion
4.1. The Response of Leaf Color Change and Pigment Content in HJWL and XJ
4.2. Effect of Chloroplast Ultrastructure on Leaf Color Change
4.3. Effects of Chlorophyll Fluorescence, Photosynthesis and Metabolites on Leaf Color
4.4. Analysis of Leaf Focal Edge in XJ
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agenda Academiae Sinicae Edita. Florae Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1959–2004. [Google Scholar]
- Tian, Y.; Rao, S.; Li, Q.; Xu, M.; Wang, A.; Zhang, H.; Chen, J. The coloring mechanism of a novel golden variety in Populus deltoides based on the RGB color mode. For. Res. 2021, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Bai, P.; Gu, K.; Yang, S.; Lin, H.; Shi, C.; Zhao, Y. Dynamic transcriptome and network-based analysis of yellow leaf mutant Ginkgo biloba. BMC Plant Biol. 2022, 22, 465–478. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, J.; Wang, T.; Gao, F.; Wang, G. Metabolomic and transcriptomic analyses of mutant yellow leaves provide insights into pigment synthesis and metabolism in Ginkgo biloba. BMC Genom. 2020, 21, 858. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; He, Z.; Yang, S.; Ye, Y.; Jiang, H.; Wang, L. Construction and analysis of a library of miRNA in gold-coloured mutant leaves of Ginkgo biloba L. Folia Hortic. 2019, 1, 81–92. [Google Scholar] [CrossRef]
- Flexas, J.; Clemente, M.J.; Bota, J.; Brodribb, T.J.; Gago, J.; Mizokami, Y.; Nadal, M.; Perera, A.V.; Roig, M.; Sugiura, D.; et al. Cell wall thickness and composition are involved in photosynthetic limitation. J. Exp. Bot. 2021, 72, 144–156. [Google Scholar] [CrossRef]
- Li, W.; Yang, S.; Lu, Z.; He, Z.; Ye, Y.; Zhao, B.; Wang, L.; Jin, B. Publisher correction: Cytological, physiological, and transcriptomic analyses of golden leaf coloration in Ginkgo biloba L. Hortic. Res. 2018, 5, 5–32. [Google Scholar] [CrossRef]
- Yang, W.; Xu, H.; Xiao, Q.; Li, X.; Shao, Q. Combined analysis of metabolome and transcriptome provides insights into metabolisms of chlorophylls, carotenoids, and flavonoids in the yellowing leaves of ‘HAES344′ macadamia. Sci. Hortic. 2023, 308, 111600. [Google Scholar] [CrossRef]
- Li, S.; Li, Q.; Tang, L.; Wen, J. Pigment comparison and expression of chlorophyll metabolism genes in yellow and green Acer palmatum Thunb. ex Murray leaves. Can. J. Plant Sci. 2017, 97, 775–783. [Google Scholar] [CrossRef]
- Shao, G.; Liu, R.; Qian, Z.; Zhang, H.; Hu, Q.; Zhu, Y.; Chen, S.; Chen, F.; Jiang, J.; Wang, L. Transcriptome analysis reveals genes respond to chlorophyll deficiency in green and yellow leaves of Chrysanthemum morifolium Ramat. Horticulturae 2022, 8, 14. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, P.; Fan, X.; Yang, H. Characterization and RNA-Seq analysis of yellow-green leaf mutants in Tomato. Agronomy. 2024, 14, 828. [Google Scholar] [CrossRef]
- Li, B.; Zhang, J.; Tian, P.; Gao, X.; Song, X.; Pan, X.; Wu, Y. Cytological, physiological, and transcriptomic analyses of the leaf color mutant yellow leaf 20 (yl20) in eggplant (Solanum melongena L.). Plants 2024, 13, 855. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Wang, Y.; Xiao, Y.; Yang, J.; Wang, R.; Jiang, Y.; Huang, R.; Liu, X.; Jiang, Y. Relationships between leaf color changes, pigment levels, enzyme activity, photosynthetic fluorescence characteristics and chloroplast ultrastructure of Liquidambar formosana Hance. J. For. Res. 2022, 33, 1559–1572. [Google Scholar] [CrossRef]
- Van, K.O.; Snel, J.F. The use of chlorophyll fluorescence nomenclature in plant stress physiology. Photosynth. Res. 1990, 25, 147–150. [Google Scholar] [CrossRef]
- Junker, L.V.; Ensminger, I. Relationship between leaf optical properties, chlorophyll fluorescence and pigment changes in senescing Acer saccharum leaves. Tree Physiol. 2016, 36, 694–711. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, Y.; Xu, M.; Zhao, M.; Chen, H.; Lu, Y.; Pang, S.; Xu, W. Characterization of low-temperature sensitivity and chlorophyll fluorescence in yellow leaf mutants of Tomato. Agronomy 2024, 14, 2382. [Google Scholar] [CrossRef]
- Zhang, K.; Liu, Z.; Shan, X.; Li, C.; Tang, X.; Chi, M.; Feng, H. Physiological properties and chlorophyll biosynthesis in a Pak-choi (Brassica rapa L. ssp. chinensis) yellow leaf mutant, pylm. Acta Physiol. Plant. 2017, 39, 22. [Google Scholar] [CrossRef]
- Guo, J.; Wu, Y.; Jiang, M.; Wu, C.; Wang, G. An LC-MS-based metabolomic approach provides insights into the metabolite profiles of Ginkgo biloba L. at different developmental stages and in various organs. Food Res. Int. 2022, 159, 111644. [Google Scholar] [CrossRef]
- Jurčević Šangut, I.; Šamec, D. Seasonal variation of polyphenols and pigments in Ginkgo (Ginkgo biloba L.) leaves: Focus on 3′,8″-Biflavones. Plants 2024, 13, 3044. [Google Scholar] [CrossRef]
- Hu, Y.; Šmarda, P.; Liu, G.; Wang, B.; Gao, X.; Guo, Q. High-depth transcriptome reveals differences in natural haploid Ginkgo biloba L. due to the effect of reduced gene dosage. Int. J. Mol. Sci. 2022, 23, 8958. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, Y.; Šmarda, P.; Bureš, P.; Guo, Q. Transcriptome and proteome associated analysis of flavonoid metabolism in haploid Ginkgo biloba. Int. J. Biol. Macromol. 2023, 224, 306–318. [Google Scholar] [CrossRef]
- Shi, J.; Zou, X.; Zhao, J.; Holmes, M.; Wang, K.; Wang, X.; Chen, H. Determination of total flavonoids content in fresh Ginkgo biloba leaf with different colors using near infrared spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2012, 94, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, J.; Tang, C.; Anthony, U.V.; Xu, L.; Zhang, F.; Sun, J.; Chen, Y. Flavonoids-targeted metabolomic analysis following rice yellowing. Food Chem. 2023, 430, 136984. [Google Scholar] [CrossRef] [PubMed]
- Lenucci, M.S.; Leucci, M.R.; Piro, G.; Dalessandro, G. Variability in the content of soluble sugars and cell wall polysaccharides in red-ripe cherry and high-pigment tomato cultivars. J. Sci. Food Agric. 2008, 88, 1837–1844. [Google Scholar] [CrossRef]
- Simões, R.; Rodrigues, A.; Ferreira-Dias, S.; Miranda, I.; Pereira, H. Chemical composition of cuticular waxes and pigments and morphology of leaves of Quercus suber trees of different provenance. Plants 2020, 9, 1165. [Google Scholar] [CrossRef]
- Hwang, K.; Oh, S.; Kim, K.; Han, H.; Oh, Y.; Lim, H.; Kim, Y.; Kim, D. Genotyping-by-sequencing approaches using optimized two-enzyme combinations in Asian pears (Pyrus spp.). Mol. Breed. 2019, 39, 484–501. [Google Scholar] [CrossRef]
- Hu, Y.; Yu, Z.; Gao, X.; Liu, G.; Zhang, Y.; Šmarda, P.; Guo, Q. Genetic diversity, population structure, and genome-wide association analysis of ginkgo cultivars. Hortic. Res. 2023, 10, 136. [Google Scholar] [CrossRef]
- Hu, Y.; Hu, Q.; Ma, X.; Zhang, X.; Zheng, J. Population genetics and origin of horticultural germplasm in Clematis via genotyping-by-sequencing. Hortic. Res. 2024, 336, uhae336. [Google Scholar] [CrossRef]
- Yuan, G.; Sun, B.; Yuan, J.; Wang, Q. Effect of 1-methylcyclopropene on shelf life, visual quality, antioxidant enzymes and health-promoting compounds in broccoli florets. Food Chem. 2010, 118, 774–781. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, J.; Shen, N.; Li, N.; Cui, Y.; Xin, H. Phytochemical constituents from Ginkgo biloba exocarp. Chem. Nat. Compd. 2022, 58, 926–928. [Google Scholar] [CrossRef]
- Sun, B.; Tian, Y.; Jiang, M.; Yuan, Q.; Chen, Q.; Zhang, Y.; Luo, Y.; Zhang, F.; Tang, H. Variation in the main health-promoting compounds and antioxidant activity of whole and individual edible parts of baby mustard (Brassica juncea var. gemmifera). RSC Adv. 2018, 8, 33845–33854. [Google Scholar] [CrossRef]
- Carelli, A.A.; Frizzera, L.M.; Forbito, P.R.; Crapiste, G.H. Wax composition of sunflower seed oils. J. Am. Oil Chem. Soc. 2002, 79, 763–768. [Google Scholar] [CrossRef]
- Doyle, J.J. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Wang, G.; Cui, P.; Wu, S.; Ai, C.; Hu, N.; Li, A.; He, B.; Shao, X. The nearly complete genome of Ginkgo biloba illuminates gymnosperm evolution. Nat. Plants 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Van, D.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del, A.G.; Levy, M.A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From fastQ data to high confidence variant calls: The genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 101–103. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. Annovar: Functional annotation of genetic variants from high throughput sequencing data. Nucleic Acids Res. 2010, 38, 164–174. [Google Scholar] [CrossRef]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Li, H.; Durbin, R.; Birney, E. Ensembl compara genetrees: Complete, duplicationaware phylogenetic trees in vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Wu, Y.; Li, X.; Wang, T.; Guo, J.; Cao, F.; Wang, G. Ginkgo biloba microRNA profiling reveals new insight into leaf color mutation. Sci. Hortic. 2020, 265, 109189. [Google Scholar] [CrossRef]
- Li, J.; Wu, K.; Li, L.; Wang, M.; Fang, L.; Zeng, S. Cytological, biochemical, and transcriptomic analyses of a novel yellow leaf variation in a Paphiopedilum (Orchidaceae) SCBG COP15. Genes 2022, 13, 71. [Google Scholar] [CrossRef]
- Murray, D.L.; Kohorn, B.D. Chloroplasts of Arabidopsis thaliana homozygous for the ch-1 locus lack chlorophyll b, lack stable LHCPII and have stacked thylakoids. Plant Mol. Biol. 1991, 16, 71–79. [Google Scholar] [CrossRef]
- Falbel, T.G.; Meehl, J.B.; Staehelin, L.A. Severity of mutant phenotype in a series of chlorophyll deficient wheat mutants depends on light intensity and the severity of the block in chlorophyll synthesis. Plant Physiol. 1996, 112, 821–832. [Google Scholar] [CrossRef]
- Terao, T.; Katoh, S. Antenna sizes of photosystem I and photosystem II in chlorophyll b-deficient mutants of rice. evidence for an antenna function of photosystem II centers that are inactive in electron transport. Plant Cell Physiol. 1996, 37, 307–312. [Google Scholar] [CrossRef]
- Ladygin, V.G. Pigment composition and photosynthetic activity of pea chlorophyll mutants. Izv. Akad. Nauk. Ser. Biol. 2003, 30, 370–376. [Google Scholar] [CrossRef]
- Zou, Y.; Huang, Y.; Zhang, D.; Chen, H.; Liang, Y.; Hao, M.; Yin, Y. Molecular mechanisms of chlorophyll deficiency in Ilex×attenuata ‘Sunny Foster’ Mutant. Plants 2024, 13, 1284. [Google Scholar] [CrossRef]
- Aguilar-Méndez, E.D.; Monribot-Villanueva, J.L.; Guerrero-Analco, J.A.; De-la-Peña, C. Chlorophyll deficiency in Agave angustifolia Haw.: Unveiling the impact on secondary metabolite production. Planta 2024, 260, 77. [Google Scholar] [CrossRef]
- Zhu, P.; Zeng, M.; Lin, Y.; Tang, Y.; He, T.; Zheng, Y.; Chen, L. Variability in Leaf Color Induced by Chlorophyll Deficiency: Transcriptional Changes in Bamboo Leaves. Curr. Issues Mol. Biol. 2024, 46, 1503–1515. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, X.; Wang, J.; Wan, J. Leaf chloroplast ultrastructure and photosynthetic properties of a chlorophyll-deficient mutant of rice. Int. J. Photosynth. Res. 2014, 52, 217–222. [Google Scholar] [CrossRef]
- Nie, L.; Zheng, Y.; Zhang, L.; Wu, Y.; Zhu, S.; Hou, J.; Chen, G.; Tang, X.; Wang, C.; Yuan, L. Characterization and transcriptomic analysis of a novel yellow-green leaf wucai (Brassica campestris L.) germplasm. BMC Genom. 2021, 22, 258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ruan, R.; Zhao, F.; Li, C.; Fu, Q. Comprehensive transcriptome analysis discovers genes related to leaf color in Cymbidium ‘Sakura’ etiolated mutant. Acta Physiol. Plant 2023, 45, 12. [Google Scholar] [CrossRef]
- Qi, X.; Chen, S.; Wang, H.; Feng, J.; Chen, Z.; Qin, Z.; Deng, Y. Comparative physiology and transcriptome analysis reveals that chloroplast development influences silver-white leaf color formation in Hydrangea macrophylla var. maculata. BMC Plant Biol. 2022, 22, 345. [Google Scholar] [CrossRef]
- Schmitz-Linneweber, C.; Williams-Carrier, R.E.; Williams-Voelker, P.M.; Kroeger, T.S.; Vichas, A.; Barkan, A. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell. 2006, 18, 2650–2663. [Google Scholar] [CrossRef]
- Zubo, Y.O.; Blakley, I.; Franco-Zorrilla, J.M.; Yamburenko, M.; Solano, R.; Kieber, J.J.; Ann, E.; Loraine, A.E.; Schaller, G.E. Coordination of chloroplast development through the action of the GNC and GLK transcription factor families. Plant Physiol. 2018, 178, 130–147. [Google Scholar] [CrossRef] [PubMed]
- Hikosaka, K. Photosynthesis, chlorophyll fluorescence and photochemical reflectance index in photoinhibited leaves. Funct. Plant Biol. 2021, 48, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Najar, R.; Aydi, S.; Sassi-Aydi, S.; Zarai, A.; Abdelly, C. Effect of salt stress on photosynthesis and chlorophyll fluorescence in Medicago truncatula. Plant Biosyst. 2018, 153, 88–97. [Google Scholar] [CrossRef]
- Pleban, J.R.; Guadagno, C.R.; Mackay, D.S.; Weinig, C.; Ewers, B.E. Rapid chlorophyll a fluorescence light response curves mechanistically inform photosynthesis modeling. Plant Physiol. 2020, 183, 602–619. [Google Scholar] [CrossRef]
- Xing, C.; Wang, S.; Zhang, C.; Guo, T.; Hao, H.; Zhang, Z.; Wang, S.; Shu, J. Effects of leaf scorch on chlorophyll fluorescence characteristics of walnut leaves. J. Plant. Dis. Prot. 2023, 130, 115–124. [Google Scholar] [CrossRef]
- Li, Y.; Gao, H.; Zhang, Z. Effects of environmental and non-environmental factors on dynamic photosynthetic carbon assimilation in leaves under changing light. Plants 2023, 12, 2015. [Google Scholar] [CrossRef]
- Huo, J.; Zhang, N.; Gong, Y.; Bao, Y.; Li, Y.; Zhang, L.; Nie, S. Effects of different light intensity on leaf color changes in a Chinese cabbage yellow cotyledon mutant. Front. Plant Sci. 2024, 15, 1371451. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Tian, X.; Fu, Y.; Jiang, X.; Li, Y.; Wang, G. Influence of light intensity on chloroplast development and pigment accumulation in the wild-type and etiolated mutant plants of Anthurium andraeanum ‘Sonate’. Plant Signal. Behav. 2018, 13, e1482174. [Google Scholar] [CrossRef] [PubMed]
- Yue, C.; Wang, Z.; Yang, P. Review: The effect of light on the key pigment compounds of photosensitive etiolated tea plant. Bot. Stud. 2021, 62, 21. [Google Scholar] [CrossRef]
- Yang, Y.; Wei, F.; Jia, R.; Zhao, X.; Gan, Y.; Ge, H. Differences in leaf anatomy, photosynthesis, and photoprotective strategies in the yellow-green leaf mutant and wild type of Rosa beggeriana Schrenk. Photosynthetica 2020, 58, 1167–1177. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, Y.; Mao, X.; Yu, W.; Lu, J.; Wang, L. Integration of morphological, physiological, cytological, metabolome and transcriptome analyses reveal age inhibited accumulation of flavonoid biosynthesis in Ginkgo biloba leaves. Ind. Crops Prod. 2022, 187, 115405. [Google Scholar] [CrossRef]
- Gao, X.; Hu, Y.; Li, F.; Cao, F.; Guo, Q. Sex identification and male-female differences in Ginkgo biloba hybrid F1 generation seedlings. Forests 2024, 15, 1636. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, F.; Hu, Y.; Jing, W.; Wang, Y.; Gao, X.; Guo, Q. Cytological, Physiological and Genotyping-by-Sequencing Analysis Revealing Dynamic Variation of Leaf Color in Ginkgo biloba L. Horticulturae 2025, 11, 395. https://doi.org/10.3390/horticulturae11040395
Li F, Hu Y, Jing W, Wang Y, Gao X, Guo Q. Cytological, Physiological and Genotyping-by-Sequencing Analysis Revealing Dynamic Variation of Leaf Color in Ginkgo biloba L. Horticulturae. 2025; 11(4):395. https://doi.org/10.3390/horticulturae11040395
Chicago/Turabian StyleLi, Fangdi, Yaping Hu, Wenxuan Jing, Yirui Wang, Xiaoge Gao, and Qirong Guo. 2025. "Cytological, Physiological and Genotyping-by-Sequencing Analysis Revealing Dynamic Variation of Leaf Color in Ginkgo biloba L." Horticulturae 11, no. 4: 395. https://doi.org/10.3390/horticulturae11040395
APA StyleLi, F., Hu, Y., Jing, W., Wang, Y., Gao, X., & Guo, Q. (2025). Cytological, Physiological and Genotyping-by-Sequencing Analysis Revealing Dynamic Variation of Leaf Color in Ginkgo biloba L. Horticulturae, 11(4), 395. https://doi.org/10.3390/horticulturae11040395




