Genome-Wide Identification of the SmHD-zip Genes That Respond to Multiple Ripening-Related Signals in Eggplant Fruit
Abstract
1. Introduction
2. Results
2.1. Identification of SmHD-zip Genes in Solanum melongena
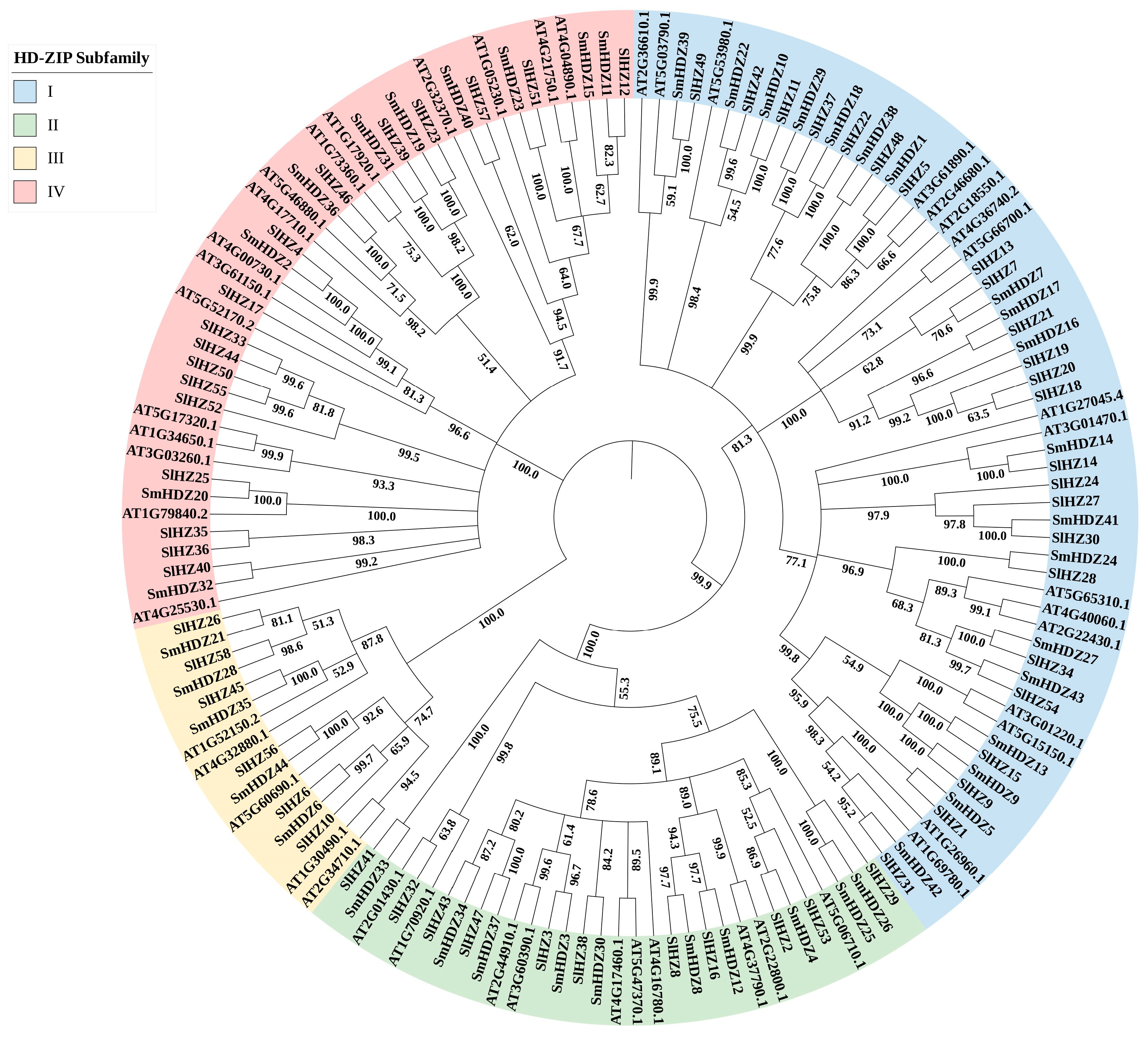
2.2. Characterization and Phylogenetic Reconstruction of the SmHD-zip Gene Family
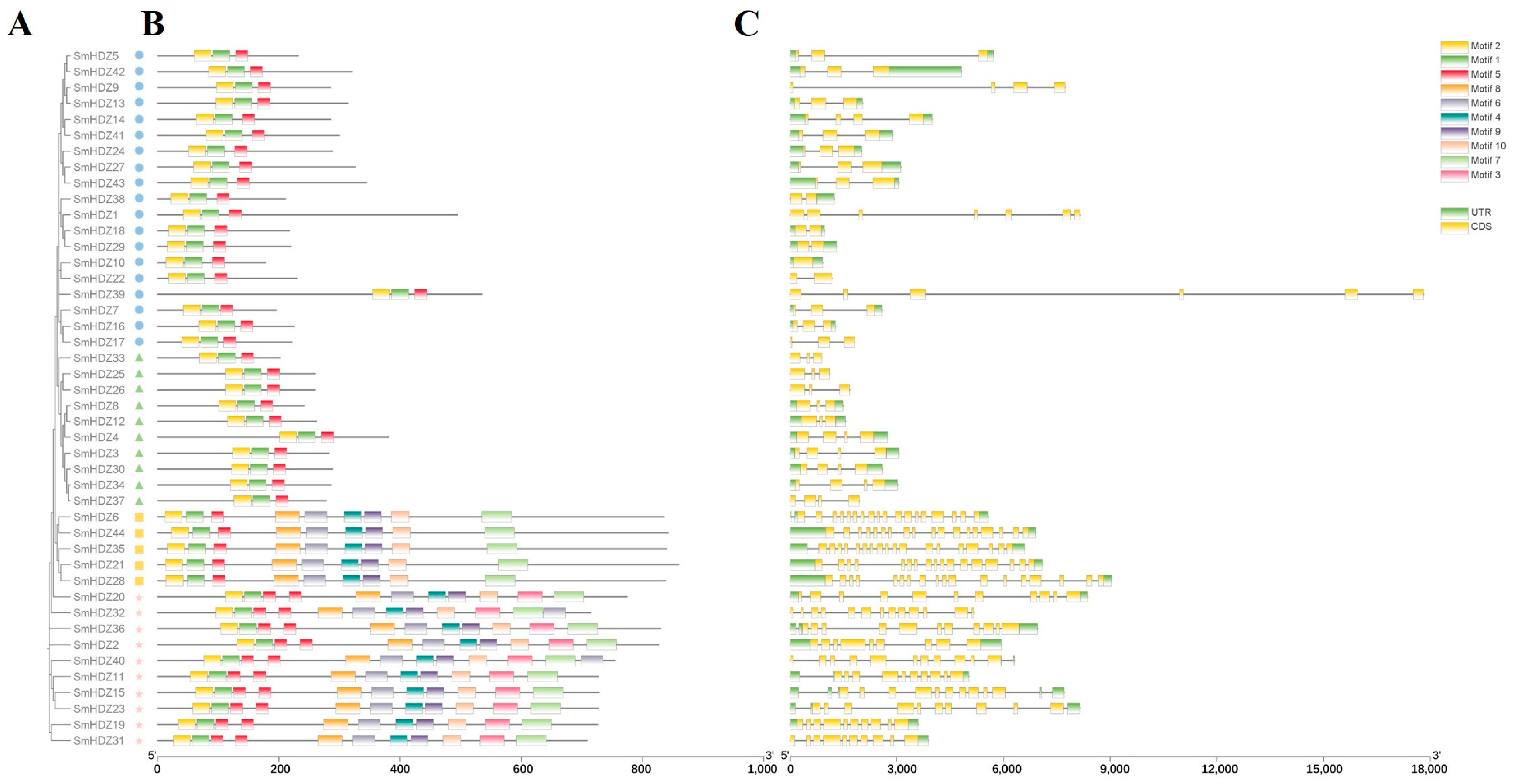
2.3. Gene Structure and Conserved Motif Composition Analysis of SmHD-zip Family
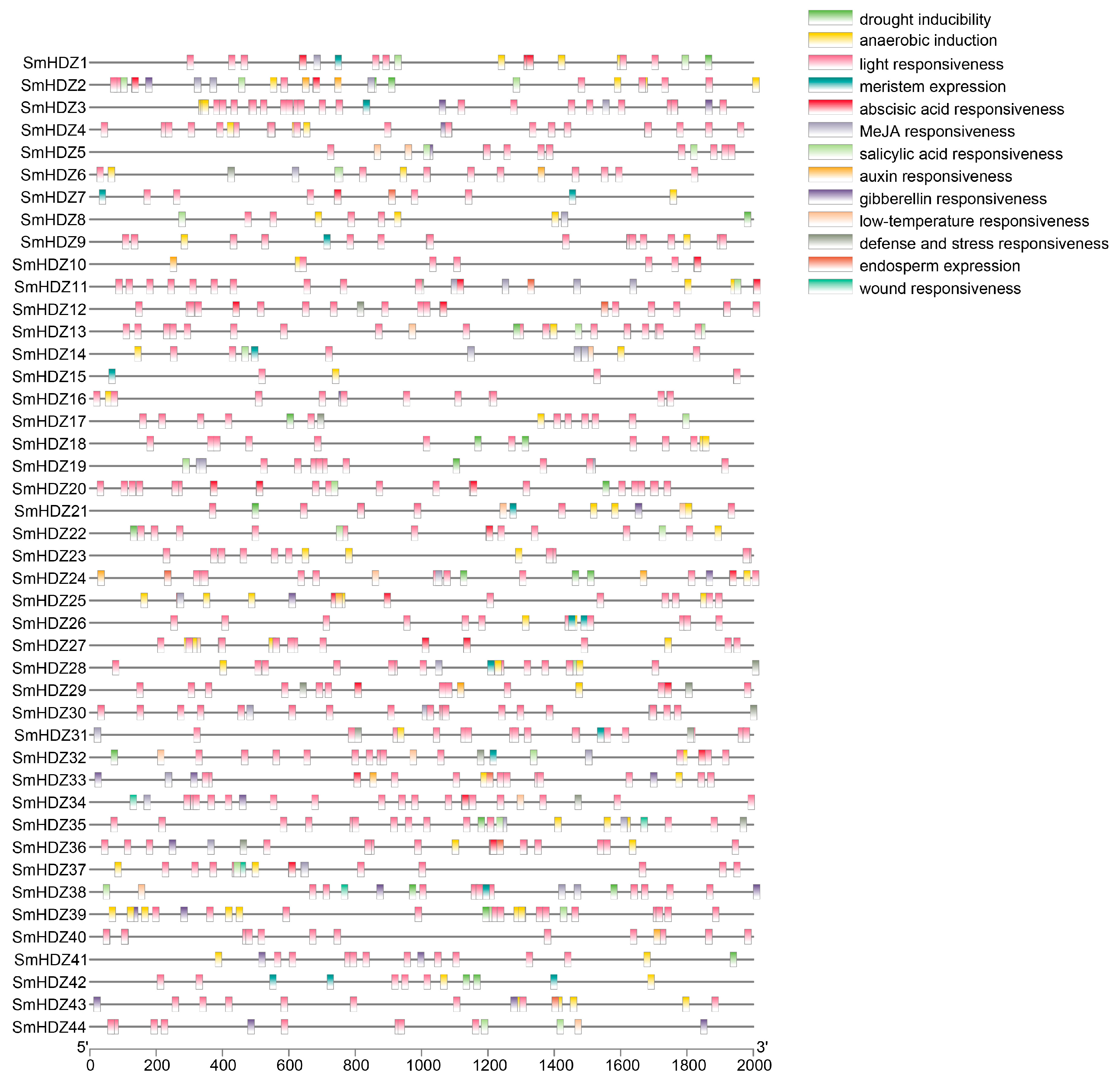
2.4. Cis-Acting Regulatory Elements Analysis in the Promoter Region of SmHD-zips
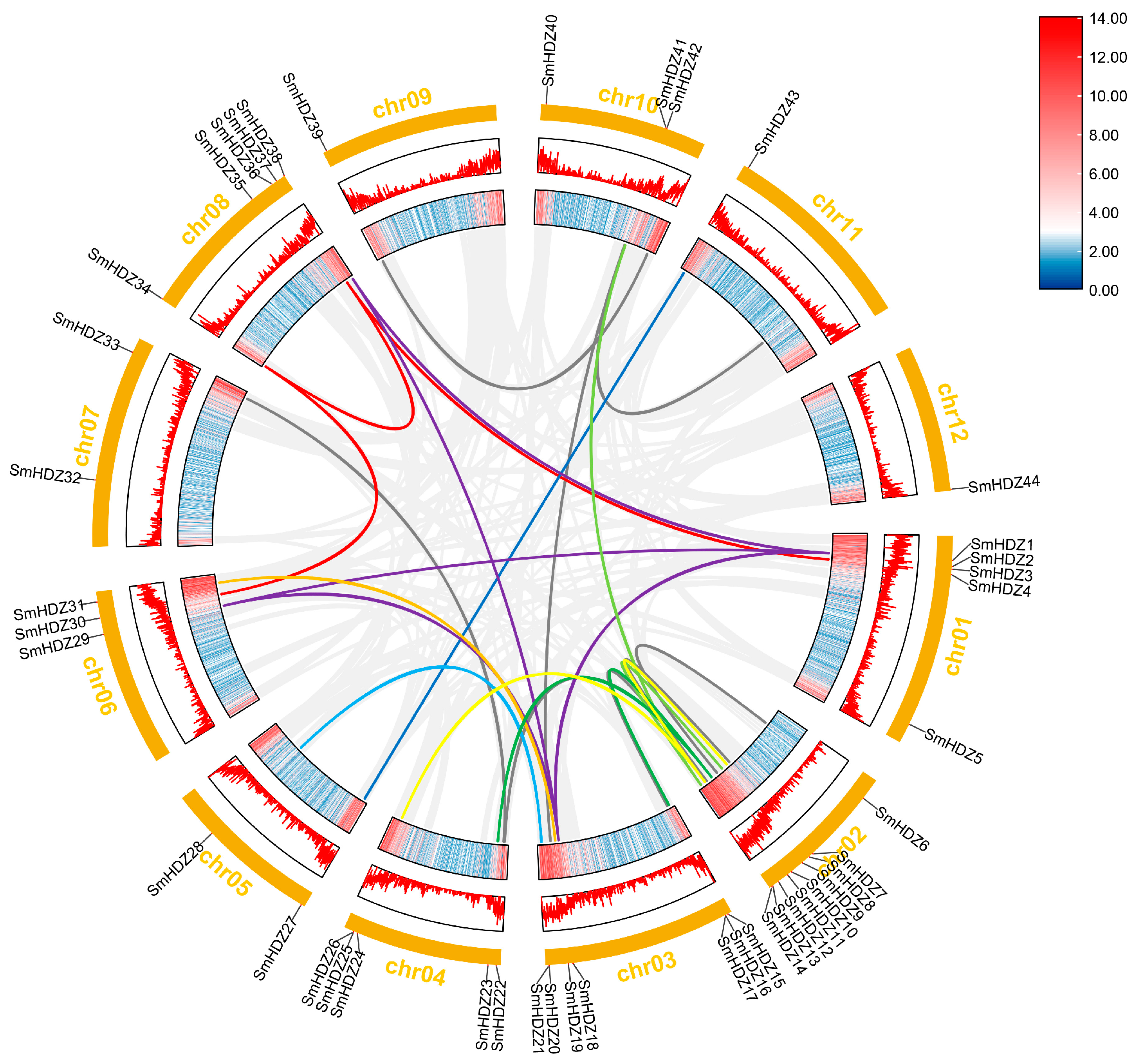
2.5. Gene Duplication Events Analysis of SmHD-zips
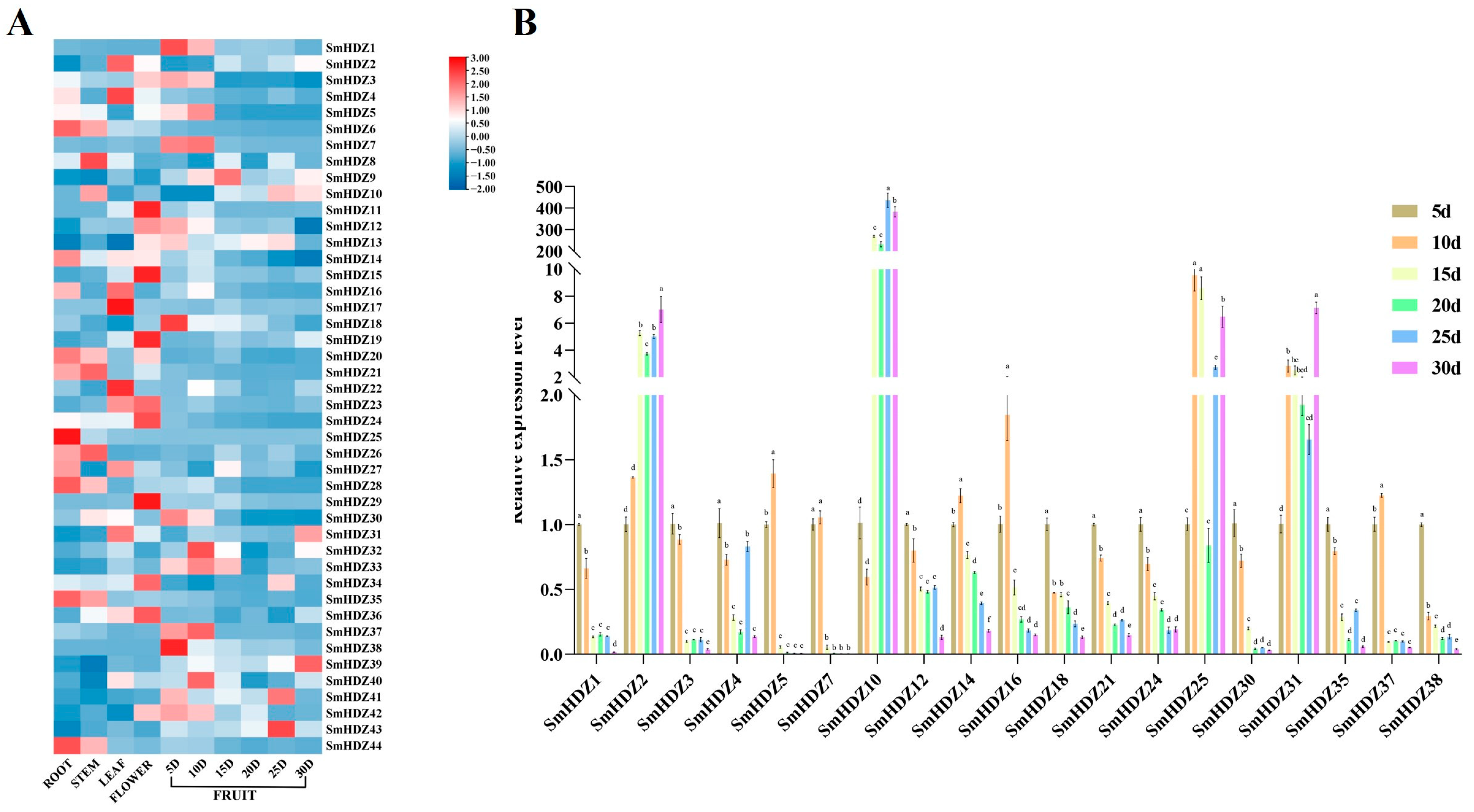
2.6. Expression Analysis of SmHD-zip Genes in Different Tissues and Fruit Ripening
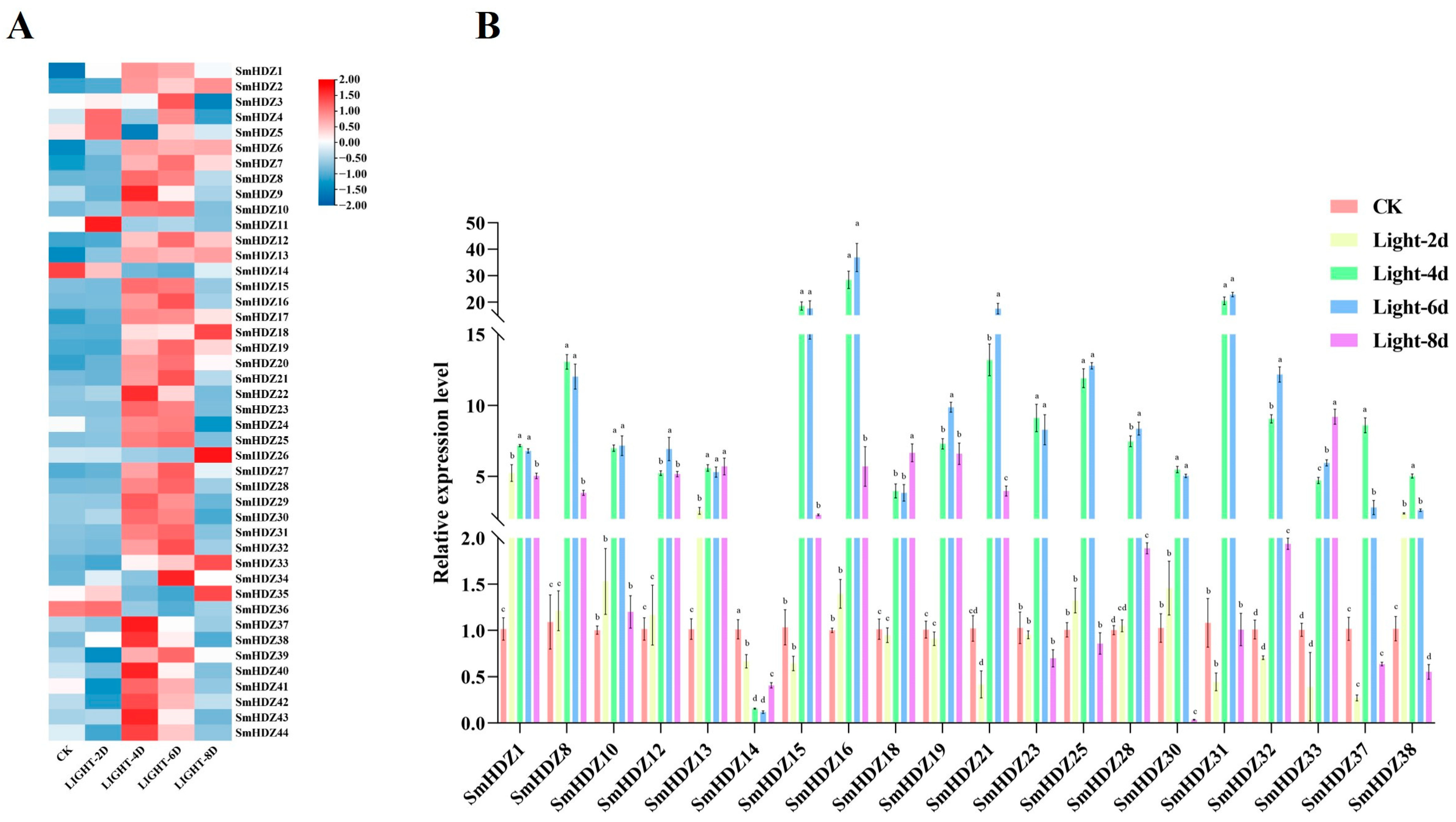
2.7. Regulation of SmHD-zip Gene Expression Under Light Induction
2.8. Expression Analysis of SmHD-zip Genes in Response to Hormone Treatments
3. Discussion
3.1. Evolutionary Analysis of Eggplant HD-zip Genes
3.2. Potential Role of SmHD-zip Genes in Fruit Development and Ripening of Eggplant
3.3. Potential Role of SmHD-zip Genes in Mediating Response to Light and Multiple Hormones
4. Materials and Methods
4.1. Plant Growth Conditions, Light, and Multiple Hormone Treatments
4.2. Genome-Wide Identification of SmHD-zip Genes in Eggplant
4.3. Phylogenetic Tree Analysis and Gene Structure Analysis
4.4. Chromosome Duplication Events and Cis-Acting Elements Analysis
4.5. Quantitative Real-Time RT-PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, W.; Dong, J.Y.; Cao, M.X.; Gao, X.X.; Wang, D.D.; Liu, B.L.; Chen, Q. Genome-wide identification and characterization of HD-ZIP genes in potato. Gene 2019, 697, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis Among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wei, X.B.; Lai, D.L.; Yang, H.; Feng, L.; Li, L.; Niu, K.X.; Chen, L.; Xiang, D.B.; Ruan, J.J.; et al. Genome-wide investigation of the GRAS transcription factor family in foxtail millet (Setaria italica L.). BMC Plant Biol. 2021, 21, 508. [Google Scholar] [CrossRef]
- He, G.H.; Liu, P.; Zhao, H.X.; Sun, J.Q. The HD-ZIP II Transcription Factors Regulate Plant Architecture through the Auxin Pathway. Int. J. Mol. Sci. 2020, 21, 3250. [Google Scholar] [CrossRef]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Sessa, G.; Carabelli, M.; Possenti, M.; Morelli, G.; Ruberti, I. Multiple Links between HD-zip Proteins and Hormone Networks. Int. J. Mol. Sci. 2018, 19, 4047. [Google Scholar] [CrossRef]
- Mukherjee, K.; Bürglin, T.R. MEKHLA, a novel domain with similarity to PAS domains, is fused to plant homeodomain-leucine zipper III proteins. Plant Physiol. 2006, 140, 1142–1150. [Google Scholar] [CrossRef]
- Mukherjee, K.; Brocchieri, L.; Bürglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Sessa, G.; Morelli, G.; Ruberti, I. The Athb-1 and -2 HD-zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993, 12, 3507–3517. [Google Scholar] [CrossRef]
- Tron, A.E.; Bertoncini, C.W.; Palena, C.M.; Chan, R.L.; Gonzalez, D.H. Combinatorial interactions of two amino acids with a single base pair define target site specificity in plant dimeric homeodomain proteins. Nucleic Acids Res. 2001, 29, 4866–4872. [Google Scholar] [CrossRef]
- Chan, R.L.; Gago, G.M.; Palena, C.M.; Gonzalez, D.H. Homeoboxes in plant development. Biochim. Biophys. Acta 1998, 1442, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Schrick, K.; Nguyen, D.; Karlowski, W.M.; Mayer, K.F.X. START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 2004, 5, R41. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.E. Shoot meristem function and leaf polarity: The role of class III HD-ZIP genes. PLoS Genet. 2006, 2, e89. [Google Scholar] [CrossRef] [PubMed]
- Sessa, G.; Steindler, C.; Morelli, G.; Ruberti, I. The Arabidopsis Athb-8, -9 and -14 genes are members of a small gene family coding for highly related HD-ZIP proteins. Plant Mol. Biol. 1998, 38, 609–622. [Google Scholar] [CrossRef]
- Abe, M.; Takahashi, T.; Komeda, Y. Identification of a cis-regulatory element for L1 layer-specific gene expression, which is targeted by an L1-specific homeodomain protein. Plant J. 2001, 26, 487–494. [Google Scholar] [CrossRef]
- Jiang, H.; Jin, J.; Liu, H.; Dong, Q.; Yan, H.; Gan, D.; Zhang, W.; Zhu, S. Genome-wide analysis of HD-zip genes in grape (Vitis vinifera). Tree Genet. Genomes 2015, 11, 827. [Google Scholar] [CrossRef]
- Wei, M.Y.; Liu, A.L.; Zhang, Y.J.; Zhou, Y.; Li, D.H.; Dossa, K.; Zhou, R.; Zhang, X.R.; You, J. Genome-wide characterization and expression analysis of the HD-zip gene family in response to drought and salinity stresses in sesame. BMC Genom. 2019, 20, 748. [Google Scholar] [CrossRef]
- Li, Y.X.; Yang, Z.R.; Zhang, Y.Y.; Guo, J.J.; Liu, L.L.; Wang, C.F.; Wang, B.S.; Han, G.L. The roles of HD-ZIP proteins in plant abiotic stress tolerance. Front. Plant Sci. 2022, 13, 1027071. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Shan, W.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. Four HD-ZIPs are involved in banana fruit ripening by activating the transcription of ethylene biosynthetic and cell wall-modifying genes by activating the transcription of ethylene biosynthetic and cell wall-modifying genes. Plant Cell Rep. 2020, 39, 351–362. [Google Scholar] [CrossRef]
- Li, C.Q.; Zhao, M.L.; Ma, X.S.; Wen, Z.X.; Ying, P.Y.; Peng, M.J.; Ning, X.P.; Xia, R.; Wu, H.; Li, J.G. The HD-zip transcription factor LcHB2 regulates litchi fruit abscission through the activation of two cellulase genes. J. Exp. Bot. 2019, 70, 5189–5203. [Google Scholar] [CrossRef]
- Gu, C.; Guo, Z.H.; Cheng, H.Y.; Zhou, Y.H.; Qi, K.J.; Wang, G.M.; Zhang, S.L. A HD-ZIP II HOMEBOX transcription factor, PpHB.G7, mediates ethylene biosynthesis during fruit ripening in peach. Plant Sci. 2019, 278, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Bor, J.Y.; Chen, H.Y.; Yen, G.C. Evaluation of antioxidant activity and inhibitory effect on nitric oxide production of some common vegetables. J. Agric. Food Chem. 2006, 54, 1680–1686. [Google Scholar] [CrossRef] [PubMed]
- Wei, Q.Z.; Wang, J.L.; Wang, W.H.; Hu, T.H.; Hu, H.J.; Bao, C.L. A high-quality chromosome-level genome assembly reveals genetics for important traits in eggplant. Hortic. Res. 2020, 7, 153. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, J.; Zhou, X.H.; Liu, S.Y.; Zhuang, Y. Identification of WRKY gene family and characterization of cold stress-responsive WRKY genes in eggplant. PeerJ 2020, 8, e8777. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Li, X.Y.; Mo, Y.R.; Jiang, C.Q.; Zhou, Y.; Hu, J.Y.; Zhang, Y.L.; Lv, J.H.; Zhao, K.; Lu, Z.Y. Identification and expression profiling of SmGATA genes family involved in response to light and phytohormones in eggplant. Front. Plant Sci. 2024, 15, 1415921. [Google Scholar] [CrossRef]
- Li, D.L.; He, Y.J.; Li, S.H.; Shi, S.L.; Li, L.Z.; Liu, Y.; Chen, H.Y. Genome-wide characterization and expression analysis of AP2/ERF genes in eggplant (Solanum melongena L.). Plant Physiol. Biochem. 2021, 167, 492–503. [Google Scholar] [CrossRef]
- He, Y.J.; Chen, H.; Zhou, L.; Liu, Y.; Chen, H.Y. Comparative transcription analysis of photosensitive and non-photosensitive eggplants to identify genes involved in dark regulated anthocyanin synthesis. BMC Genom. 2019, 20, 678. [Google Scholar] [CrossRef]
- Ohyama, A.; Yamaguchi, H.; Miyatake, K.; Negoro, S.; Nunome, T.; Saito, T.; Fukuoka, H. Genetic mapping of simply inherited categorical traits, including anthocyanin accumulation profiles and fruit appearance, in eggplant (Solanum melongena). Mol. Biol. Rep. 2022, 49, 9147–9157. [Google Scholar] [CrossRef]
- Sharif, R.; Xie, C.; Wang, J.; Cao, Z.; Zhang, H.Q.; Chen, P.; Li, Y.H. Genome wide identification, characterization and expression analysis of HD-ZIP gene family in Cucumis sativus L. under biotic and various abiotic stresses. Int. J. Biol. Macromol. 2020, 158, 502–520. [Google Scholar] [CrossRef]
- Li, Y.; Xiong, H.Y.; Cuo, D.J.; Wu, X.X.; Duan, R.J. Genome-wide characterization and expression profiling of the relation of the HD-zip gene family to abiotic stress in barley (Hordeum vulgare L.). Plant Physiol. Biochem. 2019, 141, 250–258. [Google Scholar] [CrossRef]
- Zhang, Z.Z.; Chen, X.L.; Guan, X.; Liu, Y.; Chen, H.Y.; Wang, T.T.; Mouekouba, L.D.O.; Li, J.F.; Wang, A.X. A genome-wide survey of homeodomain-leucine zipper genes and analysis of cold-responsive HD-zip I members’ expression in tomato. Biosci. Biotechnol. Biochem. 2014, 78, 1337–1349. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.B.; Chi, X.Y.; Chai, G.H.; Kong, Y.Z.; He, G.; Wang, X.Y.; Shi, D.C.; Zhang, D.Y.; Zhou, G.K. Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS ONE 2017, 7, e31149. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Li, H.; Teng, R.M.; Wang, Y.X.; Wang, W.L.; Zhuang, J. Genomic and transcriptomic analyses of HD-zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Genomics 2019, 111, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.H.; Wang, J.; Bao, X.X.; Liang, M.Y.; Lou, H.M.; Zhao, J.W.; Sun, M.T.; Liang, J.; Jin, L.S.; Li, G.L.; et al. Genome-wide identification and expression profile of HD-ZIP genes in physic nut and functional analysis of the JcHDZ16 gene in transgenic rice. BMC Plant Biol. 2019, 19, 298. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Sun, X.F.; Zhang, Q.; Song, P.Y.; Hu, Q.M.; Zhang, X.J.; Li, X.; Hu, J.B.; Pan, J.S.; Sun, S.R.; et al. GLABROUS (CmGL) encodes a HD-ZIP IV transcription factor playing roles in multicellular trichome initiation in melon. Theor. Appl. Genet. 2018, 131, 569–579. [Google Scholar] [CrossRef]
- Zhang, Z.R.; Zhu, R.R.; Ji, X.H.; Li, H.J.; Lv, H.; Zhang, H.Y. Genome-Wide Characterization and Expression Analysis of the HD-ZIP Gene Family in Response to Salt Stress in Pepper. Int. J. Genom. 2021, 2021, 8105124. [Google Scholar] [CrossRef]
- Yang, G.B.; Li, L.J.; Wei, M.; Li, J.; Yang, F.J. SmMYB113 is a key transcription factor responsible for compositional variation of anthocyanin and color diversity among eggplant peels. Front. Plant Sci. 2022, 13, 843996. [Google Scholar] [CrossRef]
- Lin, Z.F.; Hong, Y.G.; Yin, M.A.; Li, C.Y.; Zhang, K.; Grierson, D. A tomato HD-zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008, 55, 301–310. [Google Scholar] [CrossRef]
- Jiang, Y.H.; Liu, C.H.; Yan, D.; Wen, X.H.; Liu, Y.L.; Wang, H.J.; Dai, J.Y.; Zhang, Y.J.; Liu, Y.F.; Zhou, B.; et al. MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J. Exp. Bot. 2017, 68, 1055–1069. [Google Scholar] [CrossRef]
- Kubo, H.; Peeters, A.J.M.; Aarts, M.G.M.; Pereira, A.; Koornneef, M. ANTHOCYANINLESS2, a Homeobox Gene Affecting Anthocyanin Distribution and Root Development in Arabidopsis. Plant Cell 1999, 11, 1217–1226. [Google Scholar] [CrossRef]
- Li, F.; Fu, M.; Zhou, S.; Xie, Q.; Chen, G.; Chen, X.; Hu, Z. A tomato HD-zip I transcription factor, VAHOX1, acts as a negative regulator of fruit ripening. Hortic. Res. 2023, 10, uhac236. [Google Scholar] [CrossRef] [PubMed]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.; Puente, P.; Deng, X.W.; Wei, N. Combinatorial interaction of light-responsive elements plays a critical role in determining the response characteristics of light-regulated promoters in Arabidopsis. Plant J. 1998, 15, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Han, Q.; Kang, X.; Tan, W.; Yao, X.; Zhang, Y.; Shi, H.; Xia, R.; Wu, X.; Lin, H.; et al. The HAT1 transcription factor regulates photomorphogenesis and skotomorphogenesis via phytohormone levels. Plant Physiol. 2024, 197, kiae542. [Google Scholar] [CrossRef]
- Zheng, T.; Tan, W.R.; Yang, H.; Zhang, L.E.; Li, T.T.; Liu, B.H.; Zhang, D.W.; Lin, H.H. Regulation of anthocyanin accumulation via MYB75/HAT1/TPL-mediated transcriptional repression. PLoS Genet. 2019, 15, e1007993. [Google Scholar] [CrossRef]
- Li, S.H.; Dong, Y.X.; Li, D.L.; Shi, S.L.; Zhao, N.; Liao, J.L.; Liu, Y.; Chen, H.Y. Eggplant transcription factor SmMYB5 integrates jasmonate and light signaling during anthocyanin biosynthesis. Plant Physiol. 2024, 194, 1139–1165. [Google Scholar] [CrossRef]
- Wang, Q.Q.; Zha, K.Y.; Chai, W.B.; Wang, Y.; Liu, B.; Jiang, H.Y.; Cheng, B.J.; Zhao, Y. Functional analysis of the HD-zip I gene ZmHDZ1 in ABA-mediated salt tolerance in rice. J. Plant Biol. 2017, 60, 207–214. [Google Scholar] [CrossRef]
- Himmelbach, A.; Hoffmann, T.; Leube, M.; Höhener, B.; Grill, E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002, 21, 3029–3038. [Google Scholar] [CrossRef]
- Zhang, M.X.; Shi, Y.N.; Liu, Z.M.; Zhang, Y.J.; Yin, X.R.; Liang, Z.H.; Huang, Y.Q.; Grierson, D.; Chen, K. An EjbHLH14-EjHB1-EjPRX12 module is involved in methyl jasmonate alleviation of chilling-induced lignin deposition in loquat fruit. J. Exp. Bot. 2022, 73, 1668–1682. [Google Scholar] [CrossRef]
- Cai, Y.L.; Bartholomew, E.S.; Dong, M.M.; Zhai, X.L.; Yin, S.; Zhang, Y.Q.; Feng, Z.X.; Wu, L.C.; Liu, W.; Shan, N.; et al. The HD-ZIP IV transcription factor CsGL2-LIKE regulates male flowering time and fertility in cucumber. J. Exp. Bot. 2020, 71, 5425–5437. [Google Scholar] [CrossRef]
- Rombolá-Caldentey, B.; Rueda-Romero, P.; Iglesias-Fernández, R.; Carbonero, P.; Oñate-Sánchez, L. Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element. Plant Cell 2014, 26, 2905–2919. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Wu, Y.; Xia, R. A painless way to customize Circos plot: From data preparation to visualization using TBtools. iMeta 2022, 1, e35. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]

| Name | Genome ID | Protein Length/aa | Chrom | Chr Start | Chr End | MW (Da) | pI | Aliphatic Index | GRAVY |
|---|---|---|---|---|---|---|---|---|---|
| SmHDZ1 | Smechr0101299.1 | 257 | Chr1 | 12,588,851 | 12,597,004 | 29,162.42 | 5.52 | 63.42 | −0.833 |
| SmHDZ2 | Smechr0101591.1 | 826 | Chr1 | 15,597,095 | 15,603,032 | 90,131.25 | 5.97 | 79.58 | −0.322 |
| SmHDZ3 | Smechr0101697.1 | 282 | Chr1 | 16,935,525 | 16,938,582 | 31,495.45 | 5.97 | 72.98 | −0.743 |
| SmHDZ4 | Smechr0101915.1 | 380 | Chr1 | 19,575,004 | 19,577,744 | 41,258.04 | 8.08 | 62.42 | −0.637 |
| SmHDZ5 | Smechr0103648.1 | 118 | Chr1 | 96,300,845 | 96,306,567 | 13,893.92 | 8.93 | 79.41 | −0.887 |
| SmHDZ6 | Smechr0200129.1 | 907 | Chr2 | 11,092,838 | 11,098,401 | 100,094.30 | 6.58 | 85.71 | −0.226 |
| SmHDZ7 | Smechr0200761.1 | 201 | Chr2 | 47,977,301 | 47,979,883 | 23,518.64 | 6.87 | 64.03 | −0.989 |
| SmHDZ8 | Smechr0200863.1 | 241 | Chr2 | 50,396,475 | 50,397,965 | 27,163.64 | 8.98 | 70.41 | −0.820 |
| SmHDZ9 | Smechr0201080.1 | 255 | Chr2 | 54,789,348 | 54,797,075 | 29,281.66 | 5.76 | 63.49 | −0.922 |
| SmHDZ10 | Smechr0201659.1 | 177 | Chr2 | 61,727,689 | 61,728,610 | 20,516.98 | 7.74 | 77.68 | −0.876 |
| SmHDZ11 | Smechr0201902.1 | 726 | Chr2 | 64,140,549 | 64,145,567 | 79,843.27 | 5.68 | 78.97 | −0.363 |
| SmHDZ12 | Smechr0202551.1 | 261 | Chr2 | 69,757,778 | 69,759,330 | 29,121.72 | 7.60 | 69.16 | −0.731 |
| SmHDZ13 | Smechr0202951.1 | 305 | Chr2 | 73,206,063 | 73,208,102 | 34,869.77 | 5.95 | 56.59 | −0.970 |
| SmHDZ14 | Smechr0203037.1 | 776 | Chr2 | 73,893,789 | 73,897,785 | 85,826.59 | 5.10 | 80.04 | −0.459 |
| SmHDZ15 | Smechr0300196.1 | 728 | Chr3 | 2,373,704 | 2,381,403 | 79,776.37 | 5.55 | 83.83 | −0.305 |
| SmHDZ16 | Smechr0300320.1 | 167 | Chr3 | 3,712,936 | 3,714,206 | 19,768.71 | 9.58 | 80.00 | −0.854 |
| SmHDZ17 | Smechr0300325.1 | 224 | Chr3 | 3,794,580 | 3,796,394 | 25,965.22 | 6.62 | 75.71 | −0.776 |
| SmHDZ18 | Smechr0302263.1 | 216 | Chr3 | 82,786,950 | 82,787,919 | 25,224.17 | 5.17 | 67.27 | −0.992 |
| SmHDZ19 | Smechr0302477.1 | 666 | Chr3 | 85,078,273 | 85,081,875 | 72,747.09 | 5.93 | 83.30 | −0.316 |
| SmHDZ20 | Smechr0303487.1 | 773 | Chr3 | 94,511,653 | 94,520,019 | 86,185.61 | 6.27 | 76.08 | −0.475 |
| SmHDZ21 | Smechr0303520.1 | 782 | Chr3 | 94,710,348 | 94,717,438 | 86,430.14 | 6.14 | 87.20 | −0.120 |
| SmHDZ22 | Smechr0400235.1 | 544 | Chr4 | 2,479,449 | 2,480,634 | 60,623.15 | 6.03 | 116.95 | 0.398 |
| SmHDZ23 | Smechr0400520.1 | 615 | Chr4 | 6,580,098 | 6,588,250 | 68,910.76 | 6.13 | 77.54 | −0.514 |
| SmHDZ24 | Smechr0401924.1 | 287 | Chr4 | 72,800,591 | 72,802,605 | 32,958.41 | 5.16 | 67.25 | −0.956 |
| SmHDZ25 | Smechr0402063.1 | 259 | Chr4 | 74,835,105 | 74,836,211 | 29,803.59 | 8.89 | 79.42 | −0.839 |
| SmHDZ26 | Smechr0402065.1 | 259 | Chr4 | 74,847,989 | 74,849,667 | 29,636.33 | 8.76 | 79.42 | −0.815 |
| SmHDZ27 | Smechr0500181.1 | 348 | Chr5 | 2,131,446 | 2,134,561 | 39,782.86 | 5.00 | 62.79 | −0.851 |
| SmHDZ28 | Smechr0501670.1 | 955 | Chr5 | 58,518,203 | 58,527,247 | 104,964.19 | 6.56 | 89.76 | −0.145 |
| SmHDZ29 | Smechr0601407.1 | 217 | Chr6 | 68,139,079 | 68,140,392 | 25,398.24 | 5.25 | 62.90 | −1.134 |
| SmHDZ30 | Smechr0601803.1 | 278 | Chr6 | 76,285,853 | 76,288,445 | 31,697.12 | 8.49 | 65.61 | −0.826 |
| SmHDZ31 | Smechr0602556.1 | 708 | Chr6 | 84,297,656 | 84,301,539 | 78,026.79 | 6.27 | 85.49 | −0.261 |
| SmHDZ32 | Smechr0700782.1 | 613 | Chr7 | 33,240,754 | 33,245,920 | 67,709.46 | 4.90 | 87.93 | −0.247 |
| SmHDZ33 | Smechr0702033.1 | 212 | Chr7 | 98,824,686 | 98,825,578 | 24,263.76 | 9.28 | 69.01 | −0.867 |
| SmHDZ34 | Smechr0800044.1 | 1484 | Chr8 | 687,257 | 690,287 | 168,338.26 | 5.88 | 91.15 | −0.313 |
| SmHDZ35 | Smechr0801392.1 | 789 | Chr8 | 66,884,601 | 66,891,197 | 87,156.14 | 5.99 | 89.26 | −0.109 |
| SmHDZ36 | Smechr0801916.1 | 819 | Chr8 | 79,167,138 | 79,174,098 | 91,110.10 | 5.14 | 75.91 | −0.403 |
| SmHDZ37 | Smechr0802130.1 | 277 | Chr8 | 81,852,289 | 81,854,237 | 31,346.17 | 7.58 | 62.67 | −0.909 |
| SmHDZ38 | Smechr0802554.1 | 210 | Chr8 | 86,509,323 | 86,510,568 | 24,514.54 | 5.31 | 64.57 | −0.951 |
| SmHDZ39 | Smechr0900151.1 | 490 | Chr9 | 2,257,356 | 2,275,167 | 56,179.77 | 8.03 | 75.98 | −0.483 |
| SmHDZ40 | Smechr1000249.1 | 754 | Chr10 | 2,991,779 | 2,998,093 | 83,615.53 | 5.70 | 79.11 | −0.383 |
| SmHDZ41 | Smechr1001559.1 | 257 | Chr10 | 63,092,964 | 63,095,845 | 30,004.23 | 4.72 | 68.64 | −0.972 |
| SmHDZ42 | Smechr1001580.1 | 123 | Chr10 | 63,609,600 | 63,614,425 | 14,318.16 | 8.55 | 63.58 | −1.166 |
| SmHDZ43 | Smechr1100234.1 | 394 | Chr11 | 2,684,707 | 2,687,766 | 44,368.20 | 4.89 | 66.80 | −0.722 |
| SmHDZ44 | Smechr1201882.1 | 835 | Chr12 | 73,207,126 | 73,214,030 | 91,373.12 | 6.07 | 86.24 | −0.120 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, C.; Mo, Y.; Zhang, H.; Chen, K.; Zhou, Y.; Ma, Z.; Jing, Y.; Liu, Y.; Wang, Y.; Zhao, K. Genome-Wide Identification of the SmHD-zip Genes That Respond to Multiple Ripening-Related Signals in Eggplant Fruit. Horticulturae 2025, 11, 261. https://doi.org/10.3390/horticulturae11030261
Jiang C, Mo Y, Zhang H, Chen K, Zhou Y, Ma Z, Jing Y, Liu Y, Wang Y, Zhao K. Genome-Wide Identification of the SmHD-zip Genes That Respond to Multiple Ripening-Related Signals in Eggplant Fruit. Horticulturae. 2025; 11(3):261. https://doi.org/10.3390/horticulturae11030261
Chicago/Turabian StyleJiang, Caiqian, Yunrong Mo, Haoran Zhang, Kaiyun Chen, Ying Zhou, Zushuai Ma, Yuhao Jing, Yu Liu, Yanyan Wang, and Kai Zhao. 2025. "Genome-Wide Identification of the SmHD-zip Genes That Respond to Multiple Ripening-Related Signals in Eggplant Fruit" Horticulturae 11, no. 3: 261. https://doi.org/10.3390/horticulturae11030261
APA StyleJiang, C., Mo, Y., Zhang, H., Chen, K., Zhou, Y., Ma, Z., Jing, Y., Liu, Y., Wang, Y., & Zhao, K. (2025). Genome-Wide Identification of the SmHD-zip Genes That Respond to Multiple Ripening-Related Signals in Eggplant Fruit. Horticulturae, 11(3), 261. https://doi.org/10.3390/horticulturae11030261




