JmjC Protein-Mediated Histone Demethylation: Regulating Growth, Development, and Stress Adaptation in Brassica rapa
Abstract
1. Introduction
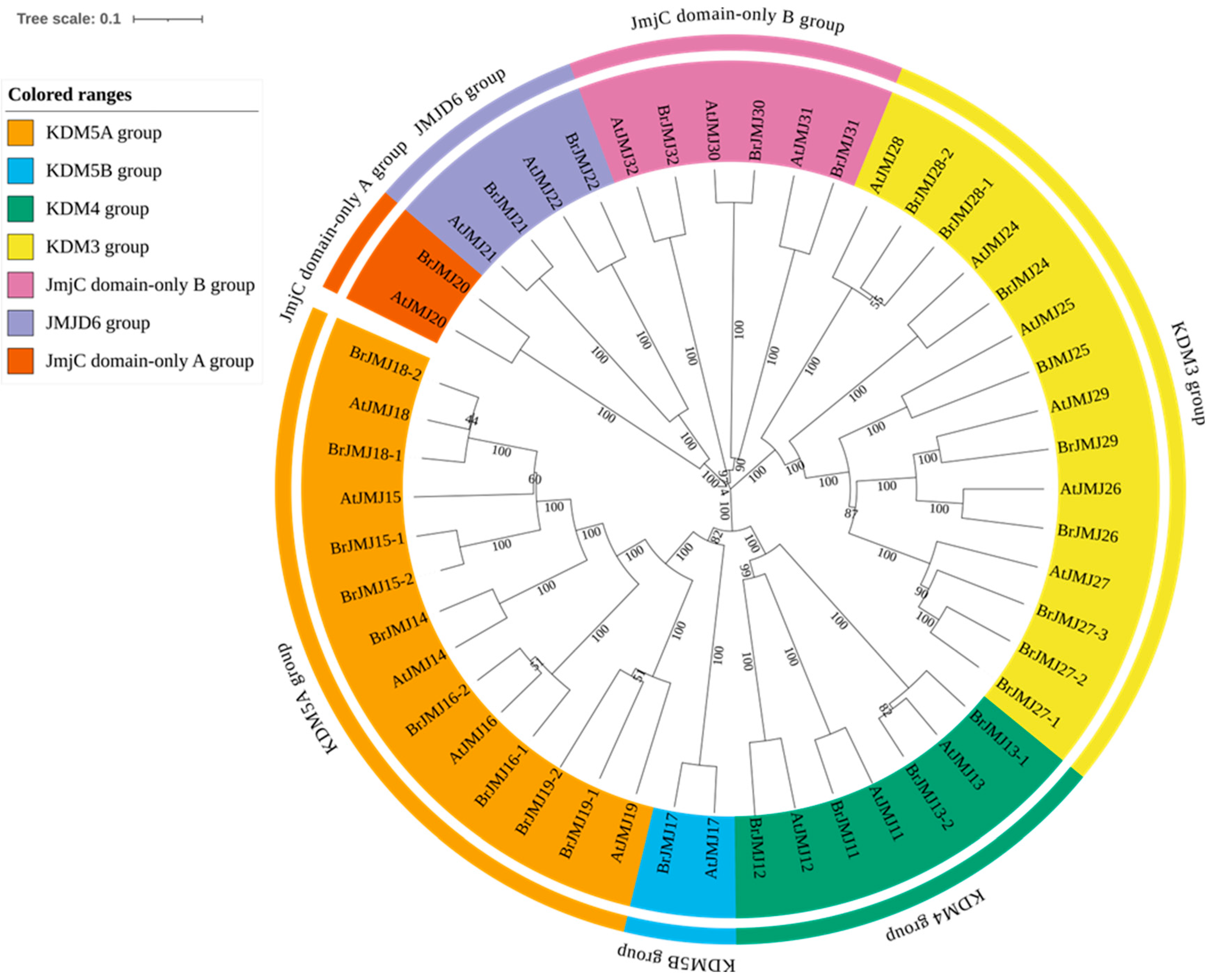
2. Key Features and Mutation Impacts of JmjC Proteins in Arabidopsis and B. rapa
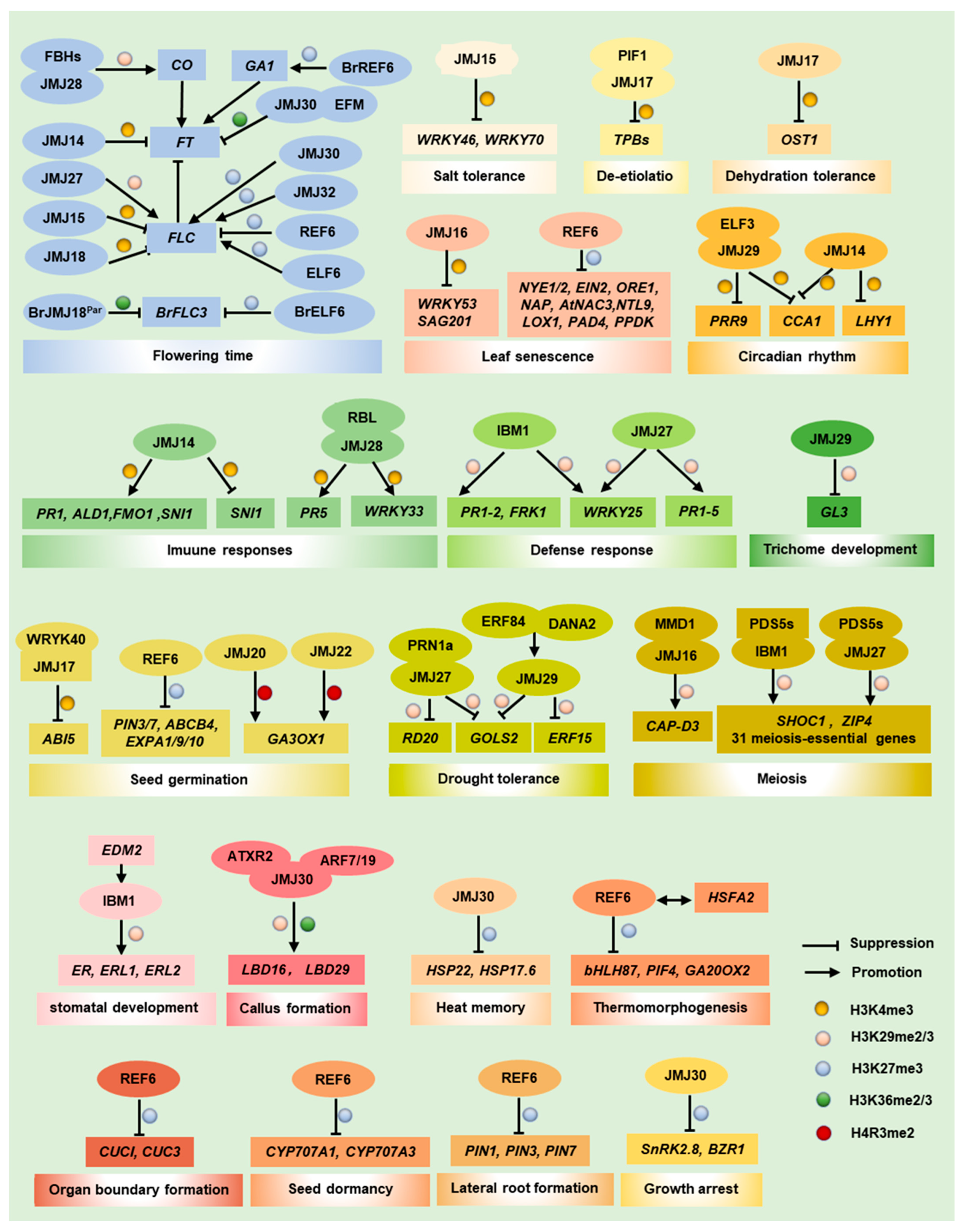
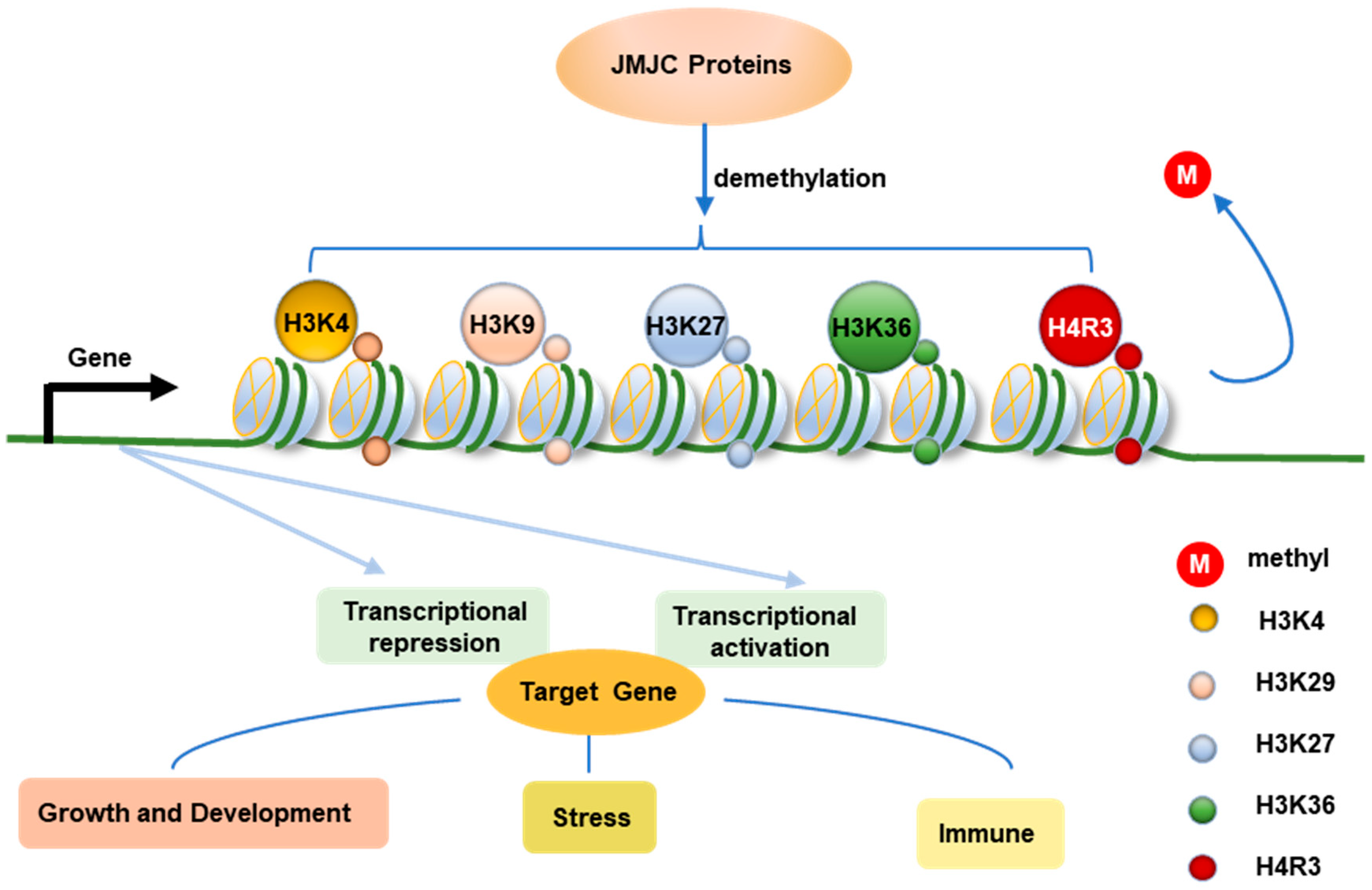
3. The H3K4me Demethylation Function of JmjC Proteins
| Gene | Interaction | Target Gene | Function |
|---|---|---|---|
| JMJ14 | FT | Flowering time [29] | |
| NAC050/052, TRBs | Gene silence [56,57,58] | ||
| PR1, ALD1, FMO1, SNI1 | Immune responses [52] | ||
| CCA1, LHY | Circadian rhythms [44] | ||
| JMJ15 | FLC | Flowering time [31] | |
| WRKY46, WRKY70 | Salt tolerance [45,46] | ||
| JMJ16 | WRKY53, SAG201 | Leaf Senescence [17] | |
| JMJ17 | OST1 | Dehydration stress [19] | |
| WRKY40, HY5 | ABI5 | Seed germination [50] | |
| PIF1 | TPBs | De-etiolation [51] | |
| JMJ18 | AtFLC | Flowering time [30] | |
| JMJ28 | RBL | PR5, WRKY33 | Immune responses [33] |
| JMJ29 | ELF3 | CCA1, PRR9 | Circadian rhythms [32] |
4. The H3K9me Demethylation Function of JmjC Proteins
| Gene | Interaction | Target Gene | Function |
|---|---|---|---|
| JMJ16 | MMD1 | CAP-D3 | Meiotic chromosome condensation [18] |
| JMJ24 | RDR2 | SDC, SOLTR, ATSN1 | RdDM [67] |
| CMT3 | QQS, SDC | CHG methylation and H3K9me2 [23] | |
| IBM1/JMJ25 | ASI1, EDM2, AIPP1 | Gene silencing [62,69,70] | |
| RDR2, DCL3 | RdDM, siRNA production [68] | ||
| ER, ERL1, ERL2 | Stomatal development [74] | ||
| PR1-2, FRK1, WRKY25 | Defense response [84] | ||
| PDS5s | SHOC1, ZIP4, etc. | Male meiosis [86] | |
| JMJ27 | FLC | Flowering time [21] | |
| RPN1a | GOLS2, RD20 | Drought tolerance [79] | |
| PDS5s | SHOC1, ZIP4. etc. | Male meiosis [86] | |
| PR1-5, WRKY25 | Defense response [21] | ||
| JMJ28 | FBHs | CO | Flowering time [64] |
| JMJ29 | GL3 | Trichome development [66] | |
| ERF15, GOLS2 | Drought tolerance [80] | ||
| JMJ30 | ARF7, ARF19, ATXR2 | LBD16, LBD29 | Callus formation [65] |
5. The H3K27me Demethylation Function of JmjC Proteins
| Gene | Target Gene | Function |
|---|---|---|
| AtJMJ11/AtELF6 | Self-fertility [115] | |
| AtFLC | Flowering time [110] | |
| BrELF6 | BrFLC3 | Flowering time [111] |
| AtREF6 | CUCs | Organ boundary formation [90] |
| NYE1/2, EIN2, ORE1, NAP, AtNNAC3, NTL9, LOX1, PAD4, PPDK | Leaf senescence [95] | |
| PIN1/3/7 | Lateral root formation [96] | |
| PIN3/7, ABCB4, EXPA1/9/10 | Seed germination [98,101] | |
| CYP707A1, CYP707A3 | Seed dormancy [97] | |
| HSFA2, bHLH87, GA20OX2 | Heat response [108,109] | |
| AtFLC | Flowering time [110] | |
| BrREF6 | BrFTa, BrGA1, BrGA20OX2 | Flowering time [111] |
| JMJ13 | Flowering time [15] | |
| JAZ7, SAUR26, AGPs | Self-fertility [115] | |
| JMJ30 | FLC | Flowering time [22] |
| HSP22, HSP17.6C | Heat memory [107] | |
| SnRK2.8, BZR1 | Growth arrest [103] | |
| JMJ32 | FLC | Flowering time [22] |
6. The H3K36me Demethylation Function of JmjC Proteins
7. The H4R3me2s Demethylation Function of JmjC Proteins
8. Conclusions and Outlook
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, F.; Cui, X.; Cao, X. Histone Methylation in Higher Plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Prakash, K.; Fournier, D. Evidence for the implication of the histone code in building the genome structure. Biosystems 2018, 164, 49–59. [Google Scholar] [CrossRef]
- Jenuwein, T.; Allis, C.D. Translating the Histone Code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Berger, S.L. The complex language of chromatin regulation during transcription. Nature 2007, 447, 407–412. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, W.; He, Y.; Amasino, R.M. Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 2007, 19, 2975–2987. [Google Scholar] [CrossRef]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jia, Q.; Jia, X.; Li, J.; Sun, X.; Min, L.; Liu, Z.; Ma, W.; Zhao, J. Brassica vegetables-an undervalued nutritional goldmine. Hortic. Res. 2025, 12, uhae302. [Google Scholar] [CrossRef]
- Lu, F.; Li, G.; Cui, X.; Liu, C.; Wang, X.J.; Cao, X. Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 2008, 50, 886–896. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, D.; Liu, C.; Shen, W.; Ruan, Y. Evolution and conservation of JmjC domain proteins in the green lineage. Mol. Genet. Genom. 2016, 291, 33–49. [Google Scholar] [CrossRef]
- Lu, F.; Cui, X.; Zhang, S.; Jenuwein, T.; Cao, X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 2011, 43, 715–719. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Hu, H.; Ren, H.; Yang, Z.; Qiu, Q.; Qi, W.; Liu, X.; Chen, X.; Cui, X.; Li, S.; et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 2019, 10, 1303. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Cui, X.; Zhang, S.; Liu, C.; Cao, X. JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res. 2010, 20, 387–390. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, S.; Zhou, B.; Luo, X.; Zhou, X.F.; Cai, B.; Jin, Y.H.; Niu, D.; Lin, J.; Cao, X.; et al. The Histone H3K4 Demethylase JMJ16 Represses Leaf Senescence in Arabidopsis. Plant Cell 2019, 31, 430–443. [Google Scholar] [CrossRef]
- Wang, J.; Yu, C.; Zhang, S.; Ye, J.; Dai, H.; Wang, H.; Huang, J.; Cao, X.; Ma, J.; Ma, H.; et al. Cell-type-dependent histone demethylase specificity promotes meiotic chromosome condensation in Arabidopsis. Nat. Plants 2020, 6, 823–837. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, A.; Jin, J.B.; Zhao, B.; Wang, T.J.; Wu, Y.; Wang, S.; Liu, Y.; Wang, J.; Guo, P.; et al. Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 2019, 223, 1372–1387. [Google Scholar] [CrossRef]
- Inagaki, S.; Miura-Kamio, A.; Nakamura, Y.; Lu, F.; Cui, X.; Cao, X.; Kimura, H.; Saze, H.; Kakutani, T. Autocatalytic differentiation of epigenetic modifications within the Arabidopsis genome. EMBO J. 2010, 29, 3496–3506. [Google Scholar] [CrossRef]
- Dutta, A.; Choudhary, P.; Caruana, J.; Raina, R. JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. Plant J. 2017, 91, 1015–1028. [Google Scholar] [CrossRef]
- Gan, E.S.; Xu, Y.; Wong, J.Y.; Goh, J.G.; Sun, B.; Wee, W.Y.; Huang, J.; Ito, T. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 2014, 5, 5098. [Google Scholar] [CrossRef]
- Deng, S.; Jang, I.C.; Su, L.; Xu, J.; Chua, N.H. JMJ24 targets CHROMOMETHYLASE3 for proteasomal degradation in Arabidopsis. Genes. Dev. 2016, 30, 251–256. [Google Scholar] [CrossRef]
- Shang, J.Y.; Lu, Y.J.; Cai, X.W.; Su, Y.N.; Feng, C.; Li, L.; Chen, S.; He, X.J. COMPASS functions as a module of the INO80 chromatin remodeling complex to mediate histone H3K4 methylation in Arabidopsis. Plant Cell 2021, 33, 3250–3271. [Google Scholar] [CrossRef]
- Zhang, X.; Bernatavichute, Y.V.; Cokus, S.; Pellegrini, M.; Jacobsen, S.E. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009, 10, R62. [Google Scholar] [CrossRef] [PubMed]
- Leng, X.; Thomas, Q.; Rasmussen, S.H.; Marquardt, S. A G(enomic)P(ositioning)S(ystem) for Plant RNAPII Transcription. Trends Plant Sci. 2020, 25, 744–764. [Google Scholar] [CrossRef]
- Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Schones, D.E.; Wang, Z.; Wei, G.; Chepelev, I.; Zhao, K. High-resolution profiling of histone methylations in the human genome. Cell 2007, 129, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Cenik, B.K.; Shilatifard, A. COMPASS and SWI/SNF complexes in development and disease. Nat. Rev. Genet. 2020, 22, 38–58. [Google Scholar] [CrossRef]
- Jeong, J.H.; Song, H.R.; Ko, J.H.; Jeong, Y.M.; Kwon, Y.E.; Seol, J.H.; Amasino, R.M.; Noh, B.; Noh, Y.S. Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 2009, 4, e8033. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Han, Z.; Cao, Y.; Fan, D.; Li, H.; Mo, H.; Feng, Y.; Liu, L.; Wang, Z.; Yue, Y.; et al. A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet. 2012, 8, e1002664. [Google Scholar] [CrossRef]
- Yang, H.; Mo, H.; Fan, D.; Cao, Y.; Cui, S.; Ma, L. Overexpression of a histone H3K4 demethylase, JMJ15, accelerates flowering time in Arabidopsis. Plant Cell Rep. 2012, 31, 1297–1308. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Seo, P.J. The Arabidopsis JMJ29 Protein Controls Circadian Oscillation through Diurnal Histone Demethylation at the CCA1 and PRR9 Loci. Genes 2021, 12, 529. [Google Scholar] [CrossRef]
- Xie, S.S.; Zhang, Y.Z.; Peng, L.; Yu, D.T.; Zhu, G.; Zhao, Q.; Wang, C.H.; Xie, Q.; Duan, C.G. JMJ28 guides sequence-specific targeting of ATX1/2-containing COMPASS-like complex in Arabidopsis. Cell Rep. 2023, 42, 112163. [Google Scholar] [CrossRef]
- Yang, Z.; Qiu, Q.; Chen, W.; Jia, B.; Chen, X.; Hu, H.; He, K.; Deng, X.; Li, S.; Tao, W.A.; et al. Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases. Plant Cell 2018, 30, 167–177. [Google Scholar] [CrossRef]
- Pien, S.; Fleury, D.; Mylne, J.S.; Crevillen, P.; Inze, D.; Avramova, Z.; Dean, C.; Grossniklaus, U. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 2008, 20, 580–588. [Google Scholar] [CrossRef]
- Tamada, Y.; Yun, J.Y.; Woo, S.C.; Amasino, R.M. ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 2009, 21, 3257–3269. [Google Scholar] [CrossRef]
- He, Y. Control of the Transition to Flowering by Chromatin Modifications. Mol. Plant 2009, 2, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Berry, S.; Olsson, T.S.G.; Hartley, M.; Howard, M.; Dean, C. Distinct phases of Polycomb silencing to hold epigenetic memory of cold in Arabidopsis. Science 2017, 357, 1142–1145. [Google Scholar] [CrossRef]
- Bastow, R.; Mylne, J.S.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef]
- Yang, H.; Howard, M.; Dean, C. Antagonistic Roles for H3K36me3 and H3K27me3 in the Cold-Induced Epigenetic Switch at Arabidopsis FLC. Curr. Biol. 2014, 24, 1793–1797. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Chua, N.H. Inverted-Repeat RNAs Targeting FT Intronic Regions Promote FT Expression in Arabidopsis. Plant Cell Physiol. 2015, 56, 1667–1678. [Google Scholar] [CrossRef]
- He, Y. Chromatin regulation of flowering. Trends Plant Sci. 2012, 17, 556–562. [Google Scholar] [CrossRef]
- Bu, Z.; Yu, Y.; Li, Z.; Liu, Y.; Jiang, W.; Huang, Y.; Dong, A.W. Regulation of arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet. 2014, 10, e1004617. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Huang, T.Y.; Yu, H.H.; Ando, A.; Mas, P.; Ha, M.; Chen, Z.J. Diurnal regulation of SDG2 and JMJ14 by circadian clock oscillators orchestrates histone modification rhythms in Arabidopsis. Genome Biol. 2019, 20, 170. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Conde, E.S.N.; Audonnet, L.; Servet, C.; Wei, W.; Zhou, D.X. Over-expression of histone H3K4 demethylase gene JMJ15 enhances salt tolerance in Arabidopsis. Front. Plant Sci. 2014, 5, 290. [Google Scholar] [CrossRef]
- Shen, Y.; Chi, Y.; Lu, S.; Lu, H.; Shi, L. Involvement of JMJ15 in the dynamic change of genome-wide H3K4me3 in response to salt stress. Front. Plant Sci. 2022, 13, 1009723. [Google Scholar] [CrossRef]
- Acharya, B.R.; Jeon, B.W.; Zhang, W.; Assmann, S.M. Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol. 2013, 200, 1049–1063. [Google Scholar] [CrossRef] [PubMed]
- Skubacz, A.; Daszkowska-Golec, A.; Szarejko, I. The Role and Regulation of ABI5 (ABA-Insensitive 5) in Plant Development, Abiotic Stress Responses and Phytohormone Crosstalk. Front. Plant Sci. 2016, 7, 1884. [Google Scholar] [CrossRef]
- Shang, Y.; Yan, L.; Liu, Z.Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.Q.; Wang, X.F.; Du, S.Y.; Jiang, T.; et al. The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef]
- Wang, T.J.; Huang, S.; Zhang, A.; Guo, P.; Liu, Y.; Xu, C.; Cong, W.; Liu, B.; Xu, Z.Y. JMJ17-WRKY40 and HY5-ABI5 modules regulate the expression of ABA-responsive genes in Arabidopsis. New Phytol. 2021, 230, 567–584. [Google Scholar] [CrossRef]
- Islam, M.T.; Wang, L.C.; Chen, I.J.; Lo, K.L.; Lo, W.S. Arabidopsis JMJ17 promotes cotyledon greening during de-etiolation by repressing genes involved in tetrapyrrole biosynthesis in etiolated seedlings. New Phytol. 2021, 231, 1023–1039. [Google Scholar] [CrossRef]
- Li, D.; Liu, R.; Singh, D.; Yuan, X.; Kachroo, P.; Raina, R. JMJ14 encoded H3K4 demethylase modulates immune responses by regulating defence gene expression and pipecolic acid levels. New Phytol. 2020, 225, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Butel, N.; Yu, A.; Le Masson, I.; Borges, F.; Elmayan, T.; Taochy, C.; Gursanscky, N.R.; Cao, J.; Bi, S.; Sawyer, A.; et al. Contrasting epigenetic control of transgenes and endogenous genes promotes post-transcriptional transgene silencing in Arabidopsis. Nat. Commun. 2021, 12, 2787. [Google Scholar] [CrossRef] [PubMed]
- Le Masson, I.; Jauvion, V.; Bouteiller, N.; Rivard, M.; Elmayan, T.; Vaucheret, H. Mutations in the Arabidopsis H3K4me2/3 demethylase JMJ14 suppress posttranscriptional gene silencing by decreasing transgene transcription. Plant Cell 2012, 24, 3603–3612. [Google Scholar] [CrossRef]
- Butel, N.; Le Masson, I.; Bouteiller, N.; Vaucheret, H.; Elmayan, T. sgs1: A neomorphic nac52 allele impairing post-transcriptional gene silencing through SGS3 downregulation. Plant J. 2017, 90, 505–519. [Google Scholar] [CrossRef]
- Ning, Y.Q.; Ma, Z.Y.; Huang, H.W.; Mo, H.; Zhao, T.T.; Li, L.; Cai, T.; Chen, S.; Ma, L.; He, X.J. Two novel NAC transcription factors regulate gene expression and flowering time by associating with the histone demethylase JMJ14. Nucleic Acids Res. 2015, 43, 1469–1484. [Google Scholar] [CrossRef]
- Zhang, S.; Zhou, B.; Kang, Y.; Cui, X.; Liu, A.; Deleris, A.; Greenberg, M.V.; Cui, X.; Qiu, Q.; Lu, F.; et al. C-terminal domains of a histone demethylase interact with a pair of transcription factors and mediate specific chromatin association. Cell Discov. 2015, 1, 15003. [Google Scholar] [CrossRef]
- Wang, M.; Zhong, Z.; Gallego-Bartolome, J.; Feng, S.; Shih, Y.H.; Liu, M.; Zhou, J.; Richey, J.C.; Ng, C.; Jami-Alahmadi, Y.; et al. Arabidopsis TRB proteins function in H3K4me3 demethylation by recruiting JMJ14. Nat. Commun. 2023, 14, 1736. [Google Scholar] [CrossRef]
- Searle, I.R.; Pontes, O.; Melnyk, C.W.; Smith, L.M.; Baulcombe, D.C. JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev. 2010, 24, 986–991. [Google Scholar] [CrossRef]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications—Writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Stroud, H.; Do, T.; Du, J.; Zhong, X.; Feng, S.; Johnson, L.; Patel, D.J.; Jacobsen, S.E. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 2013, 21, 64–72. [Google Scholar] [CrossRef]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Shiraishi, A.; Miura, A.; Kakutani, T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 2008, 319, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Hung, F.Y.; Lai, Y.C.; Wang, J.; Feng, Y.R.; Shih, Y.H.; Chen, J.H.; Sun, H.C.; Yang, S.; Li, C.; Wu, K. The Arabidopsis histone demethylase JMJ28 regulates CONSTANS by interacting with FBH transcription factors. Plant Cell 2021, 33, 1196–1211. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.S.; Seo, P.J. JMJ30-mediated demethylation of H3K9me3 drives tissue identity changes to promote callus formation in Arabidopsis. Plant J. 2018, 95, 961–975. [Google Scholar] [CrossRef]
- Hung, F.Y.; Chen, J.H.; Feng, Y.R.; Lai, Y.C.; Yang, S.; Wu, K. Arabidopsis JMJ29 is involved in trichome development by regulating the core trichome initiation gene GLABRA3. Plant J. 2020, 103, 1735–1743. [Google Scholar] [CrossRef]
- Deng, S.; Xu, J.; Liu, J.; Kim, S.H.; Shi, S.; Chua, N.H. JMJ24 binds to RDR2 and is required for the basal level transcription of silenced loci in Arabidopsis. Plant J. 2015, 83, 770–782. [Google Scholar] [CrossRef]
- Fan, D.; Dai, Y.; Wang, X.; Wang, Z.; He, H.; Yang, H.; Cao, Y.; Deng, X.W.; Ma, L. IBM1, a JmjC domain-containing histone demethylase, is involved in the regulation of RNA-directed DNA methylation through the epigenetic control of RDR2 and DCL3 expression in Arabidopsis. Nucleic Acids Res. 2012, 40, 8905–8916. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Kitayama, J.; Takashima, K.; Miura, S.; Harukawa, Y.; Ito, T.; Kakutani, T. Mechanism for full-length RNA processing of Arabidopsis genes containing intragenic heterochromatin. Nat. Commun. 2013, 4, 2301. [Google Scholar] [CrossRef]
- Duan, C.G.; Wang, X.; Zhang, L.; Xiong, X.; Zhang, Z.; Tang, K.; Pan, L.; Hsu, C.C.; Xu, H.; Tao, W.A.; et al. A protein complex regulates RNA processing of intronic heterochromatin-containing genes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E7377–E7384. [Google Scholar] [CrossRef]
- Coustham, V.; Vlad, D.; Deremetz, A.; Gy, I.; Cubillos, F.A.; Kerdaffrec, E.; Loudet, O.; Bouché, N. SHOOT GROWTH1 maintains Arabidopsis epigenomes by regulating IBM1. PLoS ONE 2014, 9, e84687. [Google Scholar] [CrossRef]
- Lei, M.; La, H.; Lu, K.; Wang, P.; Miki, D.; Ren, Z.; Duan, C.G.; Wang, X.; Tang, K.; Zeng, L.; et al. Arabidopsis EDM2 promotes IBM1 distal polyadenylation and regulates genome DNA methylation patterns. Proc. Natl. Acad. Sci. USA 2014, 111, 527–532. [Google Scholar] [CrossRef]
- Rigal, M.; Kevei, Z.; Pélissier, T.; Mathieu, O. DNA methylation in an intron of the IBM1 histone demethylase gene stabilizes chromatin modification patterns. EMBO J. 2012, 31, 2981–2993. [Google Scholar] [CrossRef]
- Wang, Y.; Xue, X.; Zhu, J.K.; Dong, J. Demethylation of ERECTA receptor genes by IBM1 histone demethylase affects stomatal development. Development 2016, 143, 4452–4461. [Google Scholar] [CrossRef]
- Lee, S.-B.; Lee, S.-J.; Kim, S.Y. AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 2014, 34, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Aubert, Y.; Vile, D.; Pervent, M.; Aldon, D.; Ranty, B.; Simonneau, T.; Vavasseur, A.; Galaud, J.-P. RD20, a Stress-Inducible Caleosin, Participates in Stomatal Control, Transpiration and Drought Tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2010, 51, 1975–1987. [Google Scholar] [CrossRef]
- Taji, T.; Ohsumi, C.; Iuchi, S.; Seki, M.; Kasuga, M.; Kobayashi, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002, 29, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, P.; Jing, H.; Zhou, X.F.; Zhao, B.; Li, Y.; Jin, J.B. JMJ27-mediated histone H3K9 demethylation positively regulates drought-stress responses in Arabidopsis. New Phytol. 2021, 232, 221–236. [Google Scholar] [CrossRef]
- Zhang, P.; He, R.; Yang, J.; Cai, J.; Qu, Z.; Yang, R.; Gu, J.; Wang, Z.Y.; Adelson, D.L.; Zhu, Y.; et al. The long non-coding RNA DANA2 positively regulates drought tolerance by recruiting ERF84 to promote JMJ29-mediated histone demethylation. Mol. Plant 2023, 16, 1339–1353. [Google Scholar] [CrossRef]
- Wang, S.; Hubbard, L.; Chang, Y.; Guo, J.; Schiefelbein, J.; Chen, J.-G. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008, 8, 81. [Google Scholar] [CrossRef]
- Wester, K.; Digiuni, S.; Geier, F.; Timmer, J.; Fleck, C.; Hülskamp, M. Functional diversity of R3 single-repeat genes in trichome development. Development 2009, 136, 1487–1496. [Google Scholar] [CrossRef]
- Lee, K.; Park, O.S.; Seo, P.J. Arabidopsis ATXR2 deposits H3K36me3 at the promoters of LBD genes to facilitate cellular dedifferentiation. Sci. Signal 2017, 10, eaan0316. [Google Scholar] [CrossRef]
- Chan, C.; Zimmerli, L. The Histone Demethylase IBM1 Positively Regulates Arabidopsis Immunity by Control of Defense Gene Expression. Front. Plant Sci. 2019, 10, 1587. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Mosher, S.L.; Fan, B.; Klessig, D.F.; Chen, Z. Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 2007, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Xu, L.; Bergér, V.; Bruckmann, A.; Yang, C.; Schubert, V.; Grasser, K.D.; Schnittger, A.; Zheng, B.; Jiang, H. H3K9 demethylases IBM1 and JMJ27 are required for male meiosis in Arabidopsis thaliana. New Phytol. 2022, 235, 2252–2269. [Google Scholar] [CrossRef] [PubMed]
- Jacob, Y.; Bergamin, E.; Donoghue, M.T.A.; Mongeon, V.; LeBlanc, C.; Voigt, P.; Underwood, C.J.; Brunzelle, J.S.; Michaels, S.D.; Reinberg, D.; et al. Selective Methylation of Histone H3 Variant H3.1 Regulates Heterochromatin Replication. Science 2014, 343, 1249–1253. [Google Scholar] [CrossRef]
- Xiao, J.; Lee, U.-S.; Wagner, D. Tug of war: Adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 2016, 34, 41–53. [Google Scholar] [CrossRef]
- Crevillén, P.; Yang, H.; Cui, X.; Greeff, C.; Trick, M.; Qiu, Q.; Cao, X.; Dean, C. Epigenetic reprogramming that prevents transgenerational inheritance of the vernalized state. Nature 2014, 515, 587–590. [Google Scholar] [CrossRef]
- Cui, X.; Lu, F.; Qiu, Q.; Zhou, B.; Gu, L.; Zhang, S.; Kang, Y.; Cui, X.; Ma, X.; Yao, Q.; et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 2016, 48, 694–699. [Google Scholar] [CrossRef]
- Li, C.; Gu, L.; Gao, L.; Chen, C.; Wei, C.Q.; Qiu, Q.; Chien, C.W.; Wang, S.; Jiang, L.; Ai, L.F.; et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016, 48, 687–693. [Google Scholar] [CrossRef]
- Qiu, Q.; Mei, H.; Deng, X.; He, K.; Wu, B.; Yao, Q.; Zhang, J.; Lu, F.; Ma, J.; Cao, X. DNA methylation repels targeting of Arabidopsis REF6. Nat. Commun. 2019, 10, 2063. [Google Scholar] [CrossRef] [PubMed]
- Hibara, K.-i.; Karim, M.R.; Takada, S.; Taoka, K.-i.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 Regulates Postembryonic Shoot Meristem and Organ Boundary Formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef]
- Tian, Z.; Li, X.; Li, M.; Wu, W.; Zhang, M.; Tang, C.; Li, Z.; Liu, Y.; Chen, Z.; Yang, M.; et al. Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms. Cell Discov. 2020, 6, 17. [Google Scholar] [CrossRef]
- Wang, X.; Gao, J.; Gao, S.; Song, Y.; Yang, Z.; Kuai, B. The H3K27me3 demethylase REF6 promotes leaf senescence through directly activating major senescence regulatory and functional genes in Arabidopsis. PLoS Genet. 2019, 15, e1008068. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gao, J.; Gao, S.; Li, Z.; Kuai, B.; Ren, G. REF6 promotes lateral root formation through de-repression of PIN1/3/7 genes. J. Integr. Plant Biol. 2018, 61, 383–387. [Google Scholar] [CrossRef]
- Chen, H.; Tong, J.; Fu, W.; Liang, Z.; Ruan, J.; Yu, Y.; Song, X.; Yuan, L.; Xiao, L.; Liu, J.; et al. The H3K27me3 Demethylase RELATIVE OF EARLY FLOWERING6 Suppresses Seed Dormancy by Inducing Abscisic Acid Catabolism. Plant Physiol. 2020, 184, 1969–1978. [Google Scholar] [CrossRef]
- Pan, J.; Zhang, H.; Zhan, Z.; Zhao, T.; Jiang, D. A REF6-dependent H3K27me3-depleted state facilitates gene activation during germination in Arabidopsis. J. Genet. Genom. 2023, 50, 178–191. [Google Scholar] [CrossRef]
- Kushiro, T.; Okamoto, M.; Nakabayashi, K.; Yamagishi, K.; Kitamura, S.; Asami, T.; Hirai, N.; Koshiba, T.; Kamiya, Y.; Nambara, E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 2004, 23, 1647–1656. [Google Scholar] [CrossRef]
- Okamoto, M.; Kuwahara, A.; Seo, M.; Kushiro, T.; Asami, T.; Hirai, N.; Kamiya, Y.; Koshiba, T.; Nambara, E. CYP707A1 and CYP707A2, Which Encode Abscisic Acid 8′-Hydroxylases, Are Indispensable for Proper Control of Seed Dormancy and Germination in Arabidopsis. Plant Physiol. 2006, 141, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, D.; Deng, L.; He, C.; Zheng, F.; Liu, X. The Histone H3K27 Demethylase REF6 Is a Positive Regulator of Light-Initiated Seed Germination in Arabidopsis. Cells 2023, 12, 295. [Google Scholar] [CrossRef]
- Lopez-Molina, L.; Mongrand, S.; Chua, N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 2001, 98, 4782–4787. [Google Scholar] [CrossRef]
- Wu, J.; Ichihashi, Y.; Suzuki, T.; Shibata, A.; Shirasu, K.; Yamaguchi, N.; Ito, T. Abscisic acid-dependent histone demethylation during postgermination growth arrest in Arabidopsis. Plant Cell Environ. 2019, 42, 2198–2214. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, H.; Zhao, Y.; Feng, Z.; Li, Q.; Yang, H.-Q.; Luan, S.; Li, J.; He, Z.-H. Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 15485–15490. [Google Scholar] [CrossRef]
- Wu, J.; Yamaguchi, N.; Ito, T. Histone demethylases control root elongation in response to stress-signaling hormone abscisic acid. Plant Signal Behav. 2019, 14, 1604019. [Google Scholar] [CrossRef]
- Wu, J.; Yan, M.; Zhang, D.; Zhou, D.; Yamaguchi, N.; Ito, T. Histone Demethylases Coordinate the Antagonistic Interaction Between Abscisic Acid and Brassinosteroid Signaling in Arabidopsis. Front. Plant Sci. 2020, 11, 596835. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Matsubara, S.; Yoshimizu, K.; Seki, M.; Hamada, K.; Kamitani, M.; Kurita, Y.; Nomura, Y.; Nagashima, K.; Inagaki, S.; et al. H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. 2021, 12, 3480. [Google Scholar] [CrossRef]
- Liu, J.; Feng, L.; Gu, X.; Deng, X.; Qiu, Q.; Li, Q.; Zhang, Y.; Wang, M.; Deng, Y.; Wang, E.; et al. An H3K27me3 demethylase-HSFA2 regulatory loop orchestrates transgenerational thermomemory in Arabidopsis. Cell Res. 2019, 29, 379–390. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Mei, H.; Zhu, J.; Qiu, Q.; Cao, X.; Deng, X. The histone H3K27 demethylase REF6/JMJ12 promotes thermomorphogenesis in Arabidopsis. Natl. Sci. Rev. 2022, 9, nwab213. [Google Scholar] [CrossRef]
- Noh, B.; Lee, S.H.; Kim, H.J.; Yi, G.; Shin, E.A.; Lee, M.; Jung, K.J.; Doyle, M.R.; Amasino, R.M.; Noh, Y.S. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 2004, 16, 2601–2613. [Google Scholar] [CrossRef]
- Poza-Viejo, L.; Payá-Milans, M.; San Martín-Uriz, P.; Castro-Labrador, L.; Lara-Astiaso, D.; Wilkinson, M.D.; Piñeiro, M.; Jarillo, J.A.; Crevillén, P. Conserved and distinct roles of H3K27me3 demethylases regulating flowering time in Brassica rapa. Plant Cell Environ. 2022, 45, 1428–1441. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Shen, L.; Chen, Y.; Bao, S.; Thong, Z.; Yu, H. A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 2014, 30, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Knowles, S.M.; Webb, C.J.; Celaya, R.B.; Cha, C.; Siu, J.P.; Tobin, E.M. The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 2011, 155, 906–915. [Google Scholar] [CrossRef]
- Yan, W.; Chen, D.; Smaczniak, C.; Engelhorn, J.; Liu, H.; Yang, W.; Graf, A.; Carles, C.C.; Zhou, D.X.; Kaufmann, K. Dynamic and spatial restriction of Polycomb activity by plant histone demethylases. Nat. Plants 2018, 4, 681–689. [Google Scholar] [CrossRef]
- Keyzor, C.; Mermaz, B.; Trigazis, E.; Jo, S.; Song, J. Histone Demethylases ELF6 and JMJ13 Antagonistically Regulate Self-Fertility in Arabidopsis. Front. Plant Sci. 2021, 12, 640135. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, Z.; Dong, A.; Soubigou-Taconnat, L.; Renou, J.-P.; Steinmetz, A.; Shen, W.-H. Di- and Tri- but Not Monomethylation on Histone H3 Lysine 36 Marks Active Transcription of Genes Involved in Flowering Time Regulation and Other Processes in Arabidopsis thaliana. Mol. Cell. Biol. 2023, 28, 1348–1360. [Google Scholar] [CrossRef]
- Shim, S.; Lee, H.G.; Lee, H.; Seo, P.J. H3K36me2 is highly correlated with m6A modifications in plants. J. Integr. Plant Biol. 2020, 62, 1455–1460. [Google Scholar] [CrossRef]
- Xin, X.; Li, P.; Zhao, X.; Yu, Y.; Wang, W.; Jin, G.; Wang, J.; Sun, L.; Zhang, D.; Zhang, F.; et al. Temperature-dependent jumonji demethylase modulates flowering time by targeting H3K36me2/3 in Brassica rapa. Nat. Commun. 2024, 15, 5470. [Google Scholar] [CrossRef]
- Jones, M.A.; Harmer, S. JMJD5 Functions in concert with TOC1 in the arabidopsis circadian system. Plant Signal Behav. 2011, 6, 445–448. [Google Scholar] [CrossRef]
- Jones, M.A.; Morohashi, K.; Grotewold, E.; Harmer, S.L. Arabidopsis JMJD5/JMJ30 Acts Independently of LUX ARRHYTHMO Within the Plant Circadian Clock to Enable Temperature Compensation. Front. Plant Sci. 2019, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.N.; Ryu, J.Y.; Jeong, Y.M.; Park, J.; Song, J.J.; Amasino, R.M.; Noh, B.; Noh, Y.S. Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 2012, 22, 736–748. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, R.; Wang, Q.; Wang, J.; Wang, X.; Zhao, J.; Li, N.; Yang, L. JmjC Protein-Mediated Histone Demethylation: Regulating Growth, Development, and Stress Adaptation in Brassica rapa. Horticulturae 2025, 11, 1424. https://doi.org/10.3390/horticulturae11121424
Yang R, Wang Q, Wang J, Wang X, Zhao J, Li N, Yang L. JmjC Protein-Mediated Histone Demethylation: Regulating Growth, Development, and Stress Adaptation in Brassica rapa. Horticulturae. 2025; 11(12):1424. https://doi.org/10.3390/horticulturae11121424
Chicago/Turabian StyleYang, Rui, Qianyun Wang, Jiajie Wang, Xiaona Wang, Jianjun Zhao, Na Li, and Lei Yang. 2025. "JmjC Protein-Mediated Histone Demethylation: Regulating Growth, Development, and Stress Adaptation in Brassica rapa" Horticulturae 11, no. 12: 1424. https://doi.org/10.3390/horticulturae11121424
APA StyleYang, R., Wang, Q., Wang, J., Wang, X., Zhao, J., Li, N., & Yang, L. (2025). JmjC Protein-Mediated Histone Demethylation: Regulating Growth, Development, and Stress Adaptation in Brassica rapa. Horticulturae, 11(12), 1424. https://doi.org/10.3390/horticulturae11121424






